Abstract
Free full text

New Equations to Estimate GFR in Children with CKD
Abstract
The Schwartz formula was devised in the mid-1970s to estimate GFR in children. Recent data suggest that this formula currently overestimates GFR as measured by plasma disappearance of iohexol, likely a result of a change in methods used to measure creatinine. Here, we developed equations to estimate GFR using data from the baseline visits of 349 children (aged 1 to 16 yr) in the Chronic Kidney Disease in Children (CKiD) cohort. Median iohexol-GFR (iGFR) was 41.3 ml/min per 1.73 m2 (interquartile range 32.0 to 51.7), and median serum creatinine was 1.3 mg/dl. We performed linear regression analyses assessing precision, goodness of fit, and accuracy to develop improvements in the GFR estimating formula, which was based on height, serum creatinine, cystatin C, blood urea nitrogen, and gender. The best equation was

This formula yielded 87.7% of estimated GFR within 30% of the iGFR, and 45.6% within 10%. In a test set of 168 CKiD patients at 1 yr of follow-up, this formula compared favorably with previously published estimating equations for children. Furthermore, with height measured in cm, a bedside calculation of 0.413*(height/serum creatinine), provides a good approximation to the estimated GFR formula. Additional studies of children with higher GFR are needed to validate these formulas for use in screening all children for CKD.
GFR is the most useful indicator of kidney function and kidney disease progression; however, determination of true GFR is time-consuming, costly, and difficult to perform for regular clinical use in children. Thus, there is considerable interest in developing formulas to estimate GFR using endogenous surrogate markers such as creatinine1,2 or the low molecular weight protein cystatin C.3–5 The Schwartz formula, devised for children in the mid-1970s,1,6 estimates GFR from an equation that uses serum creatinine (Scr), height, and an empirical constant.1,2,6 GFR as estimated by the Schwartz formula has been used as one of the enrollment criteria for the Chronic Kidney Disease in Children (CKiD) study, an National Institutes of Health–funded North American cohort study whose goal is to recruit children and adolescents with mild to moderate chronic kidney disease (CKD) and characterize progression and the effects of CKD on cardiovascular, growth, and behavioral indices.
We used iohexol plasma disappearance as a gold standard in measuring GFR (iGFR) on the basis of its success as such an agent in Scandinavian adults7,8 and children,9,10 as well as in our recently published pilot study.11 Iohexol is used as a safe, nonionic, low-osmolar contrast agent of molecular weight 821 Da (Omnipaque). It is not secreted, metabolized, or reabsorbed by the kidney,7,12,13 and has <2% plasma protein binding7,14 and nearly negligible extrarenal clearance.15–17
In our pilot study,11 measured iGFR was compared with that estimated using the Schwartz formula,6 and there was a substantial positive bias by the Schwartz formula. This overestimation of GFR has been attributed to the change in creatinine methods since the development of the original formula. The more recent enzymatic creatinine method results in lower determinations compared with the older Jaffe method, even when the latter was improved with a dialysis step and elimination of interfering samples.18 A more accurate estimate of GFR was crucial to the goals of the CKiD study, because iGFR was measured only biennially after the first two visits; therefore, an accurate assessment of GFR is needed during the annual study visits when iGFR is not measured.
Accordingly, a primary goal of CKiD was to develop a formula to estimate GFR using demographic variables and endogenous biochemical markers of renal function, including creatinine, cystatin C, and blood urea nitrogen (BUN). A secondary goal was to determine how well such a formula estimated GFR in the CKiD participants who had a second iGFR, with the idea that such an estimate might substitute for iGFR at visits where, by design, iGFR is not measured. A tertiary goal was to develop a formula that could be applied to the clinical treatment of children with CKD and to generate in clinical laboratories an estimated GFR (eGFR) from endogenous serum markers.
RESULTS
Characteristics of Study Population
Of the 349 children studied, 61% were male, 69% were white, 15% were black, 79% were Tanner stages I through III, and only 20% had a form of glomerulonephritis (Table 1). The median age was 10.8 yr. Body habitus showed notable growth retardation, in that the median age- and gender-specific height percentile was 22.8 compared with the median age- and gender-specific weight percentile of 45.3. The median values of biochemical predictors of kidney function were 1.3 mg/dl, 1.8 mg/L, and 27 mg/dl for Scr, cystatin C, and BUN, respectively. The median iGFR was 41.3 ml/min per 1.73 m2 with an interquartile range from 32.0 to 51.7. Ninety-five percent of the iGFR values were between 21.1 and 75.9 ml/min per 1.73 m2, indicating that the distribution was positively skewed. Fewer than 10% had nephrotic-range proteinuria (urine protein-to-creatinine ratio >2.0), and the median serum albumin was 3.7: none had nephrotic syndrome.
Table 1.
