Abstract
Free full text

THE SERINE PROTEASE MATRIPTASE-2 (TMPRSS6) INHIBITS HEPCIDIN ACTIVATION BY CLEAVING MEMBRANE HEMOJUVELIN
Associated Data
Summary
The liver peptide hepcidin regulates body iron, is upregulated in iron overload and inflammation and downregulated in iron deficiency/hypoxia. The transmembrane serine protease matriptase-2 (TMPRSS6) inhibits the hepcidin response and its mutational inactivation causes iron-deficient anemia in mice and humans. Here we confirm the inhibitory effect of matriptase-2 on hepcidin promoter; we show that matriptase-2 lacking the serine protease domain, identified in the anemic Mask mouse (matriptase-2MASK), is fully inactive and that mutant R774C found in patients with genetic iron deficiency has decreased inhibitory activity. Matriptase-2 cleaves hemojuvelin (HJV), a regulator of hepcidin, on plasma membrane; matriptase-2MASK shows no and the human mutant only partial cleavage capacity. Matriptase-2 interacts with HJV through the ectodomain since the interaction is conserved in matriptase-2MASK. The expression of matriptase-2 mutants in zebrafish results in anemia, confirming the matriptase-2 role in iron metabolism and its interaction with HJV.
Introduction
The liver peptide hepcidin is the master regulator of intestinal and macrophage iron release to circulating transferrin and thus of the body iron supply, through its ability to bind, internalize and degrade the cellular iron exporter ferroportin (Nemeth et al., 2004). Hepcidin expression increases in inflammation and iron overload to limit iron availability. The inflammatory cytokines, especially IL-6, activate hepcidin through STAT3-mediated signaling (Wrighting and Andrews, 2006) (Verga Falzacappa et al., 2007; Pietrangelo et al., 2007) to sequester iron in macrophages. Low/inappropriate hepcidin production occurs in most types of hemochromatosis (Camaschella, 2005), but especially in the severe juvenile form due to hemojuvelin (HJV) mutations (Papanikolaou et al., 2004; Lanzara et al., 2004). Genes (HFE, TFR2, HJV) have been identified that affect hepcidin regulation, although the molecular mechanisms are not fully understood. The main activator of hepcidin transcription is HJV, a glycosilphosphatidylinositol (GPI)-anchored protein that behaves as coreceptor of Bone Morphogenetic Proteins (BMPs) (Babitt et al., 2006), which signal through son of mothers against decapentaplegic (SMAD) proteins. SMAD4 liver conditional knock out mice develop iron overload due to downregulation of hepcidin (Wang et al., 2005), supporting the SMAD role in the pathway and linking the reduction of BMP signaling to iron overload. Soluble HJV (s-HJV) in vitro acts as a “decoy-receptor” competing with m-HJV for the BMP ligand (Lin et al., 2005). Chronic injection of s-HJV causes hemochromatosis in mice (Babitt et al., 2007), indicating that s-HJV can inhibit hepcidin and increase iron absorption in vivo. HJV is cleaved by furin at position 335 (Valore and Ganz, 2008; Silvestri et al., 2008), releasing s-HJV as a major (42 kDa) and a minor (33 kDa) component into the cell culture supernatant. We have shown that furin cleavage in vitro occurs in conditions of iron deficiency and chemically induced hypoxia (Silvestri et al., 2008).
Recently a genetic inability to downregulate hepcidin has been recognized, due to TMPRSS6 mutations (Du et al., 2008; Finberg et al., 2008). TMPRSS6 encodes a type II plasma membrane serine protease, also known as matriptase-2, which belongs to a family of cell surface proteolytic enzymes (type II transmembrane serine protease, TTPS) (Velasco et al., 2002), highly conserved in mammals (Ramsay et al., 2008). Matriptase-2 is mainly expressed in liver both in humans and in mice. The protein has a short cytoplasmic tail and a transmembrane domain. The extracellular domain contains multiple motifs and domains including: two complement protein subcomponents C1r/C1 motifs, urchin embryonic growth factor and bone morphogenetic protein 1 (CUB) domains; three low density lipoprotein receptor class A (LDLR) repeats; a prodomain region that contains the cleavage site for protease activation; a carboxy-terminal catalytic domain (Velasco et al., 2002). Matriptase-2 has a strong effect on hepcidin suppression as shown by the Mask mouse model, which results from a deletion of the serine protease domain. Mask mice have abnormal hair distribution and iron deficiency anemia, due to decreased iron absorption because of inappropriately high hepcidin levels (Du et al., 2008). A similar phenotype is present in the Tmprss6 deficient mice (Folgueras et al., 2008). In transfected HepG2 cells matriptase-2 inhibits hepcidin activation by multiple stimuli (BMP, HJV, SMAD1, IL-6) (Du et al., 2008). The mechanism of hepcidin inactivation by matriptase-2 remains speculative and was proposed to occur through a novel pathway, independent from known hepcidin regulatory pathways (Du et al., 2008). The function of matriptase-2 is maintained in humans. Mutations of matriptase-2 lead to iron-refractory iron-deficient anemia (IRIDA). IRIDA patients have remarkably high hepcidin levels, in spite of iron deficiency. Nonsense/frameshift/splice mutations lead to truncated variants reminiscent of matriptase-2MASK. Missense changes in matriptase-2 have also been identified, which affect conserved residues: G442R in the second CUB domain, D521N in the second LDLR domain and R774C in the protease domain (Finberg et al., 2008). Results in mice and humans indicate that matriptase-2 is required to sense iron deficiency in mammals (Du et al., 2008).
In this paper we studied wild type and matriptase-2 mutants for their ability to downregulate hepcidin expression. We provide evidence that matriptase-2 inhibits hepcidin by cleaving HJV from plasma membrane. Mutants of matriptase, that are inactive (matriptase-2MASK) or partially active (matriptase-2R774C) in cleaving HJV, permit the continued expression of hepcidin.
Results
Matriptase-2 processing
To analyze the maturation and processing of wild type and mutant matriptase-2, we mutagenized pcDNA 3.1-matriptase-2-FLAG expressing vector (kindly provided by Prof. C. Lopez-Otin) to obtain the human homologue of “Mask” lacking the catalytic domain (Du et al., 2008) and a mutation (R774C) described in a IRIDA patient, replacing arginine with an extra cysteine at position 774 in the serine protease domain (Finberg et al., 2008) (Fig. 1A). Immunofluorescence of cells transfected with plasmids expressing either wild type or mutant matriptase-2 revealed that the expressed proteins were localized in structures resembling the endoplasmic reticulum in permeabilized cells (Fig. S1, P). In non-permeabilized conditions the expressed proteins were present on the plasma membrane (Fig. S1, UP). When quantified by a binding assay the amount of matriptase-2R774C was reduced on the cell surface, suggesting that the amino acid substitution interfered with the correct localization of the protein. In contrast, the amount of surface matriptase-2MASK was greater than that of the wild type protein (Fig. 1B), likely due to its inability to undergo the autocleavage of the serine protease domain. To study the processing and maturation of matriptase-2 variants, electron microscopy and morphometric analysis were performed on transfected HeLa cells. As shown in Figure 1C, 18 hours after transfection matriptase-2wt and matriptase-2MASK localized mainly to the plasma membrane and in lesser amounts to the Golgi (G) and the endoplasmic reticulum (ER). In contrast, matriptase-2R774C predominantly accumulated in the ER and showed lower expression in the plasma membrane.
