Abstract
Free full text

Essential Role of Nuclear Localization for Yeast Ulp2 SUMO Protease Function
Associated Data
Abstract
The SUMO protein is covalently attached to many different substrates throughout the cell. This modification is rapidly reversed by SUMO proteases. The Saccharomyces cerevisiae SUMO protease Ulp2 is a nuclear protein required for chromosome stability and cell cycle restart after checkpoint arrest. Ulp2 is related to the human SENP6 protease, also a nuclear protein. All members of the Ulp2/SENP6 family of SUMO proteases have large but poorly conserved N-terminal domains (NTDs) adjacent to the catalytic domain. Ulp2 also has a long C-terminal domain (CTD). We show that CTD deletion has modest effects on yeast growth, but poly-SUMO conjugates accumulate. In contrast, the NTD is essential for Ulp2 function and is required for nuclear targeting. Two short, widely separated sequences within the NTD confer nuclear localization. Efficient Ulp2 import into the nucleus requires the β-importin Kap95, which functions on classical nuclear-localization signal (NLS)-bearing substrates. Remarkably, replacement of the entire >400-residue NTD by a heterologous NLS results in near-normal Ulp2 function. These data demonstrate that nuclear localization of Ulp2 is crucial in vivo, yet only small segments of the NTD provide the key functional elements, explaining the minimal sequence conservation of the NTDs in the Ulp2/SENP6 family of enzymes.
INTRODUCTION
Small ubiquitin-related modifier (SUMO) is a highly conserved eukaryotic protein that becomes covalently attached to target proteins (Johnson, 2004; Geiss-Friedlander and Melchior, 2007). Sumoylation, the addition of SUMO to a substrate protein, changes the functional properties of the substrate, such as its localization, activity, or interaction with other macromolecules. The sumoylated proteome includes hundreds of different proteins from both the nucleus and cytoplasm, exceeding in complexity all other ubiquitin-like protein (Ubl)-modified proteomes except that of ubiquitin. Protein sumoylation is also under tight temporal and spatial control (Li and Hochstrasser, 1999; Bossis and Melchior, 2006; Xu and Peng, 2006). In the yeast Saccharomyces cerevisiae, SUMO is encoded by a single essential gene (SMT3), whereas mammals typically express three paralogs: SUMO1 and the very similar SUMO2 and SUMO3 proteins (Saitoh and Hinchey, 2000). SUMO1 is ~45% identical to SUMO2 and SUMO3.
Protein modification by SUMO is reversible, and SUMO is synthesized in a C-terminally extended precursor form that must also be processed (Kerscher et al., 2006; Mukhopadhyay and Dasso, 2007). Both the processing of the SUMO precursor to form conjugation-competent SUMO, which ends with two glycines, and the cleavage of SUMO from SUMO–protein conjugates is carried out by a set of specialized cysteine proteases. In the ubiquitin pathway, an organism may have over a hundred different deubiquitylating enzymes (DUBs); these fall into multiple protein families. Even the unicellular eukaryote S. cerevisiae has on the order of 20 different DUBs (Hochstrasser, 1996; Amerik and Hochstrasser, 2004; Nijman et al., 2005). In contrast, only two desumoylating enzymes are known in yeast, Ubl-specific protease-1 (Ulp1) and Ulp2, and only six in humans (Li and Hochstrasser, 2000; Mukhopadhyay and Dasso, 2007). Although additional SUMO proteases from distinct sequence families may yet be discovered, it is unlikely that the number will be anywhere near the number of DUBs. This suggests that each SUMO protease must act on a broad range of substrates and that these enzymes are subject to complex regulation of both their activity and localization. Relatively little is known about how the SUMO proteases are regulated (Mukhopadhyay and Dasso, 2007).
The yeast SUMO proteases provide a useful model for studying the regulation of this important class of enzymes. Ulp1 and Ulp2 share ~27% identity between their catalytically active ULP domains (UDs), but their noncatalytic domains share no obvious similarity (Li and Hochstrasser, 1999, 2000; Strunnikov et al., 2001). Ulp1 is required for cell cycle progression, and it both processes the SUMO precursor and cleaves SUMO from target proteins (Li and Hochstrasser, 1999). Ulp2 is not essential and is not necessary for SUMO precursor processing, but it is required for cell cycle restart after a DNA damage–induced checkpoint arrest (Schwartz et al., 2007). Loss of Ulp2 causes slow growth, increased rates of chromosome loss, and sensitivity to a number of stress conditions (Li and Hochstrasser, 2000; Schwienhorst et al., 2000; Strunnikov et al., 2001; Bachant et al., 2002). A nonidentical set of SUMO conjugates accumulate upon mutation of either ULP. Hence, Ulp1 and Ulp2 have distinct substrates and nonredundant functions. The most abundant of the sumoylated species that accumulate in ulp2Δ cells contain high-molecular-mass poly-SUMO chains, which upon SDS-PAGE are largely retained in the stacking gel (Bylebyl et al., 2003). It is not known whether these chains are free poly-SUMO chains or are anchored to substrates or both.