Baseline characteristics of 349 children of the CKiD studya
| Variable | Value |
|---|---|
| Male gender (%) | 61 |
| Race (%) | |
    white white | 69 |
    black black | 15 |
    other other | 8 |
    >1 race >1 race | 8 |
| Hispanic ethnicity (%) | 15 |
| Tanner stage (%) | |
    I I | 60 |
    II II | 11 |
    III III | 8 |
    IV IV | 14 |
    V V | 7 |
| CKD cause (%) | |
    GN diagnosis GN diagnosis | 20 |
    non-GN (genito-urinary, cystic, hereditary) non-GN (genito-urinary, cystic, hereditary) | 73 |
    non-GN other non-GN other | 7 |
| Age (yr; median [IQR]) | 10.8 (7.7 to 14.3) |
| Height (m; median [IQR]) | 1.4 (1.2 to 1.6) |
| Height percentile (median [IQR])b | 22.8 (5.5 to 51.3) |
| Weight (kg; median [IQR]) | 35.0 (23.6 to 53.0) |
| Weight percentile (median [IQR])b | 45.3 (15.0 to 78.2) |
| BSA (m2; median [IQR]) | 1.2 (0.9 to 1.5) |
| BMI (kg/m2; median [IQR]) | 18.1 (16.2 to 21.6) |
| BMI percentile (median [IQR])b | 64.4 (35.3 to 88.4) |
| Serum creatinine (mg/dl; median [IQR]) | 1.3 (1.0 to 1.8) |
| Height (m)/serum creatinine (mg/dl; median [IQR]) | 1.0 (0.8 to 1.3) |
| Cystatin C (mg/L; median [IQR]) | 1.8 (1.5 to 2.3) |
| BUN (mg/dl; median [IQR]) | 27.0 (22.0 to 38.0) |
| Iohexol GFR (ml/min per 1.73 m2; median [IQR]) | 41.3 (32.0 to 51.7) |
Univariate Linear Regression Analyses
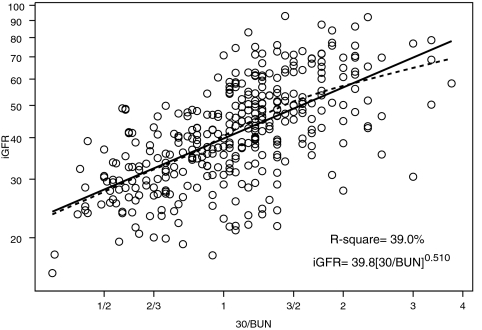
We performed univariate analyses of body surface area (BSA)-unadjusted iGFR on markers of body size and biochemical markers of kidney function (Table 2). BSA and weight had the highest R2 values (57.4 and 56.9%, respectively). Furthermore, the regression coefficient for log(BSA) was 1.074 (not statistically different from 1, P = 0.140); therefore, the classical calibration to a BSA of 1.73 m2 corresponds to the residuals of the regression, and adjusting iGFR for BSA essentially removed the variability in GFR that was attributable to the high variation in body size in our pediatric population. After adjustment for BSA, none of the body size variables had any additional predictive power, and the ability of the endogenous kidney markers to explain the variability of individuals of similar body sizes substantially increased (Table 2). Specifically, height/Scr, as previously shown by Schwartz et al.1 explained the greatest proportion of the variability (R2 = 65.0%) in iGFR when compared with the reciprocals of Scr (R2 = 44.4%), cystatin C (R2 = 47.3%), and BUN (R2 = 39.0%).
Table 2.
Values from univariate linear regression models used to determine the predictability of BSA-unadjusted and BSA-adjusted iGFR for 349 childrena
| Variables | Dependent Variable
| |||
|---|---|---|---|---|
| BSA-Unadjusted iGFRb
| BSA-Adjusted iGFRc
| |||
| Regression Coefficient ± SE | R2 (%) | Regression Coefficient ± SE | R2 (%) | |
| Markers of body size | ||||
    male gender male gender | 0.034 ± 0.059 | 0.1 | 0.035 ± 0.039 | 0.2 |
    age (yr) age (yr) | 0.621 ± 0.042 | 38.7 | −0.007 ± 0.035 | <0.1 |
    weight (kg) weight (kg) | 0.725 ± 0.034 | 57.4 | 0.055 ± 0.034 | 0.8 |
    height (m) height (m) | 1.870 ± 0.102 | 49.0 | 0.075 ± 0.094 | 0.2 |
    BSA (m2) BSA (m2) | 1.074 ± 0.050 | 56.9 | 0.074 ± 0.050 | 0.6 |
| Markers of kidney function | ||||
    1/serum creatinine (mg/dl) 1/serum creatinine (mg/dl) | 0.138 ± 0.066 | 1.2 | 0.545 ± 0.033 | 44.4 |
    height (m)/Scr (mg/dl) height (m)/Scr (mg/dl) | 0.748 ± 0.067 | 26.3 | 0.775 ± 0.031 | 65.0 |
    1/cystatin C (mg/L) 1/cystatin C (mg/L) | 0.745 ± 0.083 | 18.9 | 0.777 ± 0.044 | 47.3 |
    1/BUN (mg/dl) 1/BUN (mg/dl) | 0.539 ± 0.060 | 18.9 | 0.510 ± 0.034 | 39.0 |
Figures 1 through through33 show the scatter plots (in logarithmic scale) of iGFR versus height/Scr, 1.8/cystatin C, and 30/BUN, respectively. The relationships between iGFR and each of these biochemical markers were appropriately described by regression lines, because there was good agreement with nonparametric splines depicted by dashed curves in the figures.