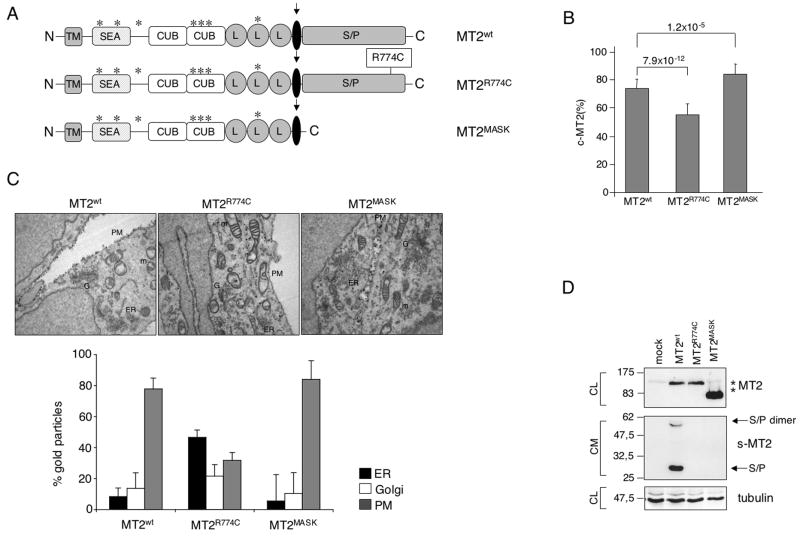
(A) Schematic representation of matriptase-2 (MT2) functional domains and localization of the studied mutations [modified from (Ramsay et al., 2008)]. TM: transmembrane domain. SEA: sea urchin sperm protein, enteropeptidase agrin. CUB: complement protein subcomponents C1r/C1s, urchin embryonic growth factor and bone morphogenetic protein 1 domain. L: low density lipoprotein receptor class A domain. S/P: serine protease domain. The arrow indicates the cleavage activation site. Asterisks indicate the predicted consensus N-glycosylation sites. (B) Quantification of membrane bound matriptase-2 (m-MT2) by binding assay. Hela cells were transiently transfected with the wild type and mutant expressing vectors, or the empty vector, and analyzed for the amount of matriptase-2 on the cell surface. The amount was calculated as the ratio between the absorbance of unpermeabilized and permeabilized cells. Error bars indicate standard deviation. Statistical significance was calculated on a total of three experiments, made in triplicate. Exact P-values are shown above bars. (C) Electron microscopy and morphometric analysis of MT2wt, MT2R774C and MT2MASK. (D) Characterization of MT2. Whole cell extracts and concentrated media of transiently transfected HeLa cells were analyzed by 10% SDS-PAGE. Western blot was performed following standard procedures; MT2 was revealed by the anti-FLAG antibody. s-MT2: soluble MT2. S/P: serine protease domain. CL: cellular lysates; CM: conditioned medium. The equal loading was verified by α-tubulin.
Expression of matriptase-2wt results in the release into the medium of two species, migrating at about 30 and 60 kDa. These shorter forms originate from the cleavage of the ectodomain, which releases the C-terminus of the protein (Fig. 1D). The molecular weight of the bands, their reactivity to the anti-FLAG antibody and their insensitivity to peptide N-Glycosidase F (PNGase) treatment (data not shown), suggest that they represent the serine protease domain which lacks N-linked-carbohydrates (Ramsay et al., 2008) and migrates as a single (30 kDa) or dimeric (60 kDa) species. The same fragments are undetectable in the supernatant of cells transfected with matriptase-2MASK, which lacks the proteolytic domain, supporting the interpretation. There are relatively fewer soluble fragments in medium from cells expressing matriptase-2R774C, suggesting that an active serine-protease domain and/or a correct protein folding are essential to release the serine protease domain.
Matriptase-2 inhibits HJV-mediated hepcidin activation
To study the effect of matriptase-2 on hepcidin expression, we transfected Hep3B cells with a hepcidin promoter/firefly luciferase reporter construct (Hep-Luc) and a renilla luciferase vector (Pagani et al., 2008). The expression of matriptase-2wt or matriptase-2 mutants did not alter hepcidin promoter activity in basal conditions nor after BMP2 stimulation (Fig. 2A). As expected, the presence of HJV enhanced hepcidin activation. Co-expression of HJV and matriptase-2wt, however, significantly decreased hepcidin reporter gene activation. No decrease in hepcidin expression resulted when HJV was coexpressed with matriptase-2MASK, while expression of matriptase-2R774C led to a partial inhibition. A comparable trend was observed when HJV-matriptase-2 cotransfected cells were treated with BMP2 (Fig. 2A), suggesting that matriptase-2 specifically blocks the HJV-mediated effect. To verify the minimal effective concentration of matriptase-2, Hep3B cells were transfected with escalating concentrations of the serine protease. Matriptase-2wt inhibited hepcidin activation in a dose-dependent manner, matriptase-2R774C was less active and reached an inhibitory effect comparable to that of the wild type protein only at high concentrations but matriptase-2MASK was inactive at all concentrations tested (Fig. 2B).
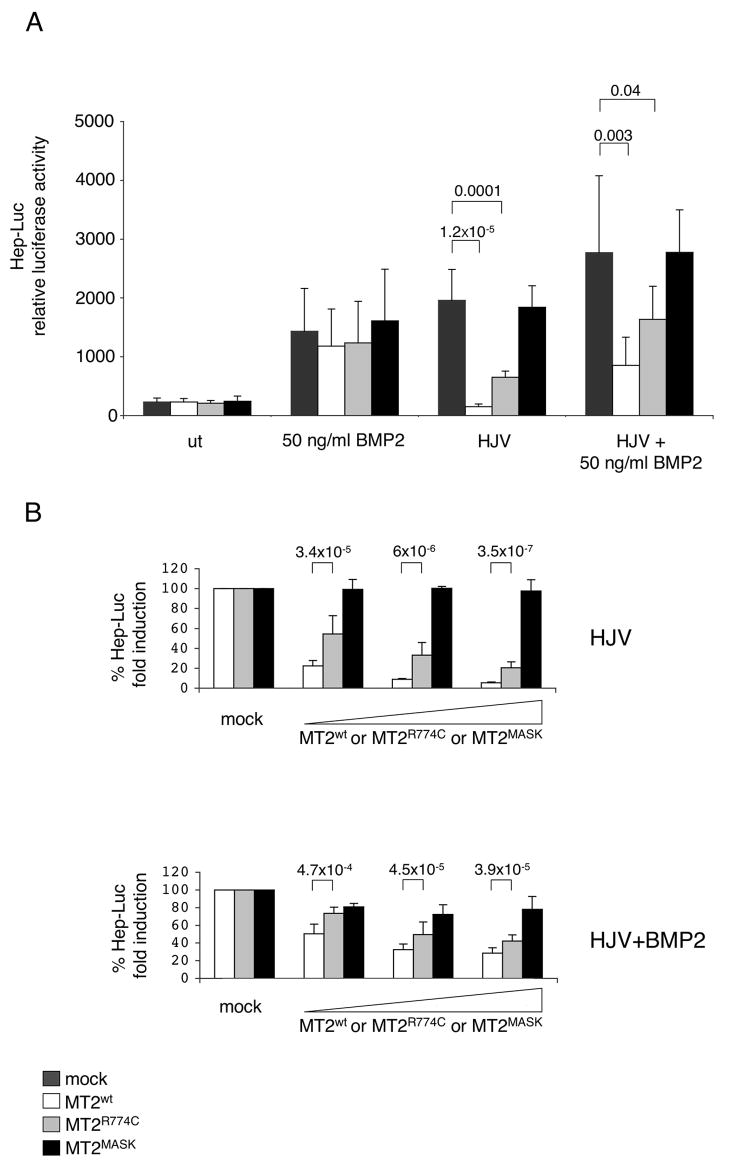
(A) Hepcidin promoter responses by BMP2 and HJV, in the presence of matriptase-2 (MT2). A firefly luciferase reporter driven by 2.9 kb of the proximal hepcidin promoter was cotransfected into Hep3B cells with pRL-TK, either alone or with HJV and/or MT2 expressing vectors. Relative luciferase activity is calculated as reported in material and method and expressed as a multiple of the activity of cells transfected with the reporter alone. (B) Dose dependent modulation of the hepcidin activity by MT2. Hep3B cells were transfected with increasing concentrations of wild type or mutant MT2 expressing vectors in the presence of fixed amount of HJV expressing vector and treated or not with BMP2 (50 ng/ml). The fold induction is calculated as a ratio between the MT2-mediated and the mock-mediated hepcidin promoter activity, calculated as described in (A). Experiments, made in triplicate, were performed three times. Error bars indicate SD. Exact P-values are shown above bars.