As noted, six human SUMO proteases of the ULP family have been identified based on their similarity to yeast Ulp1. They have diverged into two distinct branches: SENP1, 2, 3, and 5 are most closely related to yeast Ulp1, and SENP6 and 7 are closest to yeast Ulp2 (Mukhopadhyay and Dasso, 2007). A third, more divergent branch encodes a family of NEDD8-cleaving enzymes; NEDD8 is another Ubl. Like Ulp2, SENP6/SUSP1 and SENP7 both concentrate in the nucleus (Mukhopadhyay and Dasso, 2007). SENP6 is the largest of the human SUMO proteases. It requires a large portion of its ~650-residue N-terminal noncatalytic domain (NTD) for nuclear localization, and the NTD contains multiple potential nuclear localization signals (NLSs); however, the functional NLSs have not been identified nor has the import pathway(s) been determined (Mukhopadhyay et al., 2006). SENP6 is capable of desumoylating long poly-SUMO chains, a property it shares with yeast Ulp2 (Bylebyl et al., 2003). The same is true for SENP7 (Lima and Reverter, 2008).
In general, the catalytic domains of the ULP-class SUMO proteases reside near their C-terminal ends, and the noncatalytic NTDs are associated with subcellular localization and regulation of activity. The NTD of yeast Ulp1, for example, is not required for the essential function of the protein, but is necessary to localize Ulp1 to the nuclear pore complex (NPC; Li and Hochstrasser, 2003; Panse et al., 2003). NPC-localized Ulp1 may be important for desumoylating certain proteins during nucleocytoplasmic trafficking (Lewis et al., 2007). Mislocalization of a fully active Ulp1 catalytic domain to the nucleus is lethal (Mossessova and Lima, 2000; Li and Hochstrasser, 2003; Panse et al., 2003). Moderate overexpression of full-length Ulp1 causes a decrease in overall SUMO conjugates and can partially suppress the ulp2Δ phenotype, indicating that Ulp1 has activity against a broad range of substrates, including at least some substrates that are normally Ulp2 targets, which is consistent with in vitro enzyme assays (Li and Hochstrasser, 2000, 2003). These data suggest that the Ulp1 NTD restricts enzyme activity by spatial sequestration. Localization of Ulp1 to the NPC is not static, however. The septin proteins, which are located at the bud neck in dividing cells, are desumoylated by Ulp1, and the karyopherin Kap121 helps target Ulp1 to the septin ring during mitosis (Makhnevych et al., 2007).
The Ulp2 branch of the SUMO protease family is much less studied. Enzymatic activity in vitro is generally very weak (Li and Hochstrasser, 2000; Drag and Salvesen, 2008; Lima and Reverter, 2008), and little is known about the large noncatalytic domains of these proteins (Mukhopadhyay and Dasso, 2007). Yeast Ulp2 concentrates in the nucleus, although the significance of this localization is not yet established. It appears to be able to bind chromatin and may desumoylate chromatin-associated proteins such as histones and topoisomerase II (Strunnikov et al., 2001; Bachant et al., 2002; Hannich et al., 2005; Nathan et al., 2006). Other studies have correlated functional defects associated with loss of Ulp2 or its mammalian ortholog SENP6 with the failure to disassemble large poly-SUMO conjugates (Bylebyl et al., 2003; Mukhopadhyay et al., 2006). Here we show that the C-terminal noncatalytic domain (CTD) of Ulp2 is required for efficient depolymerization of large poly-SUMO conjugates in vivo, although this mutant grows much more robustly than a ulp2 null mutant.
Members of the Ulp2 subfamily of SUMO proteases all contain a large but poorly conserved NTD. The mammalian SENP6 NTD is required for nuclear localization (Mukhopadhyay et al., 2006). In the current work, we report that the NTD of yeast Ulp2 is also necessary and sufficient for nuclear localization. Such localization is critical for all tested Ulp2 functions. Surprisingly, when we replaced the entire NTD with a single heterologous 7-amino acid NLS, in vivo functionality is almost completely restored. The Ulp2 NTD has two short classical NLS-like motifs, and either is sufficient for both nuclear localization and function of Ulp2. Consistent with this, a yeast strain with a mutated Kap95 nuclear import receptor is defective for Ulp2 nuclear import and also shows an aberrant buildup of poly-SUMO structures. Our data indicate that the low sequence conservation of the NTDs of the Ulp2 family of proteases can be explained by the fact that only a pair of small elements widely dispersed within the domain is actually essential for proper cellular function.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Yeast strains and plasmids used in this study are listed in Tables 1 and and2,2, respectively. Yeast were grown and manipulated by standard methods (Guthrie and Fink, 1991). Recombinant DNA work was done in Escherichia coli strains TOP10 and MC1061 using standard techniques (Ausubel et al., 1989). For epitope-tagging of Ulp2, chromosomal ULP2 was tagged at the 3′ end of its open reading frame (ORF) in yeast by homologous recombination with a PCR fragment encoding nine repeats of a c-Myc epitope, generating the ULP2-9myc (MHY2978) strain (Li and Hochstrasser, 2000). Similarly, to create the ulp2ΔC341 (MHY3675) and ulp2ΔC631 (MHY4755) mutants, the chromosomal ULP2 allele was truncated either after residue 693 or residue 403, respectively, and a 13myc-epitope tag was placed at the C-terminal end of the resulting Ulp2 derivative.