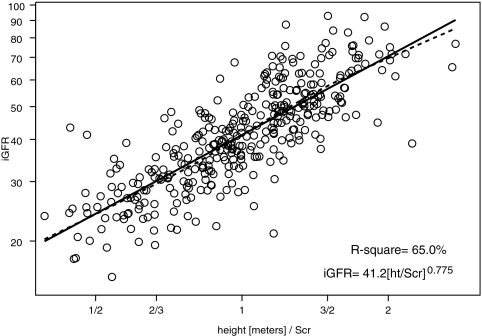
Analysis of log-transformed height/Scr and iGFR, showing that 65.0% of the variation in log(iGFR) can be explained by log(height/Scr). Regression line and nonparametric spline depicted by dashed curve are superimposed.
Model-Based eGFR Formulas
Table 3 shows the regression analyses using the overall mean as the estimate for all individuals (i.e., no model) to the model using height/Scr, cystatin C, BUN, gender, and height (model III). When no model was used, the square root of the mean square error (0.351) was simply the SD of the 349 eGFR values in the logarithmic scale. The updated Schwartz formula corresponds to the particular case of imposing the exponent of height/Scr to be 1, resulting in the equation

which yielded 79.4% of estimated GFR values within 30% of the measured iGFR. If height were reported in cm instead of meters, then this updated Schwartz formula would be

showing a 25% reduction from the previous 0.55 generated by the Jaffe-based Scr measurements, in keeping with the approximate reduction in apparent concentration by isotope dilution mass spectroscopy–referenced enzymatic creatinine determinations.1,6 Figure 1 indicates that the exponent of 1 of height/Scr in the updated Schwartz formula is not correct, because the estimate of the exponent was 0.775, significantly lower than 1. Models IA, IB, and IC in Table 3 show the three bivariate models with each of the three pairs of biochemical markers. Adding cystatin C or BUN to a model with height/Scr improved the R2 to approximately 69%. Model IC, which included only cystatin C and BUN, did not perform as well. When all three variables were incorporated into model II, root mean square error decreased to 0.185. We then tested whether there was any additional predictive power of gender, age, weight, height, BSA, Tanner stage, race, and body mass index. We found that the addition of gender and height (alone) significantly improved the eGFR. This model III,

Table 3.
Precision, goodness of fit, and agreement of eGFR derived from coefficients of indicated regression model; n = 349 children of the CKiD studya
| Model | eGFR= a [height/Scr]b [1.8/Cystatin C]c [30/BUN]d [emale] [height/1.4]f
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | RMSE | R2 (%) | % of eGFR within 30% of iGFR | % of eGFR within 10% of iGFR | |
| None | 41.0 ± 0.8 | 0 | 0 | 0 | 1 | 0 | 0.351 | 0.0 | 52.2 | 20.3 |
| Updated Schwartz | 41.3 ± 0.5 | 1 | 0 | 0 | 1 | 0 | 0.223 | 59.6 | 79.4 | 37.0 |
        IA IA | 41.6 ± 0.4 | 0.599 ± 0.038 | 0.317 ± 0.044 | 0 | 1 | 0 | 0.194 | 69.4 | 84.0 | 38.4 |
        IB IB | 40.7 ± 0.4 | 0.640 ± 0.035 | 0 | 0.202 ± 0.030 | 1 | 0 | 0.196 | 69.1 | 83.7 | 38.4 |
        IC IC | 40.9 ± 0.5 | 0 | 0.569 ± 0.045 | 0.313 ± 0.032 | 1 | 0 | 0.226 | 58.6 | 78.2 | 35.0 |
        II II | 41.1 ± 0.4 | 0.510 ± 0.039 | 0.272 ± 0.043 | 0.171 ± 0.029 | 1 | 0 | 0.185 | 72.3 | 86.3 | 38.7 |
        III III | 39.1 ± 0.6 | 0.516 ± 0.037 | 0.294 ± 0.041 | 0.169 ± 0.027 | 1.099 ± 0.021 | 0.188 ± 0.048 | 0.176 | 75.2 | 87.7 | 45.6 |
showed R2 of 75.2% with root mean square error down to 0.176, resulting in 87.7 and 45.6% of the eGFR values falling within 30 and 10%, respectively, of the measured iGFR. To assess the goodness of fit of the lognormal model [i.e., log(iGFR) as a Gaussian variate], we allowed model III to have residual error distributed as a generalized gamma variate and found that the shape parameter was −0.0098 (95% confidence interval −0.23 to 0.21), consistent with the lognormal model being appropriate to describe the distribution of iGFR as it corresponds to the case of the shape parameter being equal to 0.19
Comparison with Other GFR Prediction Equations
We examined two creatinine-based, two cystatin C–based, and two creatinine- and cystatin C–based prediction equations using published coefficients as well as coefficients derived from the CKiD training data set of 349 children (Table 4). In general, the model coefficients of the previously published formulas were different from those obtained using the CKiD data. This could be due to differing methods of measuring GFR, creatinine, or cystatin C or to CKiD's focus on children with lower levels of GFR.