Matriptase-2 cleaves HJV
Based on the observation that matriptase-2 inhibits the HJV-mediated hepcidin activation, we assessed whether it cleaved HJV. Coexpression of matriptase-2wt and HJV caused a decrease of total (both full length and 33 kDa) HJV. No decrease in HJV occurred upon coexpression of matriptase-2MASK, suggesting that matriptase-2 cleaves HJV (Fig. 3A). Since matriptase-2 is a transmembrane protease we hypothesized that it cleaves cell surface HJV (m-HJV). To test this hypothesis, HeLa cells were transfected with HJV and increasing concentrations of matriptase-2wt or the matriptase mutants. Cell surface HJV decreased in a dose dependent manner in cells expressing matriptase-2wt (Fig. 3B). In contrast, m-HJV was unchanged in cells transfected with matriptase-2MASK and partially decreased in the presence of matriptase-2R774C, indicating that the integrity of the protease domain is essential for an efficient cleavage of m-HJV (Fig. 3B). To further assess the residual amount of m-HJV after matriptase-2 cleavage, transfected cells were treated with a phosphatidylinositol-specific phospholipase (PI-PLC) that specifically cuts GPI-anchored proteins and the cleavage products in the supernatant were analyzed. HJV released by PI-PLC migrates as approximately 50 kDa and 33 kDa forms under partially reducing conditions (Fig 3A, lane 2, PI-PLC top panel) or mainly as 33 kDa fragments under strong reducing conditions (Fig. 3A, lane 2, PI-PLC bottom panel). When HJV was coexpressed with matriptase-2wt, because of matriptase-2 proteolytic activity only traces of residual m-HJV were cleaved by PI-PLC (Fig. 3A, lane 3, PI-PLC panels). As indicated by the amount of m-HJV released by PI-PLC, matriptase-2MASK was fully inactive (Fig. 3A, lane 5, PI-PLC panels), whereas matriptase-2R774C (Fig. 3A, lane 4, PI-PLC panels) had partial activity.
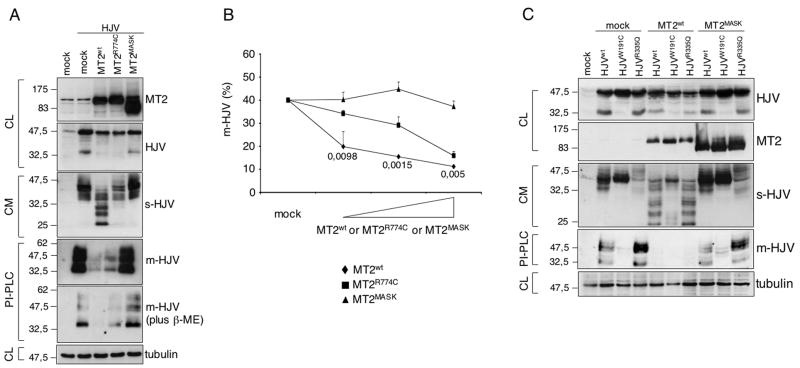
(A) HeLa cells were transfected with HJV in the presence of the empty vector (mock), matriptase-2wt (MT2wt), MT2R774C and MT2MASK. Whole cell extracts, concentrated media and PI-PLC supernatants were loaded onto a 10% SDS-PAGE and processed for western blot analysis. Anti-FLAG and anti-HJV were used to detect MT2 and HJV respectively. β-ME: beta-mercaptoethanol. (B) Binding assay was used to measure m-HJV in the presence of increasing concentrations of wild type and mutants MT2 expressing vectors and was performed essentially as described in Fig. 1B. Experiments were made in triplicate and performed three times. Error bars indicate SD. Exact P-values refer to MT2wt versus MT2R774C. (C) HeLa cells were transfected with MT2wt and MT2MASK, in the presence of HJVwt, HJVW191C and HJVR335Q. Whole cell extracts, concentrated media and PI-PLC supernatants were loaded onto a 10% SDS-PAGE and processed for western blot analyses.
s-HJV: soluble HJV; m-HJV: membrane associated HJV. CL: cellular lysates; CM: conditioned medium; PI-PLC: supernatant after PI-PLC cleavage. The equal loading was verified by α-tubulin.
Examination of the cleavage products in the culture media released by matriptase-2 revealed bands migrating between 25 and 30 kDa, in addition to decreased 42 and 33 kDa s-HJV species (Fig. 3A, lane 3, CM panel). In contrast, the former fragments were not present in the media from cells transfected with matriptase-2MASK (Fig. 3A, lane 5, CM panel) and only barely present in the media from cells expressing matriptase-2R774C (Fig. 3A, lane 4, CM panel).
To clarify the origin of the cleavage products, we took advantage of two previously developed HJV variants. W191C, which is incorrectly processed, retained into ER and is unable to reach the cell surface. This mutant form is released into the medium as a 42kDa s-HJV (Silvestri et al., 2007). A second HJV mutant R335Q is efficiently exported to plasma membrane but does not release s-HJV, since the amino acid change affects the furin consensus cleavage site (Silvestri et al., 2008). PI-PLC cleaves m-HJV only in cells transfected with HJVwt (Fig. 3C, lane 2) and HJVR335Q (Fig. 3C, lane 4) but not with HJVW191C (Fig. 3C, lane 3). When matriptase-2 was coexpressed with HJVwt or HJVR335Q PI-PLC did not release m-HJV due to the serine protease activity (Fig. 3C, lanes 5 and 7, PI-PLC panel). Moreover, cells cotransfected with matriptase-2 and HJVR335Q (Fig. 3C, lane 7, CM panel) release in the supernatant the same cleavage products as from cleavage of HJVwt (Fig. 3C, lane 5, CM panel). In contrast, when matriptase-2wt was coexpressed with HJVW191C (Fig. 3C, lane 6, CM panel), the 42 kDa s-HJV form was decreased, but no additional fragments were observed. In the same experiments matriptase-2MASK was used as an inactive control (Fig. 3C, lanes 8–10). We conclude that the 25–30 kDa fragments produced by the matriptase-2 cleavage originate from m-HJV and hypothesize that the 42 kDa s-HJV decrease follows the reduction of HJV due to matriptase-2 overexpression.
To demonstrate the specificity of the process, we tested matriptase-2wt cleavage ability on uromodulin, a GPI-anchored protein mainly expressed in the kidney (Bernascone et al., 2006). As shown in Figure S2 total, membrane-associated and soluble uromodulin were not affected by the serine protease.