Table 1.
Yeast strains
| Strain | Genotype | Origin |
|---|---|---|
| MHY500 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 | Chen et al. (1993) |
| MHY1380 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2Δ::HIS3 | Li and Hochstrasser (2000) |
| MHY2978 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ULP2-9myc::his5+ | This study |
| MHY3675 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2ΔC341-13myc::his5+ | This study |
| MHY3677 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2ΔN411-9myc::his5+ | This study |
| MHY4739 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ULP2-13myc-kanMX6::TRP1 | This study |
| MHY4741 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2ΔN411-NLSSV40-13myc-kanMX6::his5+ | This study |
| MHY4742 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2ΔN411-*NLSSV40-13myc-kanMX6::his5+ | This study |
| MHY4876 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2Δ::HIS3 ulp2-4AAA-13myc-kanMX6::TRP1 | This study |
| MHY4754 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2Δ::HIS3 ulp2-320AA-13myc-kanMX6::TRP1 | This study |
| MHY4744 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2Δ::HIS3 ulp2-4AAA/320AA-13myc-kanMX6::TRP1 | This study |
| MHY4755 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ulp2ΔC631-13myc-kanMX6 | This study |
| MHY5100 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 kap95Δ::his5+ ULP2-9myc::his5+ (pRS314kap95-14) | This study |
| MHY5101 | MATahis3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 kap95Δ::his5+ ULP2-9myc::his5+ (pRS316KAP95) | This study |
| MHY5217 | MATα/ahis3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 lys2-801/lys2-801 trp1-1/trp1-1 ULP2/ulp2Δc341::13myc::his5+ SIR2/sir2Δ::kanMX | This study |
Table 2.
Plasmids
| Plasmid | Origin |
|---|---|
| pRS315-ULP2-13myc-kanMX6 | This study |
| pRS315-ULP2Δ2-133-13myc-kanMX6 | This study |
| pRS315-ULP2Δ134-296-13myc-kanMX6 | This study |
| pRS315-ULP2Δ297-403-13myc-kanMX6 | This study |
| pRS315-ULP2Δ2-296-13myc-kanMX6 | This study |
| pRS315-ULP2Δ134-403-13myc-kanMX6 | This study |
| pRS315-ULP2Δ2-133/Δ297-403-13myc-kanMX6 | This study |
| pRS315-ULP2Δ2-403-13myc-kanMX6 | This study |
| pRS304-ULP2-13myc-kanMX6 | This study |
| pRS304-ULP2-4AAA-13myc-kanMX6 | This study |
| pRS304-ULP2-320AA-13myc-kanMX6 | This study |
| pRS304-ULP2-4AAA/320AA-13myc-kanMX6 | This study |
| p424GAL1-Flag-Ulp2 | This study |
| p424GAL1-Flag-Ulp2(1-403) | This study |
| p424GAL1-Flag-Ulp2(667-1034) | This study |
| pRS314-Ulp2 | This study |
The ulp2ΔN411 strain (MHY3677) was generated from MHY2978 by delitto perfetto (Storici et al., 2001), in which the N-terminal coding sequence of ULP2 (corresponding to amino acids 2-411) was precisely deleted, resulting in a fusion of the start codon with amino acid 412 of Ulp2. The genomic deletion was confirmed by DNA sequencing. Strains carrying ulp2ΔN-NLSSV40 alleles (MHY4741 and MHY4742) were generated directly in the yeast MHY3677 strain by inserting an NLS-encoding DNA segment at the 3′ end of the ulp2ΔN ORF. The DNA sequence, which was generated by PCR, encoded either the classical NLS of the large-T antigen of SV40 virus, the PKKKRKV heptapeptide, or an inactive derivative of this NLS (PKTKRKV; Kalderon et al., 1984), followed by a 13myc tag. The proper insertion of the segment of DNA was then confirmed by sequencing the 3′ end of the ulp2ΔN ORF. Strains MHY5100 and MHY5101 were made by crossing a ULP2-myc9 strain to a congenic kap95-14 mutant (Makhnevych et al., 2007).
For construction of plasmids encoding N-terminally deleted versions of Ulp2, the Exsite Mutagenesis protocol (Stratagene, La Jolla, CA) was used. The various deletions were generated by this PCR-based approach using a plasmid containing full length ULP2 as template DNA. 5′-phosphorylated primers were designed to hybridize adjacent to the intended deletion, priming in the direction away from the intended deletion; this resulted in an amplified linear plasmid sequence that only lacked the specific sequence between the 5′-ends of the two primers. The amplified products were self-ligated using T4 DNA ligase (NEB) and then transformed into E. coli MC1061. The deletions were confirmed by DNA sequencing. To generate a plasmid lacking both the A and C coding regions of Ulp2 (see Figure 3A), region C was deleted following the method describe above, but the template was a ULP2 plasmid that already had the A coding region deleted.