Table 4.
Comparison of GFR prediction equations using published coefficients and coefficients derived from training data set of 349 children of the CKiD studya
| Model | Data Source | N | Ageb | Agent | GFRb | Equation for eGFR |
|---|---|---|---|---|---|---|
| Scr-based formulas | ||||||
    Counahan et al.2 Counahan et al.2 | Original | 108 | 0.2 to 14 (n = 103) | 51Cr-EDTA | 4 to 200 | 43.00[height/Scr] |
| CKiD | 349 | 1.2 to 17.1 | Iohexol | 16 to 93 | 41.30[height/Scr] | |
    Leger et al.35 Leger et al.35 | Original | 97 | 0.8 to 21 | 51Cr-EDTA | 97 (31 to 200) | 0.641[weight/Scr] + 16.063[height2/Scr] |
| CKiD | 349 | 1.2 to 17.1 | Iohexol | 44 (16 to 93) | 0.542[weight/Scr] + 9.948[height2/Scr] | |
| Cystatin C–based formulas | ||||||
    Filler et al.36 Filler et al.36 | Original | 536 | 11.2 ± 4.5 | 99mTc-DTPA | 103 ± 41 | 91.62[1/CysC]1.123 |
| CKiD | 349 | 10.6 ± 4.2 | Iohexol | 44 ± 15 | 66.22[1/CysC]0.777 | |
    Grubb et al.27 Grubb et al.27 | Original | 536 | <18 (n = 85) | Iohexol | 113, 99, 63c | 84.69[1/CysC]1.680[1.3841(age < 14)] |
| CKiD | 349 | <18 | Iohexol | 41, 42, NAc | 68.06[1/CysC]0.781[0.9661(age < 14)] | |
| Scr- and cystatin C–based formulas | ||||||
    Bouvet et al.20 Bouvet et al.20 | Original | 100 | 1.4 to 22.8 | 51Cr-EDTA | 95 (18 to 200) | 63.2[1.2/CysC]0.56[(96/88.4)/(Scr)]0.35[weight/45]0.30[age/14]0.40 |
| CKiD | 349 | 1.2 to 17.1 | Iohexol | 44 (16 to 93) | 46.1[1.2/CysC]0.33[(96/88.4)/(Scr)]0.55[weight/45]0.77[age/14]0.17 | |
    Zappitelli et al.21 Zappitelli et al.21 | Original | 103 | 12.7 ± 4.7 | Iothalamate | 74 ± 36 | 43.82[1/CysC]0.635[1/Scr]0.547[1.35height] |
| CKiD | 349 | 10.6 ± 4.2 | Iohexol | 44 ± 15 | 25.38[1/CysC]0.331[1/Scr]0.602[1.88height] |
Application to the Visit 2 Testing Data Set
All formulas shown in Table 4 and model III from Table 3 were used to obtain eGFR for the 168 participants whose iGFR was again measured at visit 2, which was scheduled to occur 1 yr after the baseline visit. Table 5 shows the mean and SD of eGFR as well as the bias, 95% limits of agreement, correlation, and the percentage of eGFR values within 30 and 10% of the measured iGFR values.
Table 5.