Matriptase-2 does not cleave s-HJV
To understand whether the 42 kDa s-HJV is cleaved directly by matriptase-2, we incubated matriptase-2-transfected cells with exogenous s-HJV (Fig. 4A) and observed that matriptase-2 overexpressing cells did not cleave s-HJV in media of HJV-transfected cells. To confirm that the decrease of HJV in cell extracts was due to matriptase-2 overexpression, cells were transfected with increasing amounts of matriptase-2wt and constant amounts of HJV (Fig. 4B). At low expression levels, matriptase-2wt reduced only m-HJV, releasing the corresponding soluble fragments in the supernatant. We also observed that the serine-protease domain cleaved from matriptase-2 ectodomain was released in the cell culture supernatant. To understand whether the soluble matriptase-2 domain retained proteolytic activity on HJV, we incubated HJV-transfected cells with conditioned media from matriptase-2wt transfected cells (Fig. 4C). Neither s-HJV nor m-HJV was cleaved, indicating that the released serine-protease domain is inactive on HJV.
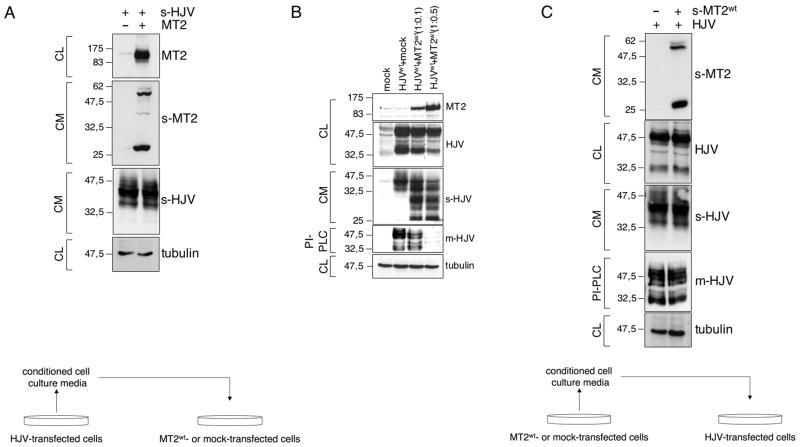
(A) HeLa cells were transfected with empty vector or matriptase-2 (MT2) expressing plasmid, and then incubated with an exogenous source of soluble HJV. Whole cell extracts and concentrated media were then analyzed by western blot. A scheme of the experiment is shown bottom left. (B) HeLa cells were transfected with HJV and increasing concentration of MT2wt expressing vectors. Whole cell extracts, concentrated media and PI-PLC derived supernatants were then analyzed by western blot. (C) HeLa cells were transfected with HJV cDNA expressing vector and incubated with exogenous soluble MT2. Whole cell extracts, concentrated media and PI-PLC derived proteins were analyzed by western blot. A scheme of the experiment is shown bottom right.
s-HJV and s-MT2 refer to soluble proteins. m-HJV indicates membrane-bound HJV. CL: cellular lysates; CM: conditioned medium; PI-PLC: supernatant after PI-PLC cleavage. The equal loading was verified by α-tubulin.
HJV interacts with matriptase-2
Our results demonstrate that matriptase-2 specifically cleaves m-HJV, generating the 25–30 kDa soluble fragments. If overexpressed, matriptase-2 causes a reduction of all HJV species including the 42 kDa soluble form.
To test whether the two proteins interact, HeLa cells were transiently transfected with matriptase-2wt, in the presence of HJV or an empty vector. Two days after transfection, cell lysates were immunoprecipitated with the anti-HJV antibody and the immunoprecipitates were analyzed by immunoblotting with anti-FLAG, which recognizes matriptase-2. The anti-HJV immunoprecipitate showed the presence of matriptase-2 (Fig. 5A). To confirm the specificity of the interaction, matriptase-2 was immunoprecipitated by the anti-FLAG, and the precipitates analyzed with anti-HJV. Matriptase-2 forms a complex with HJV. HJV also specifically coimmunoprecipitates with matriptase-2MASK (Fig. 5B), demonstrating that the matriptase-2 ectodomain is involved in HJV interaction. We observed that the amount of matriptase-2MASK precipitated with HJV was greater than that of matriptase-2wt, likely because the proteolytic activity of the overexpressed matriptase-2 cleaved HJV. The matriptase-2R774C variant also maintains the ability to interact with HJV (data not shown).
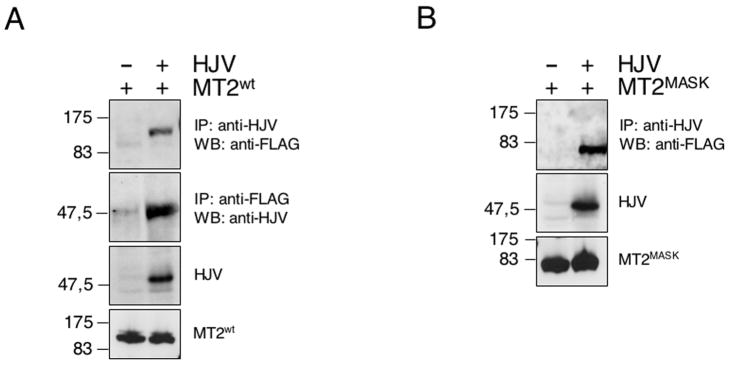
HeLa cells were cotransfected with matriptase-2wt (MT2wt)(A) and MT2MASK (B) in the presence of HJV or an empty vector. Precleared whole cell extracts were immunoprecipitated with anti-HJV and revealed with the anti-FLAG antibody, which recognizes MT2 (panels A and B) or immunoprecipitated with anti-FLAG and revealed with anti-HJV (panel A). To control for transfection, whole cell extracts were loaded and revealed with anti-HJV and anti-FLAG antibodies.
Expression of mutant matriptase-2 in zebrafish leads to a dominant negative phenotype
Mutations in matriptase-2 show a recessive phenotype in both mice and humans. Our binding data, however, show that wild type and mutant matriptase-2 binds to m-HJV. This observation suggests that highly expressed mutant matriptase-2 might act as a dominant negative. To test this possibility we transfected fertilized zebrafish eggs with plasmids expressing matriptase-2wt, matriptase-2R774C or matriptase-2MASK and examined the effect of these constructs on hematopoiesis, in comparison with matriptase-2 morpholino (Fig. 6). Increased hepcidin expression by downregulating ferroportin results in iron-limited erythropoiesis and defects in hemoglobinization in matriptase-2 mutant mice (Folgueras et al., 2008; Du et al., 2008). Results of the matriptase-2 morpholino (Fig 6 A, B) indicate that this occurs also in zebrafish. Expression of matriptase-2wt had no effect on hemoglobin production, whereas expression of matriptase-2MASK severely affected hemoglobin production as well as resulting in smaller fish. Expression of matriptase-2R774C reduced hemoglobin production although to a lesser extent than matriptase-2MASK (Fig. 6 C, D). This result is consistent with matriptase-2R774C showing partial activity. As zebrafish has an endogenous matriptase-2, our results support the hypothesis that the highly expressed mutant protein binds to the endogenous HJV. Injection of fertilized eggs with iron suppressed the hemoglobinization defect in matriptase-2R774C transfected animals, but only partially suppressed the defect when matriptase-2MASK was transfected. This result may derive from the fact that the amount of injected iron is limiting and can only correct the defect with a partially active matriptase or that a mutant matriptase-2 may have effects beyond modulation of iron metabolism.