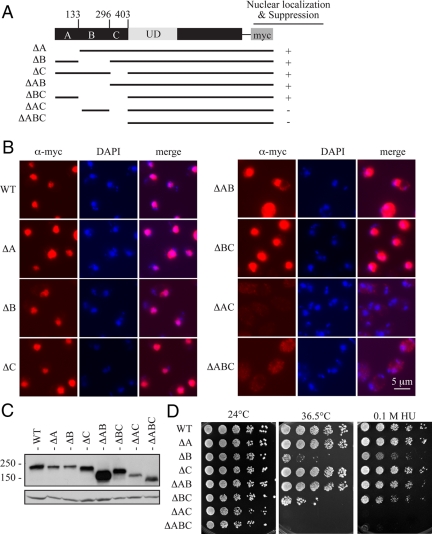
Ulp2 contains at least two distinct NLSs. (A) Schematic of plasmid-borne alleles encoding a series of myc epitope-tagged Ulp2 N-terminal deletions. Nuclear localization of the construct and its ability to suppress growth defects of the ulp2Δ strain are noted. (B) Anti-myc immunofluorescence and DNA (DAPI) images of the indicated strains are shown as in Figure 2. (C) Anti-myc immunoblot analysis of the proteins diagrammed in A. Anti-Pgk1 was used to compare sample loading. (D) Serial dilutions of yeast cultures were spotted onto plates and grown under the indicated conditions. Cells were photographed after 6 d of growth.
For the construction of ulp2-nls mutant strains, the pRS304-ULP2-13myc-kanMX6 plasmid was mutagenized by Quikchange (Stratagene) to change the sequence of NLS1 (4-RKRK-7) to AAAK and NLS2 (320-KKTK-323) to AATK. Numbers denote positions in the Ulp2 protein sequence. Mutations were confirmed by DNA sequencing. To integrate the wild-type (WT) ULP2 and ulp2-nls mutant alleles into the genome, the plasmids were linearized within the TRP1 gene by the XbaI restriction endonuclease and were transformed into ulp2Δ yeast cells, selecting for tryptophan prototrophy. Expression of the tagged Ulp2 derivatives was verified by anti-myc immunoblotting. For the construction of plasmids encoding the N-terminally Flag-tagged CTD (667-1034), NTD (1–403), or full-length Ulp2, the corresponding DNA coding sequences with the added Flag-tag sequence were cloned into the p424GAL1 expression vector between the SmaI and PstI restriction sites.
Yeast Growth Assays
Strains were grown at 24°C in liquid culture until they reached stationary phase. Cells were diluted to an OD600 of 0.4 and subsequently by fivefold serial dilutions, and the dilution series were spotted onto various tester plates. Plates containing 10 μg/ml benomyl or 0.1 M hydroxyurea were incubated at 24°C.
Immunoblotting
Total yeast cell extracts from 2.5 OD600 equivalents of cells were prepared from logarithmically growing yeast cultures grown at 24°C. Cells were lysed using 20% trichloroacetic acid (Ferris et al., 2005). Lysates were resolved by SDS-PAGE on 6–15% gradient gels. Proteins were transferred to PVDF membranes (Millipore, Bedford, MA) and analyzed by immunoblotting using various antibodies. The following antibody dilutions were used: anti-SUMO, 1:3000 (Li and Hochstrasser, 1999); anti-myc 9E10 antibody, 1:5000 (Covance); anti-Flag M2 antibody, 1:10,000 (Sigma); and anti-Pgk1, 1:40,000 (22C5 antibody, Molecular Probes, Eugene, OR) in Tris-buffered saline-0.1% Tween-20 (TBST)/1% dry milk. For anti-SUMO immunoblots, the secondary antibody used was anti-rabbit IgG-HRP–linked whole antibody from donkey (GE Healthcare, Waukesha, WI) diluted 1:4000 in TBST/1% milk, and for immunoblotting with the other antibodies, anti-mouse IgG-HRP–linked whole antibody from sheep (GE Healthcare) diluted 1:5000 in TBST/1% milk was used. ECL reagents (GE Healthcare) were used for protein visualization.
Immunofluorescence Analysis of Yeast Cells
A protocol based on that of Lewis et al. (2007) was used for immunofluorescence studies. Overnight liquid cultures were diluted to an OD600 of 0.15 and grown to midlogarithmic phase in minimal medium at 24°C. Ten to 15 OD600 equivalents of cells were collected, washed once with water, resuspended in 1 ml of 3.7% formaldehyde in water, and incubated for 90 min at 30°C. Cells were pelleted at 400 × g for 2 min and washed twice in 5 mM MgCl2, 0.1 M KPO4, pH 6.8. Cells were resuspended in 1 ml of buffer C (1.2 M sorbitol, 5 mM MgCl2, 0.1 M KPO4, pH 6.8), which also contained 10 μl of 5 mg/ml Zymolyase 100T (Seikagaku, Tokyo, Japan) and 2.5 μl β-mercaptoethanol to digest the cell wall. Cells were incubated for 40 min at 30°C, and then they were washed twice with 1 ml of buffer C, resuspended in 300 μl of buffer C, and spotted onto a pretreated 18 × 18-mm glass coverslip (Fisher Scientific, Pittsburgh, PA) or poly-l-lysine–coated glass slides (Sigma, St. Louis, MO). Coverslips were prepared by 1-h incubation in 1.0 N HCl, followed by rinsing in at least 20 volumes of water, and stored in 95% ethanol. Before use, ethanol was evaporated from the coverslips, and they were treated with 100 μl of a 1 mg/ml solution of poly-l-lysine hydro-bromide MW 70,000–150,000 (Sigma) for 5 min, washed three times with water, and air dried.