Application of published and CKiD prediction equations to testing data set of 168 children of the CKiD study at first follow-up visit with iGFR 40.47 ± 16.10 ml/min per 1.73 m2
| Model | Data Source | eGFRa | Biasb | 95% LOAc | Correlation | % of eGFR within 30% of iGFR | % of eGFR within 10% of iGFR |
|---|---|---|---|---|---|---|---|
| Scr-based formulas | |||||||
    Counahan et al.2 Counahan et al.2 | Original | 40.32 ± 17.01 | −0.15 | −18.5, 18.2 | 0.84 | 71 | 32 |
| CKiDd | 38.73 ± 16.34 | −1.75 | −19.7, 16.2 | 0.84 | 73 | 26 | |
    Leger et al.35 Leger et al.35 | Original | 54.37 ± 25.39 | 13.90 | −17.7, 45.5 | 0.79 | 45 | 13 |
| CKiD | 38.95 ± 18.06 | −1.52 | −23.3, 20.2 | 0.79 | 71 | 29 | |
| Cystatin C–based formulas | |||||||
    Filler et al.36 Filler et al.36 | Original | 42.46 ± 18.00 | 1.99 | −20.6, 24.6 | 0.78 | 65 | 26 |
| CKiD | 38.18 ± 11.21 | −2.30 | −22.5, 17.9 | 0.77 | 72 | 37 | |
    Grubb et al.27 Grubb et al.27 | Original | 35.45 ± 22.47 | −5.02 | −33.0, 22.9 | 0.78 | 43 | 14 |
| CKiD | 38.28 ± 11.34 | −2.19 | −22.6, 18.2 | 0.76 | 72 | 33 | |
| Scr and cystatin C–based formulas | |||||||
    Bouvet et al.20 Bouvet et al.20 | Original | 51.50 ± 18.70 | 11.03 | −11.1, 33.1 | 0.80 | 48 | 15 |
| CKiD | 38.31 ± 13.26 | −2.17 | −18.1, 13.8 | 0.86 | 80 | 38 | |
    Zappitelli et al.21 Zappitelli et al.21 | Original | 34.86 ± 15.65 | −5.61 | −22.3, 11.1 | 0.86 | 65 | 32 |
| CKiD | 38.27 ± 13.43 | −2.20 | −17.8, 13.4 | 0.87 | 82 | 38 | |
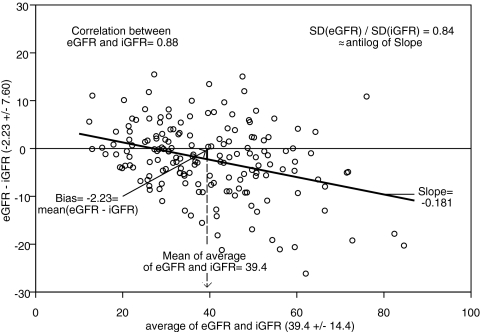
    CKiD equation IIIe CKiD equation IIIe | CKiD | 38.25 ± 13.57 | −2.23 | −17.1, 12.7 | 0.88 | 83 | 41 |
The Counahan2 and updated Schwartz formulas performed comparably and adequately, with absolute bias <2 ml/min per 1.73 m2 and correlation of 0.84. The creatinine- and cystatin C–based formulas outperformed the single endogenous marker formulas, especially when using the coefficients based on the CKiD data. The Bouvet20 and Zapitelli21 formulas based on the CKiD data had an absolute bias of approximately 2 ml/min per 1.73 m2 and correlation of 0.86, and approximately 81 and 38% of eGFR values were within 30 and 10%, respectively, of iGFR values; however, using the published coefficients,20,21 there was more imprecision and less accuracy. When model III was applied, the precision was better (limits of agreement range under 30), and 83 and 41% of eGFR values were within 30 and 10%, respectively, of measured iGFR values. Figure 4 depicts the Bland-Altman plot of eGFR values using model III and iGFR values showing a strong correlation (r = 0.88) with a small bias of −2.23 ml/min per 1.73 m2 and a significantly lower SD of the eGFR values, as they correspond to the mean values of the GFR for a given constellation of the predictors (i.e., the eGFR values do not incorporate the error of the regression coefficients or the residual error of the regression model).
DISCUSSION
Generation of formulas using endogenous serum substrates to estimate GFR is useful in clinical medicine in view of the need to adjust the dosage of nephrotoxic medication in the setting of CKD to prevent further kidney damage. Furthermore, in the CKiD study, GFR is measured directly using disappearance curves of iohexol at the first visit and at all even visits (2, 4, 6, etc.), but endogenous biochemical markers are measured at all visits. The use of the estimating equations at the odd visits will provide a means to have comparable data on GFR at all visits and thus increase the power of the study to describe the trajectories of the GFR decline. Indeed, well-established methods of multiple imputation should be implemented to account appropriately for the imprecision of the estimation at the odd visits.