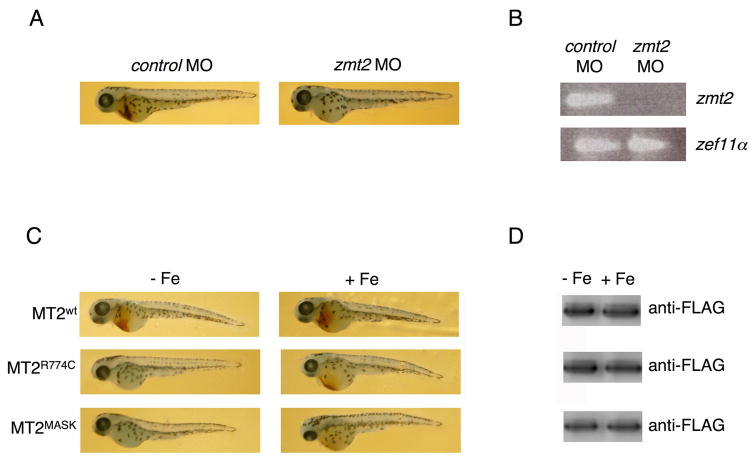
(A) Zebrafish embryos were injected with control (zcontrol MO) and matriptase-2 morpholino (zmt2 MO). Fourty eight hours post injection embryos were stained with o-dianisidine to detect hemoglobinized cells. (B) Embryos as in (A) were homogenized and RNA was extracted as described in Material and Methods. RT-PCR was performed using oligonucleotides specific for matriptase-2 (zmt2) and for the elongation factor 1α(zef1α). Four hundred fifty embryos were injected and the survival rate was 85%. (C) Zebrafish embryos were injected with matriptase-2wt (MT2wt), MT2R774C and MT2MASK expressing vectors, in the presence (+ Fe) or not (− Fe) of iron dextran. At 48 hours after fertilization embryos were stained with o-dianisidine to detect hemoglobinized cells. Representative pictures of the injected fishes (200 for each construct, made in triplicate) are shown. (D) MT2 levels were assayed by western blot analysis.
Discussion
Increasing evidence indicates that the expression of hepcidin, the key regulator of enteric iron absorption and macrophage iron release to plasma, is tightly regulated. In conditions of iron deficiency the expression of hepcidin must be suppressed to increase iron export through ferroportin to plasma transferrin. Proposed inhibitors of hepcidin are HIF-1α(Peyssonnaux et al., 2007), which is increased in hypoxia/iron deficiency and s-HJV, which downregulates hepcidin in vitro (Lin et al., 2005) and in vivo (Babitt et al., 2007). s-HJV administration in mice causes iron overload, identical in the effect to the lack of m-HJV, strengthening the fundamental role of HJV in the hepcidin pathway.
Inactivation of matriptase-2 results in excessive hepcidin production in iron deficiency both in mice and humans. We demonstrated that matriptase-2, cotransfected with HJV in Hep3B cells, was able to inhibit hepcidin activation in a Hep-Luc assay, whereas matriptase-2MASK was inactive, confirming the original observation in HepG2 cells (Du et al., 2008). The human matriptase-2R774C mutant showed decreased inhibitory activity and only inhibited hepcidin when expressed at high concentrations. Matriptase-2 was effective only in the presence of HJV and had no effect in basal conditions or in cells treated with BMP2, likely because of the extremely low endogenous HJV in Hep3B cells (Lin et al., 2005; Babitt et al., 2006). The hepcidin response to BMP maintained in cells transfected with matriptase-2 indicates the integrity of the BMP receptors, excluding BMP receptors as potential targets of matriptase-2. Consistent with their effect on hepcidin expression, matriptase-2 was able to cleave HJV exposed on cell surface, matriptase-2R774C had reduced activity and matriptase-2MASK had no cleavage activity. The residual amount of HJV on cell membrane revealed by binding assay or PI-PLC in cells transfected with matriptase-2wt and mutants agreed well with the effect of the corresponding mutant on the hepcidin promoter. Beside the 42 and 33 kDa soluble forms, cells cotransfected with HJV and matriptase-2 release other smaller size C-terminal fragments in the culture supernatants. To establish their origin we analyzed HJVW191C, which is barely present on plasma membrane but releases 42 kDa s-HJV in the supernatant (Silvestri et al., 2007) and the furin cleavage mutant R335Q, which is present on plasma membrane, but does not release soluble components (Silvestri et al., 2008). After matriptase-2 cotransfection, only HJVR335Q releases the same 25–30 kDa fragments as HJVwt, strengthening the membrane origin of these fragments. Whether these peptides have any physiological role in iron deficiency remains to be investigated.
Soluble HJV (42 kDa) is greatly decreased in HJVW191C transfected cells in the presence of matriptase-2. This is compatible with the strong decrease of HJV in whole cell extracts due to the proteolytic activity of the overexpressed matriptase-2 and strongly supports the assumption that s-HJV is secreted by an intracellular mechanism (Silvestri et al., 2008). Testing the ability of matriptase-2 transfected cells to cleave exogenous s-HJV we formally exclude a cleavage activity of matriptase-2 on s-HJV, suggesting that matriptase-2 is active only if coexpressed with HJV. As a further proof that matriptase-2 targets m-HJV, at low concentration it does not cleave the other HJV species. Although it has been shown that in vitro matriptase-2 cleaves extracellular matrix proteins (Velasco et al., 2002), the full-length protease specifically targets m-HJV. The specificity is accounted for by the physical interaction of the two proteins. The interaction occurs at the matriptase-2 ectodomain and does not require the serine protease domain, as shown by the positive results of coimmunoprecipitation of HJV with matriptase-2MASK. In agreement, matriptase-2wt does not cleave exogenous s-HJV and the released serine protease domains appear inactive on HJV.
The reduced hemoglobinization of zebrafish matriptase-2 morpholino confirms a conserved function for this serine protease and emphasizes that the iron regulatory mechanisms are highly evolutionarily conserved. Decreased hemoglobinization observed in zebrafish overexpressing mutants matriptase-2 indicates that the interaction between matriptase-2 and HJV, that we have demonstrated in vitro, occurs in vivo. Despite having a normal endogenous matriptase-2, matriptase-2MASK injected fishes are anemic, supporting the hypothesis that the highly expressed human mutant protein binds to the endogenous HJV and behaves like a dominant negative mutant.
The ability of matriptase-2 to cleave a GPI-linked protein appears analogous to that of its homologue matriptase-1, an ubiquitous transmembrane serine protease of the same TTPS family, which activates a GPI-serine protease (Leyvraz et al., 2005; Netzel-Arnett et al., 2006). Mutations of matriptase-1 in a mouse model cause ichthyosis with hypotrichosis syndrome, a disorder of the epidermal and oral barrier function (Basel-Vanagaite et al., 2007).
It has been proposed that activation of signaling through matriptase-2 depends upon the proteolytic cleavage event and occurs through the protein cytoplasmic tail (Du et al., 2008). Although no mutants have yet been found at this level, we cannot exclude that the activated cytoplasmic tail signals the iron status to a novel pathway involved in the response to iron deficiency (Du et al., 2008). However, our data emphasize that the inhibitory effect on hepcidin requires the m-HJV cleavage. Published studies suggest that the proteolytic activity of matriptase-2 is essential for its function (Du et al., 2008; Finberg et al., 2008) and the studies presented here demonstrate that HJV is the substrate for that activity. In analogy with matriptase-1 the cytoplasmic and transmembrane domains of matriptase-2 might contribute to the targeting of the protein to a particular plasma membrane side in polarized hepatocytes. Interestingly, rat matriptase-1 expression on intestinal Caco-2 cells results in protein localization almost exclusively on the basolateral side (Satomi et al., 2001).