The cell suspension in buffer C was applied to the coverslips and incubated for 7 min at room temperature. Coverslips were washed once with phosphate-buffered saline (PBS), and then blocked for 10 min in blocking solution (PBS and 5% goat serum [Sigma], 1% BSA, 0.2% Nonidet P-40, 0.02% sodium azide) at room temperature. Coverslips were then incubated with anti-myc 9E10 antibody (GE Healthcare) diluted 1:1000 in PBS, 1% BSA for 90 min at 30°C, washed four times in PBS and two times in blocking solution, and then incubated with Alexa Fluor 594–conjugated goat anti-mouse secondary antibody (Invitrogen, Carlsbad, CA) diluted 1:1000 in PBS, 1% BSA for 60 min at room temperature in the dark. Finally, coverslips were washed two times with blocking solution and once in PBS. The coverslips were then mounted onto a slide with gentle compression in Prolong Gold anti-fade reagent with DAPI (Invitrogen) and sealed with nail polish. For the experiments using poly-l-lysine–coated slides, cell suspension was applied directly to the glass slides and antibody incubation and washing were done on the slides. The slides were stored overnight in the dark at room temperature, and the samples were visualized on an Axioskop epi-fluorescence microscope (Carl Zeiss MicroImaging, Thornwood, NY) with a plan-apochromat 100× NA 1.4 objective lens at room temperature using Zeiss immersion oil IMMERSOL 518F. Pictures were taken on a Zeiss Axiocam camera using a Uniblitz shutter driver (model VMM-D1; Vincent Associates, Rochester, NY) and the program Open Lab 3.1.5 (Improvision, Lexington, MA). Pseudocolored images were merged using ImageJ software (http://rsb.info.nih.gov/ij/; Abramoff et al., 2004).
RESULTS
Roles of the Noncatalytic N- and C-terminal Domains of Ulp2 In Vivo
As shown in Figure 1A, Ulp2 has long extensions N- and C-terminal to the catalytic domain (UD). These domains contain no obvious sequence similarity to Ulp1 or other yeast proteins. To determine the potential contribution of the NTD and CTD to Ulp2 function, we removed the corresponding coding regions from the ULP2 gene and determined whether the resulting yeast strains had any phenotypic abnormalities. Chromosomal alleles were generated that lacked either the sequence for Ulp2 residues 2-411 (Ulp2ΔN) or for residues 694-1034 (Ulp2ΔC) and were tagged with a myc-epitope sequence for protein detection. Deletion of either the NTD or the CTD did not reduce the expression level of the protein relative to the full-length tagged Ulp2 protein (Figure 1B).

Strains lacking either noncatalytic domain of Ulp2 are functionally defective. (A) Schematic of terminal deletion alleles. Chromosomal copies of full-length ULP2 and ulp2ΔN included in-frame sequences encoding a C-terminal 9myc epitope tag, and ulp2ΔC encodes a C-terminal 13myc tag. (B) Anti-myc immunoblot showing expression levels of the Ulp2 derivatives. (C) Anti-SUMO immunoblot of whole cell lysates from the indicated strains. Anti-Pgk1 immunoblotting was used as a loading control. Strains were harvested during logarithmic growth at 24°C. Top panel shows a longer exposure of the stacking gel. (D) Fivefold serial dilutions of yeast cultures were spotted onto plates and grown under the indicated conditions. Cells were photographed after 4 d growth except for the hydroxyurea-containing plate, which was photographed after 7 d.
Both noncatalytic domains were required for full Ulp2 function. In the ulp2ΔN and ulp2ΔC strains, high-molecular-weight poly-SUMO conjugates accumulated; these were visualized by anti-SUMO immunoblotting as heterogeneous slow-migrating species near the top of the resolving gel and in the stacking gel (Figure 1C). This is a hallmark of cells lacking Ulp2 function (Bylebyl et al., 2003). However, there were subtle differences between the different deletion strains in the exact pattern and extent of SUMO conjugates that accumulated, particularly in the lower mass region of the gel. For example, a prominent ~40-kDa SUMO conjugate and a less distinct species of ~52 kDa were observed in WT as well as ulp2ΔC cells, but were lost in the ulp2Δ (complete deletion) and ulp2ΔN strains. These smaller sumoylated species may result from Ulp2-dependent SUMO cleavage of long poly-SUMO chains from a protein conjugate until only a mono- and di-sumoylated protein remains. In vitro evidence for such an activity for Ulp2 has been reported (Li and Hochstrasser, 2000; Bylebyl et al., 2003).