Endogenous creatinine clearance has been widely used to measure GFR, but this measurement is affected by inaccuracies in quantitatively collecting urine and by the renal tubular secretion of creatinine, which would falsely elevate the apparent GFR.22,23 Moreover, there are methodologic interferences in the measurement of the true concentration of creatinine,6,23,24 and there is a lack of availability of pediatric creatinine serum standards referenced to an isotope dilution mass spectrometry method.25 In the CKiD study, the largest prospective cohort study of CKD in children in North America, we have generated a new eGFR formula, based on an enzymatic creatinine method. The most widely used estimate of GFR is the original Schwartz formula, which was generated from a highly significant correlation between GFR and k*height/Scr.1,6 The current analysis of all of the variables in the CKiD population shows that height/Scr still provides the best correlation with iGFR (R2 = 65.0%; see Figure 1), indicating that a parameter of body habitus along with Scr provides a useful measure of kidney function. Whereas the reciprocal of cystatin C showed a somewhat weaker correlation with iGFR (R2 = 47.3%; see Figure 2), the addition of both cystatin C and BUN to the height/Scr equation (model II) substantially improved the eGFR (Table 3). When all three variables were incorporated into a gender-based equation with an added coefficient for height alone (model III), there was further improvement in root mean square error (0.176) and R2 (75.2%), and 88 and 46% of the estimates fell within 30 and 10%, respectively, of iGFR values, which is quite comparable to the best equations developed for adults; however, previous studies showed that the adult GFR estimating formulas derived from Cockcroft-Gault and the Modification of Diet in Renal Disease (MDRD) are not useful for children.26
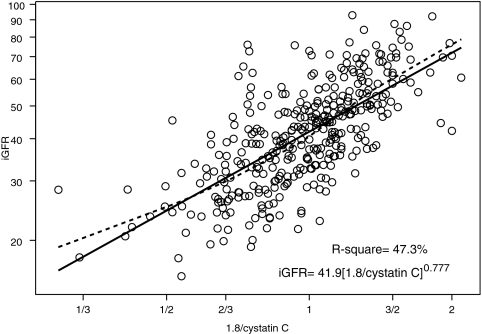
Analysis of log-transformed 1.8/cystatin C and iGFR, showing that 47.3% of the variation in log(iGFR) can be explained by the reciprocal of cystatin C. Regression line and nonparametric spline depicted by dashed curve are superimposed.
Two recent studies estimated GFR in children with higher GFR using equations including both Scr and cystatin C.20,21 We subjected the CKiD data to the models reported by Bouvet et al.20 and Zappitelli et al.21 Whereas these equations with their published coefficients did not perform optimally in our test data set, after correction of their coefficients to the CKiD data, their precision and accuracy were only slightly lower than our model III formula (Table 5). We cannot explain why their original equations did not perform well in our test data set, but differences in GFR level and methods of measuring GFR, cystatin C, and creatinine probably necessitated correcting their coefficients to the CKiD data to optimize performance.
The formulas proposed here to estimate GFR using easily obtained biochemical markers have been developed in a group of children with mild to moderate CKD. A number of limitations preclude rapid generalization of this formula to the general pediatric population for estimation of GFR. Our population with moderate CKD has a median height percentile of 22.8%. Although we collected no direct measures of muscle mass in the CKiD study, we have evidence of delayed puberty compared with normal children. The relationship between eGFR and the biochemical markers may be different in this population than in a population with more normal kidney function and without poor skeletal growth.
Similarly, although we did not observe in our population a change in formula with puberty, other populations with more normal body habitus should be examined to evaluate eGFR coefficients for adolescents. In our CKiD population, only 21% of the children were at Tanner stage IV or V, indicating that there was a very small proportion of fully developed adolescents in the study group. Examination of the estimating formulas in the children with Tanner stages IV through V failed to show substantial differences from the rest of the population (data not shown).
In addition, the method of cystatin C measurement in our study, using the Dako kit, may differ from the Siemens Dade-Behring determination, which has been reported as perhaps the most precise measurement of cystatin C available. The cystatin C–based estimate formula from Grubb et al.27 used the Dako kit as well, and this formula, regardless of whether adjusted for the CKiD training data set, did not perform well compared with our model III estimating equation.
With regard to other studies recently published using enzymatic creatinine method to estimate GFR via k[height/Scr] and measuring true GFR with a reliable method, k values in children and girls older than 13 were 0.500,28 0.430,29 0.470,30 and 0.460,31 compared with the 0.413 determined in this study. Note also that each of these other studies included individuals with normal and near-normal renal function, whereas children in CKiD were selected on the basis of reduced GFR with the mean and median approximating 40 ml/min per 1.73 m2.
In sum, we have developed new formulas to estimate GFR in children with CKD. Precision and accuracy have been optimized by including in the equation endogenous Scr and cystatin C plus BUN, which explained 75.2% of the variability of iGFR, such that 87.7 and 45.5% of all eGFR values were within 30 and 10%, respectively, of simultaneously measured iGFR values. Such formulas performed well in a test group of individuals who had a second iGFR measurement. For the clinician who provides the height of the child, our equations can estimate GFR from the standard chemistry panel, similar to what is provided for adults using the MDRD equation at most clinical chemistry laboratories. We believe that these formulas are useful in the range of GFR from 15 to 75 ml/min per 1.73 m2, but they have not been tested to estimate GFR in children with higher kidney function. Further study of children and adolescents with more normal kidney function will enhance the use of the formulas for use with most children and particularly those with mild CKD.