Matriptase-2 shows tissue-restricted, strong liver expression. It is thus quite reasonable that its proteolytic activity responds to iron deficiency, in order to downregulate the hepcidin activating pathway, based on m-HJV as BMP coreceptor. However, we suggest that the pathway may still be modulated by s-HJV, which inhibits BMP-dependent hepcidin activation (Fig. 7). Hepatocytes from Hjv−/− mice are still able to transcribe hepcidin (although to a lesser extent than wild type) in response to BMP (Babitt et al., 2006) and matriptase-2 is unable to block BMP2-dependent hepcidin activation in our model (Fig. 2A). Soluble HJV, originated from hepatocytes or skeletal muscles, might block the residual BMP-mediated hepcidin activation to fully switch off the pathway in iron deficiency. In this way, the inhibition of hepcidin will be stronger than that obtained by the effect of matriptase-2 alone. In agreement with this interpretation, s-HJV appears not to substitute for the lack of matriptase-2. In conclusion, molecular mechanisms of hepcidin downregulation by s-HJV and by matriptase-2 converge on the BMP-HJV pathway. Whether the matriptase-2 activity may be modulated by other hemochromatosis proteins is an issue for further research.
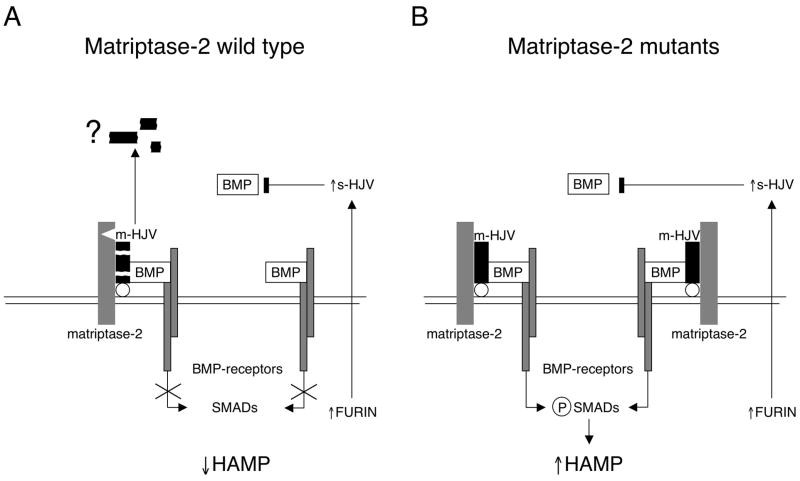
(A) Schematic representation of a model of matriptase-2 activity in iron deficiency. On the left, the serine protease cleaves m-HJV releasing soluble fragments (here simplified by the black boxes). The cleavage sites of matriptase-2 are unknown. The question mark indicates uncertainty on fragments function. The resulting hepcidin inhibition is shown. The complementary effect of s-HJV, produced by furin cleavage, to sequester BMP is shown on the right. (B) Lack of hepcidin inhibition in the presence of matriptase-2 mutations. m-HJV acts as BMP coreceptor and permits hepcidin production in iron deficiency; the effect of s-HJV cannot downregulate hepcidin in the presence of m-HJV.
Material and Methods
Matriptase-2 and HJV expressing vectors
The full-length human matriptase-2 cDNA (NM_153609; C-terminal FLAG-tagged) in pcDNA3.1 was kindly provided by Prof. Carlos Lopez-Otin (Dpto. Bioquimica y Biologia Molecular-IUOPA, Universidad de Oviedo, Spain). The matriptase-2R774C variant (Finberg et al., 2008) was obtained by mutagenesis of the wild type cDNA by using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), according to the manufacturer’s protocol. Matriptase-2MASK, lacking the serine protease domain (Du et al., 2008), was obtained by cloning the cDNA encoding the truncated protein into the EcoRI and XbaI sites of pcDNA3.1(+) (see Supplemental Method for oligonucleotide sequences).
Expressing vectors encoding HJVwt, HJVW191C HJVR335Q were obtained as described (Silvestri et al., 2007; Silvestri et al., 2008).
Cell culture and reagents
Cell culture media and reagents were from Invitrogen and Sigma-Aldrich (St Louis, MO). HeLa cells, which do not express endogenous HJV, and Hep3B cells were cultured respectively in Dulbecco’s modified Eagle’s medium (DMEM) and in Earl’s Minimal Essential Medium (EMEM), supplemented with 2 mM L-glutamine, 200 U/ml penicillin, 200 mg/ml streptomycin, 1 mM sodium pyruvate, and 10% heat-inactivated Foetal Bovine Serum (FBS) at 37°C in 95% humidifier air and 5% CO2. The rabbit polyclonal antisera directed against HJV was obtained as described (Silvestri et al., 2008). Briefly, the cDNA fragment encoding from amino acid 226 to 402 of human HJV was cloned into a pGEX (Amersham Biosciences Europe GmbH, Freiburg, Germany) vector in fusion with GST. The protein was expressed in Escherichia coli, purified by affinity chromatography, and used to elicit antibodies in rabbit. Anti–tubulin, anti-FLAG and anti-cMYC antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA) and Sigma-Aldrich, respectively.
Luciferase assay
A pGL2-basic reporter vector (Promega, Madison, WI, USA) containing a 2.9 Kb fragment of the human hepcidin promoter (Hep-Luc) (Pagani et al., 2008) was used to monitor the hepcidin promoter activation by luciferase assay.
Hep3B cells, seeded at 70–80% of confluency, were transiently transfected with 0.25 μg hepcidin promoter luciferase reporter construct in combination with pRL-TK Renilla luciferase vector (Promega) to control transfection efficiency, and with 0.1 μg/ml of expressing vectors encoding wild type or mutant HJV. Eighteen hours after transfection the medium was changed, cells were serum starved 24 hours in EMEM supplemented with 2% FBS, and then lyzed. When indicated, 50 ng/ml of BMP2 was added to the cell culture. Luciferase activity was determined according to manufacture’s instructions (Dual Luciferase Reporter Assay, Promega). Relative luciferase activity was calculated as the ratio of firefly (reporter) to Renilla luciferase activity and expressed as a multiple of the activity of cells transfected with the reporter alone. Experiments were performed in triplicate.
Western blot analysis
HeLa cells, seeded in 100-mm-diameter dishes up to 70–80% of confluency, were transiently transfected with 20 μg of plasmid DNA and 50 μl of the liposomal transfection reagent Lipofectamine 2000 (Invitrogen) in 3 ml of OptiMem (Invitrogen) according to the manufacturer’s instructions. After 18 hours the medium was replaced with 4 mL of OptiMem and 24 hours later media were collected and concentrated using 5 kDa molecular weight (MW) cutoff ultrafiltration (Amicon Ultra; Millipore, Billerica, MA), and cells were lyzed in RIPA buffer. Proteins were quantified by using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA); equal amount of total proteins (50 μg) were subjected to 10% SDS-PAGE, and then transferred to Hybond C membrane (Amersham Biosciences Europe GmbH, Freiburg, Germany) by standard western blotting technique. Blots were blocked with 2% ECL Advance Blocking Agent (Amersham Biosciences) in TBS (0.5 M Tris-Hcl pH 7.4 and 0.15 M NaCl) containing 0.1% Tween-20 (TBST), incubated 2 hours with rabbit anti-HJV (1:1000), rabbit anti-FLAG (1:1000) or mouse anti-tubulin (1:500). After washing with TBST, blots were incubated 1 hour with relevant HRP-conjugated secondary antisera and developed using a chemiluminescence detection kit (ECL, Amersham Biosciences).