We also compared growth under various conditions of the ulp2ΔN and ulp2ΔC mutants to WT and ulp2Δ strains (Figure 1D). The ulp2ΔN strain grew poorly under all growth conditions tested. Moreover, at 30°C and particularly at 33°C, the ulp2ΔN mutant grew more poorly than a congenic strain lacking the entire ULP2 sequence. It is therefore possible that removing the NTD from the Ulp2 catalytic domain causes some defects distinct from simply deleting Ulp2 from the cell, e.g., it could lead to active but mislocalized Ulp2 activity (see below). The results of Figure 1, C and D, indicate that the NTD of Ulp2 is necessary for most if not all Ulp2 functions.
Despite its accumulation of high-molecular-mass poly-SUMO conjugates, the ulp2ΔC strain grew only slightly slower than the WT strain at temperatures below 36.5°C (Figure 1D). This was clearer at shorter incubation times (Supplementary Figure S1). A growth deficit was observed in the presence of DNA damage–inducing concentrations of hydroxyurea and, to a much lesser degree, the microtubule-depolymerizing drug benomyl (Figure 1D). Loss of Ulp2 has been found to be synthetically lethal with sir2Δ, suggesting that yeast cells defective for Sir2 chromatin-mediated silencing are sensitized to defects in SUMO deconjugation (Darst et al., 2008). However, there was little or no enhancement of the ulp2ΔC growth defect by sir2Δ (Supplementary Figure S1). Thus, deletion of the CTD sequence alone clearly caused a less severe set of growth defects than did deletion of the NTD or the entire Ulp2 coding sequence. The accumulation of poly-SUMO conjugates was slightly less pronounced in ulp2ΔC cells compared with ulp2Δ (Figure 1C and not shown). Therefore, either cells can tolerate moderate levels of poly-SUMO conjugates or Ulp2 cleavage of more than just SUMO–SUMO linkages is important to its function.
We also determined whether high levels of either the NTD or the CTD (or full-length Ulp2) had any effects on the growth of WT yeast cells. Expression of FLAG epitope-tagged proteins from high-copy plasmid-borne alleles was driven by the strong galactose-inducible GAL1 promoter. As can be seen in Supplementary Figure S2A, the NTD and CTD were both expressed robustly and at slightly higher levels than the full-length Ulp2. Although strong expression of Ulp2 caused a severe impairment of cell growth and reduced SUMO conjugates, overexpression of either the NTD or CTD had no detectable effect on growth (Supplementary Figure S2B) or on SUMO-conjugate levels (Supplementary Figure S2C). This suggests that the noncatalytic Ulp2 domains overexpressed to this degree do not compete significantly for any essential target of either domain.
The N-terminal Domain of Ulp2 Is Necessary and Sufficient for Nuclear Localization
Ulp2 normally concentrates in the nucleus (Li and Hochstrasser, 2000; Schwienhorst et al., 2000). Whether such localization is required for Ulp2 function in cell growth or in the response to various stresses has not been tested, nor has the molecular basis of its subcellular distribution been examined. We first tested whether either of the noncatalytic domains of Ulp2 was important for nuclear localization. Deletion of the 341-residue CTD, which is relatively acidic (pKi = 4.4), did not affect nuclear localization of the myc-epitope–tagged protein based on immunofluorescence staining (Figure 2, compare first and third rows). In contrast, a Ulp2 derivative lacking the very basic (pKi = 10.5), 411-residue NTD did not accumulate in the nucleus, localizing instead throughout the cell (Figure 2, row 2). Conversely, the N-terminal domain of Ulp2 by itself (residues 1-403) was sufficient for localization to the nucleus (row 4). Therefore, the N-terminal domain of Ulp2 is both necessary and sufficient for the nuclear localization of the protein, and failure to localize to the nucleus correlates with a severe functional deficiency in vivo (Figure 1D).

The N-terminal domain of Ulp2 is necessary and sufficient for nuclear localization. Immunofluorescence and differential interference contrast (DIC) images were collected for cells expressing the indicated chromosomally integrated ULP2-myc derivatives. Anti-myc immunofluorescence and DNA (DAPI) images were pseudocolored red and blue, respectively, and merged to ascertain nuclear localization of the Ulp2 derivatives. The anti-myc immunofluorescence signal for Ulp2NTD was more intense than the other constructs, so the contrast and brightness of immunofluorescence images for this construct were adjusted separately from the other panels in the figure.
The N-Terminal Domain of Ulp2 Contains Multiple NLSs
The above results demonstrated that the NTD of Ulp2 is responsible for nuclear localization of the protein. Therefore we sought to identify sequences in the NTD that mediate nuclear import. To begin, we generated a set of three plasmid-borne ulp2 deletion alleles, which each lacked approximately one-third of the sequence for the NTD (ΔA, ΔB, and ΔC; Figure 3A). All three Ulp2 deletion derivatives were expressed at levels comparable to WT Ulp2 and retained the ability to localize to the nucleus, implying that the NTD has at least two separate NLSs (Figure 3, B and C). We then made a series of Ulp2 derivatives with pairwise deletions of the A, B, or C sequences and examined their subcellular distributions. The Ulp2 construct that lacked both the A and C regions (residues 2-133 and 297-403; ΔAC) no longer concentrated in the nucleus (Figure 3B). Moreover, a plasmid expressing this derivative could no longer suppress ulp2Δ (Figure 3D). A protein lacking all three regions (residues 2-403; ΔABC) was similarly defective for both localization and function, as expected. Anti-myc immunoblotting indicated that all of the Ulp2 derivatives were well expressed, although several of the proteins were present at levels slightly below that of WT Ulp2 (Figure 3C).