CONCISE METHODS
Study Participants
The CKiD study was approved by research review boards at all participating sites in the United States and Canada. Eligible individuals were 1 to 16 yr of age with mild to moderate CKD, based on GFR estimates by the Schwartz formula1,6,32 in the range of 30 to 90 ml/min per 1.73 m2 at each local site. At the study visit, demographics, height, weight, and vital signs were determined. BSA was determined using the formula of Haycock et al.33
An intravenous line or butterfly needle was used to administer 5 ml of iohexol and was removed after the injection. A second intravenous line was saline-locked and used for obtaining blood samples. Of the 503 children with an initial study visit before February 2008, 349 (69%) had a successful iGFR determined from four time points (10, 30, 120, and 300 min after infusion of iohexol) and complete data available on height, Scr, BUN, and cystatin C. Baseline sera and the serum separator tubes were shipped at room temperature to the Central Biochemistry Laboratory based in Rochester, NY, and sera for cystatin C were batched and sent quarterly to the nephrology laboratory at Children's Mercy Hospital.
Studies and Assays
Before study blood was obtained for Scr, BUN, and cystatin C determinations, an aliquot was also obtained for HPLC determination of an iohexol blank. Scr (enzymatic) and BUN were analyzed centrally at the CKiD's laboratory at the University of Rochester (G.J.S.) on an Advia 2400 (Siemens Diagnostics, Tarrytown, NY); cystatin C was measured centrally at the Children's Mercy Hospital in Kansas (S. Hellerstein) by a turbidimetric assay (Cystatin C Kit K0071; DAKO SD, Copenhagen, Demark). In a separate study, we showed that the Siemens Bayer Advia creatinine measurement closely agreed with an HPLC method traceable to reference isotope dilution mass spectroscopy developed by the National Institute of Standards and Technology.34 The method of GFR determination using the plasma disappearance of iohexol in a two-compartment system has been previously reported.11 Iohexol (Omnipaque) was provided by GE Healthcare, Amersham Division (Princeton, NJ). No serious adverse events were noted in >700 studies.
Statistical Analysis
Nonparametric statistics (e.g., median and interquartile range) were used to describe the demographics of the study population and the components used in the calculation of iGFR. Regression analyses were performed in three stages. The first stage explored the univariate associations between BSA-unadjusted GFR and various markers of body size. It turned out that BSA and weight had the highest R2 values and, more important, that BSA had a regression coefficient not different from 1. Hence, not only does the classical BSA-adjusted GFR provide a calibration to 1.73 m2, but also its logarithm corresponds to the residuals of the regression of log(GFR) on log(BSA). After this stage, we used these residuals (the GFR of individuals of similar body size) as the outcome to determine the multivariate predictability of a number of markers of kidney function (Scr, height/Scr, cystatin C and BUN). In the final stage, we determined whether other variables, including gender, race, Tanner stage, CKD cause, and body mass index, provided any additional predictive information. We explored potential interactions between variables when there were both significant main effects and biologically plausible interactions; for example, we explored the potential interaction between gender and height/Scr and between pubertal stage and Scr.
We used standard regression techniques for Gaussian data to determine the coefficients of the GFR estimating equations after logarithmic transformation of the continuous variables. All continuous independent variables were centered at the median values when entered into regression models. In this way, the models’ intercepts represent the expected value of GFR for the group of individuals with the constellation of predictors at the centering values (see the Results section).
The general regression model was of the form

where X is a constellation of continuous predictors examined in stage 2 of analyses (e.g., height[m]/Scr[mg/dl], 1.8/cystatin C[mg/L], height[m]/1.4), Z is a constellation of categorical variables (e.g., gender, race) examined in stage 3 of analyses, and  follows a normal distribution with mean 0 and variance σ2 (where σ2 corresponds to the expected value of the mean square error); therefore, a represents the expected value of iGFR for the group whose values of the continuous predictors are at the median values of the study population (e.g., height[m]/Scr[mg/dl] = 1, cystatin C[mg/L] = 1.8, height[m] = 1.4) and whose categorical variables are at the reference categories (e.g., female).
follows a normal distribution with mean 0 and variance σ2 (where σ2 corresponds to the expected value of the mean square error); therefore, a represents the expected value of iGFR for the group whose values of the continuous predictors are at the median values of the study population (e.g., height[m]/Scr[mg/dl] = 1, cystatin C[mg/L] = 1.8, height[m] = 1.4) and whose categorical variables are at the reference categories (e.g., female).
The eGFR,

was obtained by using the expected values of the regression coefficients (a, b, and c) along with specific values of the independent variables (X and Z) for each individual. To assess the properties of the estimating equations, we calculated (1) the root mean square error, which measures the unexplained variability of iGFR; (2) the R2, which measures the percentage of the variability in iGFR explained by the predictors; (3) the correlation between the observed iGFR and eGFR; and (4) the percentage of eGFR values that were within 30 and 10% of the corresponding iGFR values calculated on the training sample (i.e., the one used to develop the equations) and on a testing sample composed of the 168 participants whose iGFR was measured 1 yr after the baseline visit. In addition, we tested the appropriateness of the Gaussian distribution for log(iGFR) against the rich family provided by members of the generalized gamma distribution.19
Comparison with Published Estimating Equations
We compared the estimating equations generated using the CKiD data with published Scr-based estimating equations by Counahan et al.2 and Leger et al.,35 with cystatin C–based equations by Filler et al.36 and Grubb et al.,27 and with Scr- and cystatin C–based equations by Bouvet et al.20 and Zapitelli et al.21 using the originally published formulas and then by modifying the constants and coefficients in the published formulas after fitting them to the CKiD data. Applying each of these equations to the test group of 168 children, we determined the amount of bias from measured iGFR, 95% limits of agreement, correlation, and the percentage of estimates within 30 and 10% of measured iGFR.