Immunoprecipitation
Immunoprecipitation was performed as follows. Transfected cells were lyzed in NET/Triton buffer (150 mM NaCl, 5 mM EDTA, and 10 mM Tris (pH 7.4) with 1% Triton X-100). Total lysates were cleared with 20 μl of Protein A/G Plus Agarose (Santa Cruz) and 2 μl of irrelevant antibodies (sera from rabbit or mouse) and 500 μg of precleared lysates were incubated with anti-HJV or anti-FLAG at 4°C. After 1 hour, 20 μl of Protein A/G Plus Agarose were added and incubated at 4°C for 12 hours, followed by three washing with NET/Triton buffer. Samples were then eluted with 25 μl of Laemmli Sample Buffer 2x and subjected to SDS-PAGE on 10% acrylamide gel. Immunodetection was performed as described above, except that rabbit TrueBlot (eBioscience, 1:1000 dilution), which does not recognize denatured IgG was used as the secondary antibody.
Electron microscopy and morphometric analysis
HeLa cells were transiently transfected with Lipofectamine 2000 using pcDNA3.1-matriptase-2 (matriptase-2wt), pcDNA3.1-matriptase-2MASK (matriptase-2MASK) and pcDNA3.1-matriptase-2R774C (matriptase-2R774C) constructs. After 18 hours, cells were fixed, labeled with a polyclonal rabbit anti-FLAG using the gold-enhance protocol, embedded in Epon-812, and cut as described. Immunoelectron microscopy (EM) images were acquired from thin sections under a Philips Tecnai-12 electron microscope (Philips, Eindhoven, the Netherlands) using an ULTRA VIEW CCD digital camera (Philips). Thin sections were also used to quantify gold particles residing within different compartments of the secretory pathway.
Analysis of soluble HJV
HeLa cells were transiently transfected for 18 hours, as described in “Western blot analysis”, with HJVwt, HJVW191C or HJVR335Q expressing vectors in the presence of matriptase-2wt cDNA, matriptase-2MASK or matriptase-2R774C variants. Cells were then incubated with serum-free media, which were collected after 24 hours, concentrated using Amicon Ultra, and analyzed by SDS-PAGE. When indicated, HJV or matriptase-2 transfected cells were incubated for 24 hours with matriptase-2 or HJV-conditioned cell culture media (see Supplemental Methods for details). Soluble, membrane and whole cellular proteins were then analyzed by SDS-PAGE.
Cell surface protein quantification by binding assay
Quantification of cell surface expression of HJV and/or matriptase-2 expressing vectors were performed as described, with minor modifications. In brief, 104 HeLa cells were seeded in 48-well plates and transfected with 0.4 μg of plasmid DNA complexed with 1 μL of Lipofectamine 2000, according to the manufacturer’s instructions. After 12 hours, the medium was replaced and 24 hours later cells were fixed with 4% paraformaldehyde for 45 minutes at room temperature. Cells were washed with PBS, blocked with 5% nonfat milk in PBS, incubated with rabbit anti-HJV (1:1000) or rabbit anti-FLAG (1:1000) and then with the relative secondary HRP antibody at 37°C. For total HJV or matriptase-2 expression, cells were permeabilized with 0.1% Triton X-100 in PBS, prior to blocking and incubation with anti-HJV or anti-FLAG. Peroxidase activity was measured with an HSR substrate (o-phenylenediamine dihydrochloride), according to the manufacturer’s instructions. The amount of m-HJV or m-matriptase-2 was calculated as the ratio between the absorbance of unpermeabilized and permeabilized cells. Background absorbance was subtracted for each sample. The Student t test was used for statistical calculation.
PI-PLC cleavage of membrane HJV
A total of 106 HeLa cells, transiently transfected with HJV- and matriptase-2- expressing constructs or the empty vector were incubated in DMEM plus 0.3 U/ml phosphatidylinositol-specific phospholipase C (PI-PLC) at 37°C in a 5% CO2 incubator. After 2 hours, the supernatants were collected. Proteins were precipitated with cold acetone and resuspended in Laemmli sample buffer, in the presence of 4% v/v β-mercaptoethanol (β-ME) when indicated. Samples were then boiled for 10 minutes and loaded on a 10% SDS-PAGE.
Zebrafish embryo cultures and injection
Danio rerio (Zebrafish) were maintained as described (De Domenico et al., 2007). Matriptase-2 expressing vectors were purified using EndoFree purification Kit (Qiagen, Chatsworth, CA). Embryos at stage 1-cell were injected with 50 pg DNA and, when indicated, 100 mg/ml of iron dextran solution (PIGDEX, American Cyanamid Co, Princeton, NJ). At 48 hours after fertilization embryos were stained with o-dianisidine for erythrocytes and proteins were extracted as described previously (De Domenico et al., 2007). Western blot analyses were performed using mouse anti-FLAG (Sigma, 1:5000) followed by peroxidase-conjugated goat anti-mouse IgG (1:10.000, Jackson ImmunoResearch Labs, West Baltimore Pike West Grove, PA).
Matriptase-2 morpholino antisense oligonucleotide was synthezised (5′-GAACACCGCCATCTGAAAACAAATA-3′; intron 2:exon 3 junction) (Gene Tools LLC Corvaliss, OR). A morpholino provided by GeneTools was used as a negative control. One-cell wild type zebrafish embryos were injected with morpholino into the yolk. For each injection 450 embryos were used. After 48 hours post injection embryos were stained as above and total RNA was extracted using RNeasy (Qiagen, Valencia, CA). Zebrafish matriptase-2 (zmt2) RT-PCR was performed on RNA from dechorionated zebrafish embryos using the following primers:
zmt2-sense: 5′-CTCCTGAGAGGCTCCAACAC-3′;
zmt2-antisense: 5′-CTGCTCTCCAACTCCACCTC-3′.
Zebrafish elongation factor 1α(zef1α) was used as a loading control:
zef1α-sense: 5′-GATGCACCACGAGTCTCTGA-3′;
zef1α-antisense: 5′-TGATGACCTGAGCGTTGAAG-3′.
Acknowledgments
We gratefully acknowledge Carlos Lopez-Otin for the matriptase-2-FLAG expressing vector; Roman Polishchuk from the Telethon Electron Microscopy Core Facility (TeEMCoF; Consorzio Mario Negri Sud-Santa Maria Imbaro) for the electron microscopy and morphometric analysis on matriptase-2; Paolo Arosio for the gift of the anti-HJV antibody; Luca Rampoldi for the uromodulin cDNA expressing vector. The authors thank Barry H. Paw (Children’s Hospital Harvard, Boston, MA) who designed the morpholino oligonucleotides and provided protocols.