Based on the ulp2Δ complementation assays shown in Figure 3D, the ulp2-ΔB and ulp2-ΔBC proteins appear to be partially defective for function despite localizing to the nucleus. However, in these plasmid-based complementation assays, we encountered some variability in colony-forming ability, and upon repetition the strains expressing the ΔB and ΔBC derivatives grew better than on the plates shown, whereas strains transformed with other ulp2 deletion alleles sometimes grew slightly worse (data not shown). A likely explanation for this variability is that strains deleted for the chromosomal copy of ULP2 lose plasmids spontaneously at higher frequency than WT (Li and Hochstrasser, 2000). Therefore, the differences in suppression are probably due to variable plasmid loss under the experimental conditions used. Importantly, although colony growth fluctuated somewhat between assays, the transformants carrying NTD single-segment deletions or the ΔAB and ΔBC constructs always suppressed the ulp2Δ growth defects at least partially, whereas the ΔAC and ΔABC constructs never showed any suppression (Figures 1D and and33D).
From these results, we conclude first, that both the A and C segments of Ulp2 contain information sufficient to localize Ulp2 to the nucleus and second, that nuclear localization of Ulp2 is likely to be necessary for its function. These inferences are strongly supported by the data presented below.
Identification of Functional NLSs in Ulp2
In yeast, the classical monopartite NLS (cNLS) is composed of a stretch of approximately four basic amino acids, often beginning with an N-terminal lysine residue (Hodel et al., 2001). The cNLS binds α-importin, which moves into the nucleus as part of the α-importin/β-importin complex, allowing for nuclear import of the bound cargo through the NPC (Lange et al., 2007). The NTD of Ulp2 contains two distinct stretches of basic amino acids located at residues 4-7 (in segment A) and residues 320-323 (in segment C) that are also relatively well conserved in other Saccharomyces species.
Based on the foregoing deletion analysis, these basic elements may function as NLSs. To test this, we substituted alanines for several basic residues in each predicted NLS in the context of full-length Ulp2 expressed from a chromosomally integrated allele (Figure 4A). As can be seen in Figure 4B, mutating only one of the two putative NLSs did not affect the nuclear localization of the proteins; however, when both NLSs were mutated, the resulting Ulp2 protein no longer concentrated in the nucleus. In concordance with the localization results, the ulp2-nls1,2 double mutant, but not the ulp2-nls1 or ulp2-nls2 single mutants, accumulated an excess of poly-SUMO conjugates and displayed growth defects that were indistinguishable from those of the ulp2 null mutant (Figure 5). The single mutants were expressed at levels comparable to WT and the double mutant at a level slightly below this (Figure 5A).

Either NLS within the NTD of Ulp2 is sufficient for nuclear localization of the protein. (A) Predicted NLSs and the sequences of the mutated versions. Numbers denote position in Ulp2 amino acid sequence. (B) Anti-myc immunofluorescence images of proteins expressed from chromosomally integrated ulp2-nls mutant derivatives.
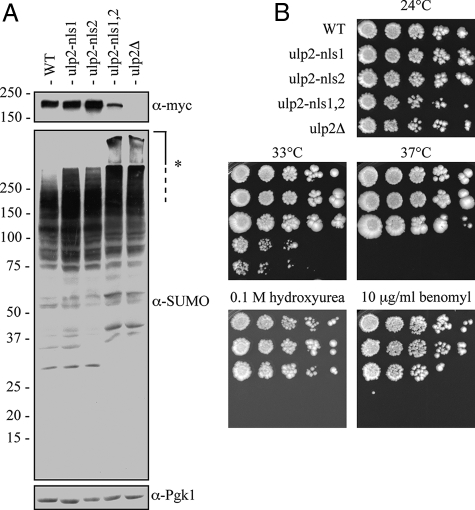
Mutation of both NLS1 and NLS2 in Ulp2 (ulp2-nls1,2) yields a null phenotype. (A) Anti-SUMO immunoblot of whole cell lysates. The membrane was reprobed with anti-myc to determine Ulp2 protein levels and anti-Pgk1 as a loading control. Strains were harvested during logarithmic growth at 24°C. Asterisk (*) denotes high-molecular-mass SUMO conjugates that remain largely in the stacking gel. (B) Serial dilutions of yeast cultures were spotted onto plates and grown under the indicated conditions. Cells were photographed after 4 d of growth.
From these data, we conclude that nuclear localization of Ulp2 is likely to be necessary for all or the majority of its cellular functions and that under the conditions tested, NLS1 and NLS2 are the only functional nuclear targeting signals in the Ulp2 protein.