Acknowledgments
Data in this article were collected by the CKiD study with clinical coordinating centers (principal investigators) at Children's Mercy Hospital and the University of Missouri–Kansas City (Bradley Warady, MD) and Johns Hopkins School of Medicine (Susan Furth, MD, PhD), and data coordinating center at the Johns Hopkins Bloomberg School of Public Health (Alvaro Muñoz, PhD) with the Central Biochemistry Laboratory at the University of Rochester (George J. Schwartz, MD). The CKiD is funded by the National Institute of Diabetes and Digestive and Kidney Diseases, with additional funding from the National Institute of Neurologic Disorders and Stroke, the National Institute of Child Health and Human Development, and the National Heart, Lung, and Blood Institute (UO1-DK-66143, UO1-DK-66174, and UO1-DK-66116). The CKID web site is located at http://www.statepi.jhsph.edu/ckid.
We are grateful to GE Healthcare, Amersham Division, for providing the CKiD study with iohexol (Omnipaque) for the GFR measurements. We are indebted to Paula Maier for coordinating the central laboratory and tracking the blood samples and to Brian Erway and Dr. Tai Kwong for skillfully developing and maintaining the iohexol HPLC assay at the University of Rochester Medical Center.
References
Articles from Journal of the American Society of Nephrology : JASN are provided here courtesy of American Society of Nephrology
Full text links
Read article at publisher's site: https://doi.org/10.1681/asn.2008030287
Read article for free, from open access legal sources, via Unpaywall:
https://jasn.asnjournals.org/content/jnephrol/20/3/629.full.pdf
Subscription required at www.jasn.org
http://www.jasn.org/cgi/content/full/20/3/629
Free to read at www.jasn.org
http://www.jasn.org/cgi/content/abstract/20/3/629
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1681/asn.2008030287
Article citations
The effect of urinary diversion on kidney function in posterior urethral valves and ureterovesical obstruction.
Int Urol Nephrol, 14 Nov 2024
Cited by: 0 articles | PMID: 39543066
Open-label, controlled, phase 2 clinical trial assessing the safety, efficacy, and pharmacokinetics of INM004 in pediatric patients with Shiga toxin-producing Escherichia coli-associated hemolytic uremic syndrome.
Pediatr Nephrol, 12 Nov 2024
Cited by: 0 articles | PMID: 39528845
Microalbuminuria in Children With Sickle Cell Disease in the Eastern Province of Saudi Arabia.
Cureus, 16(11):e73532, 12 Nov 2024
Cited by: 0 articles | PMID: 39534543
Optimizing Piperacillin Dosing in Pediatric Liver Transplant Recipients: A Case Series.
Pediatr Transplant, 28(7):e14860, 01 Nov 2024
Cited by: 0 articles | PMID: 39319995
Real-world evidence of lisinopril in pediatric hypertension and nephroprotective management: a 10-year cohort study.
Pediatr Nephrol, 28 Oct 2024
Cited by: 0 articles | PMID: 39466390
Go to all (1,955) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Relationships of Measured Iohexol GFR and Estimated GFR With CKD-Related Biomarkers in Children and Adolescents.
Am J Kidney Dis, 70(3):397-405, 24 May 2017
Cited by: 10 articles | PMID: 28549535 | PMCID: PMC5572310
Improved equations estimating GFR in children with chronic kidney disease using an immunonephelometric determination of cystatin C.
Kidney Int, 82(4):445-453, 01 Aug 2012
Cited by: 287 articles | PMID: 22622496 | PMCID: PMC3433576
Longitudinal formulas to estimate GFR in children with CKD.
Clin J Am Soc Nephrol, 4(11):1724-1730, 17 Sep 2009
Cited by: 11 articles | PMID: 19808217 | PMCID: PMC2774961
Estimating renal function in children: a new GFR-model based on serum cystatin C and body cell mass.
Dan Med J, 59(7):B4486, 01 Jul 2012
Cited by: 7 articles | PMID: 22759853
Review
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR000454
NIDDK NIH HHS (6)
Grant ID: U01 DK066116
Grant ID: U01-DK-66174
Grant ID: U01 DK066143
Grant ID: U01 DK066174
Grant ID: U01-DK-66116
Grant ID: U01-DK-66143