This work was supported by EU Contract N° LSHM-CT-2006-037296, Italian Telethon Foundation ONLUS Rome Grant GGP05024 and Grant GGP08089, Ministero Istruzione Università e Ricerca, (PRIN 2006) Rome, Italy to CC and National Institutes of Health grant DK 070947 to JK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nature genetics. 2006;38:531–539. [Abstract] [Google Scholar]
- Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. The Journal of clinical investigation. 2007;117:1933–1939. [Europe PMC free article] [Abstract] [Google Scholar]
- Basel-Vanagaite L, Attia R, Ishida-Yamamoto A, Rainshtein L, Ben Amitai D, Lurie R, Pasmanik-Chor M, Indelman M, Zvulunov A, Saban S, Magal N, Sprecher E, Shohat M. Autosomal recessive ichthyosis with hypotrichosis caused by a mutation in ST14, encoding type II transmembrane serine protease matriptase. American journal of human genetics. 2007;80:467–477. [Europe PMC free article] [Abstract] [Google Scholar]
- Bernascone I, Vavassori S, Di Pentima A, Santambrogio S, Lamorte G, Amoroso A, Scolari F, Ghiggeri GM, Casari G, Polishchuk R, Rampoldi L. Defective intracellular trafficking of uromodulin mutant isoforms. Traffic (Copenhagen, Denmark) 2006;7:1567–1579. [Abstract] [Google Scholar]
- Camaschella C. Understanding iron homeostasis through genetic analysis of hemochromatosis and related disorders. Blood. 2005;106:3710–3717. [Abstract] [Google Scholar]
- De Domenico I, Vaughn MB, Yoon D, Kushner JP, Ward DM, Kaplan J. Zebrafish as a model for defining the functional impact of mammalian ferroportin mutations. Blood. 2007;110:3780–3783. [Europe PMC free article] [Abstract] [Google Scholar]
- Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, Beutler E, Beutler B. The Serine Protease TMPRSS6 Is Required to Sense Iron Deficiency. Science. 2008;320:1088–1092. [Europe PMC free article] [Abstract] [Google Scholar]
- Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K, Andrews NC, Fleming MD. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nature genetics. 2008;40:569–571. [Europe PMC free article] [Abstract] [Google Scholar]
- Folgueras AR, Martin de Lara F, Pendas AM, Garabaya C, Rodriguez F, Astudillo A, Bernal T, Cabanillas R, Lopez-Otin C, Velasco G. The membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood. 2008 10.1182/blood-2008-04-149773. [Abstract] [CrossRef] [Google Scholar]
- Lanzara C, Roetto A, Daraio F, Rivard S, Ficarella R, Simard H, Cox TM, Cazzola M, Piperno A, Gimenez-Roqueplo AP, Grammatico P, Volinia S, Gasparini P, Camaschella C. Spectrum of hemojuvelin gene mutations in 1q-linked juvenile hemochromatosis. Blood. 2004;103:4317–4321. [Abstract] [Google Scholar]
- Leyvraz C, Charles RP, Rubera I, Guitard M, Rotman S, Breiden B, Sandhoff K, Hummler E. The epidermal barrier function is dependent on the serine protease CAP1/Prss8. The Journal of cell biology. 2005;170:487–496. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. [Abstract] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science (New York, NY) 2004;306:2090–2093. [Abstract] [Google Scholar]
- Netzel-Arnett S, Currie BM, Szabo R, Lin CY, Chen LM, Chai KX, Antalis TM, Bugge TH, List K. Evidence for a matriptase-prostasin proteolytic cascade regulating terminal epidermal differentiation. The Journal of biological chemistry. 2006;281:32941–32945. [Abstract] [Google Scholar]
- Pagani A, Silvestri L, Nai A, Camaschella C. Hemojuvelin N-terminal mutants reach the plasma membrane but do not activate the hepcidin response. Haematologica. 2008;93(10) [Abstract] [Google Scholar]
- Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nature genetics. 2004;36:77–82. [Abstract] [Google Scholar]
- Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) The Journal of clinical investigation. 2007;117:1926–1932. [Europe PMC free article] [Abstract] [Google Scholar]
- Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, Ernst M, Klein C, Trautwein C. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294–300. [Abstract] [Google Scholar]
- Ramsay AJ, Reid JC, Velasco G, Quigley JP, Hooper JD. The type II transmembrane serine protease matriptase-2--identification, structural features, enzymology, expression pattern and potential roles. Front Biosci. 2008;13:569–579. [Abstract] [Google Scholar]
- Satomi S, Yamasaki Y, Tsuzuki S, Hitomi Y, Iwanaga T, Fushiki T. A role for membrane-type serine protease (MT-SP1) in intestinal epithelial turnover. Biochemical and biophysical research communications. 2001;287:995–1002. [Abstract] [Google Scholar]
- Silvestri L, Pagani A, Camaschella C. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood. 2008;111:924–931. [Abstract] [Google Scholar]
- Silvestri L, Pagani A, Fazi C, Gerardi G, Levi S, Arosio P, Camaschella C. Defective targeting of hemojuvelin to plasma membrane is a common pathogenetic mechanism in juvenile hemochromatosis. Blood. 2007;109:4503–4510. [Abstract] [Google Scholar]
- Valore EV, Ganz T. Posttranslational processing of hepcidin in human hepatocytes is mediated by the prohormone convertase furin. Blood cells, molecules & diseases. 2008;40:132–138. [Europe PMC free article] [Abstract] [Google Scholar]
- Velasco G, Cal S, Quesada V, Sanchez LM, Lopez-Otin C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. The Journal of biological chemistry. 2002;277:37637–37646. [Abstract] [Google Scholar]
- Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–358. [Abstract] [Google Scholar]
- Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, Deng CX. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell metabolism. 2005;2:399–409. [Abstract] [Google Scholar]
- Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–3209. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cmet.2008.09.012
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1550413108003197/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cmet.2008.09.012
Article citations
Serum Iron Overload Triggers the SMAD Pathway and Induces Hepcidin Expression in Hepatocytes through SMURF1.
J Clin Transl Hepatol, 12(9):761-762, 13 Aug 2024
Cited by: 0 articles | PMID: 39280070 | PMCID: PMC11393842
Whole genome discovery of regulatory genes responsible for the response of chicken to heat stress.
Sci Rep, 14(1):6544, 19 Mar 2024
Cited by: 0 articles | PMID: 38503864 | PMCID: PMC10951342
Oral iron therapy: Current concepts and future prospects for improving efficacy and outcomes.
Br J Haematol, 204(3):759-773, 22 Jan 2024
Cited by: 1 article | PMID: 38253961
Review
Genetically determined circulating micronutrients and the risk of nonalcoholic fatty liver disease.
Sci Rep, 14(1):1105, 11 Jan 2024
Cited by: 0 articles | PMID: 38212362 | PMCID: PMC10784479
Activation of Intestinal HIF2α Ameliorates Iron-Refractory Anemia.
Adv Sci (Weinh), 11(12):e2307022, 20 Jan 2024
Cited by: 2 articles | PMID: 38243847 | PMCID: PMC10966566
Go to all (342) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Retinal expression of the serine protease matriptase-2 (Tmprss6) and its role in retinal iron homeostasis.
Mol Vis, 20:561-574, 26 Apr 2014
Cited by: 6 articles | PMID: 24791141 | PMCID: PMC4000719
Matriptase-2 and Hemojuvelin in Hepcidin Regulation: In Vivo Immunoblot Studies in Mask Mice.
Int J Mol Sci, 22(5):2650, 06 Mar 2021
Cited by: 7 articles | PMID: 33800732 | PMCID: PMC7961762
Liver hemojuvelin protein levels in mice deficient in matriptase-2 (Tmprss6).
Blood Cells Mol Dis, 47(2):133-137, 25 May 2011
Cited by: 20 articles | PMID: 21612955
Into the matrix: regulation of the iron regulatory hormone hepcidin by matriptase-2.
Nutr Rev, 67(5):284-288, 01 May 2009
Cited by: 7 articles | PMID: 19386032
Review
Funding
Funders who supported this work.
NIDDK NIH HHS (3)
Grant ID: R01 DK070947
Grant ID: DK 070947
Grant ID: R01 DK070947-04
Telethon (2)
HEMOCHROMATOSIS: FROM GENES TO CLINICS
Prof.ssa Clara Camaschella, Fondazione Centro San Raffaele
Grant ID: GGP05024
HEMOCHROMATOSIS: FROM GENES TO CLINICS AND BACK
Prof.ssa Clara Camaschella, Università Vita Salute San Raffaele
Grant ID: GGP08089