Nuclear Import of Ulp2 Requires the Importin Kap95
Both NLS1 and NLS2 have sequences similar to that of a classical NLS (cNLS), suggesting that Ulp2 may be transported into the nucleus by the heterodimeric α-β importin (karyopherin) complex, Kap60-Kap95 (Lange et al., 2007). To test this, nuclear localization of Ulp2-myc was examined in the temperature-sensitive kap95-14 mutant by immunofluorescent staining (Figure 6, A and B). In cells expressing WT Kap95, more than 80% cells displayed either strong or weak nuclear Ulp2 staining at both 24 and 37°C. The small fraction of cells that exhibited a diffuse signal throughout the cell presumably reflected the loss of the WT KAP95 plasmid from these cells, which have the chromosomal copy of KAP95 deleted (Makhnevych et al., 2007). In contrast, in the kap95-14 mutant, Ulp2-myc was diffusely distributed throughout the cell in ~55% of cells at 24°C and 80% of cells at 37°C. By Western immunoblotting, similar Ulp2 protein levels were observed for WT and kap95-14 mutant cells at both temperatures (Figure 6C).
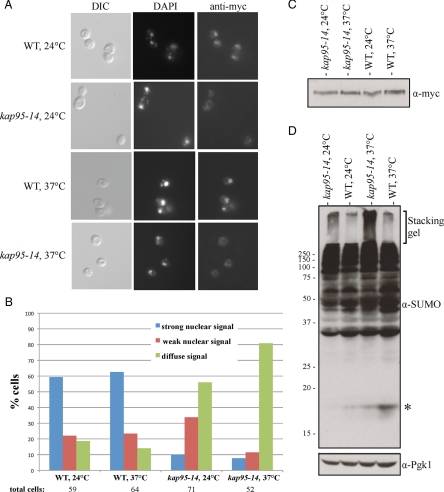
Ulp2 import into the nucleus requires the importin Kap95. (A) DIC and anti-myc immunofluorescence images were collected for kap95Δ cells carrying either a WT KAP95 (MHY5101) or mutant kap95-14 (MHY5100) plasmid. Cells were grown at 24°C or shifted from 2 to 37°C for 3 h before fixation. (B) The percentage of cells with the indicated Ulp2-myc localization in WT and kap95-14 cells at 24 and 37°C. (C) Anti-myc immunoblot of Ulp2-myc in the indicated strains. (D) Anti-SUMO immunoblot of whole cell lysates derived from WT and kap95-14 mutant strains at 24 and 37°C. * Free SUMO.
From our analysis in the preceding sections, a reduction in Ulp2 localization to the nucleus would be predicted to impair Ulp2 function. Consistent with the localization data, kap95-14 cells accumulated significantly greater amounts of poly-SUMO conjugates compared with WT cells at 24°C, and this amount increased further in kap95-14 cells shifted to 37°C (Figure 6D). The slight accumulation of poly-SUMO conjugates in WT cells correlated with the diffuse localization of Ulp2 in a fraction of these cells, which, as noted, probably had lost the KAP95 plasmid. We conclude from these results that Ulp2 is imported into the nucleus by the Kap60-Kap95 importin complex, which recognizes two cNLSs, NLS1 and NLS2, in the NTD.
Fusion to a Heterologous NLS Rescues the ulp2ΔN Mutant
The foregoing analysis indicated that the Ulp2 NTD is necessary and sufficient for Ulp2 nuclear localization and is crucial for Ulp2 function. We next asked whether the only essential function of the Ulp2 NTD is to localize the enzyme to the nucleus or whether this relatively large domain (>400 residues) has additional roles. To address this, we created a strain with a chromosomally integrated ulp2ΔN-NLSSV40 allele, which expresses a protein that fuses the classical nuclear localization sequence from the SV40 large-T antigen (NLSSV40) to the C-terminus of the Ulp2ΔN protein (Figure 7A). As a control, we introduced a point mutation into NLSSV40 that is known to block its nuclear targeting activity (denoted as *NLSSV40; Kalderon et al., 1984). As predicted, the heterologous NLSSV40 restored nuclear localization to Ulp2ΔN, whereas the mutant *NLSSV40 did not (Figure 7B). These Ulp2ΔN derivatives were expressed at levels similar to or slightly above that of Ulp2ΔN (Figure 7A). Interestingly, the immunoblot revealed that each Ulp2ΔN construct displayed a distinct pattern of bands. Ulp2 is known to be phosphorylated by the Cdc28 kinase (Ubersax et al., 2003). Our data suggest that Ulp2 modification is dependent on its subcellular localization.
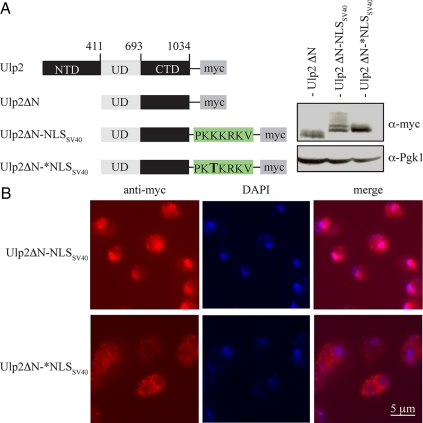
A heterologous NLS restores the nuclear localization of the Ulp2ΔN protein. (A) A schematic of the tested myc-tagged proteins, all expressed from chromosomally integrated alleles, and anti-myc immunoblot analysis. (B) Anti-myc immunofluorescent localization of Ulp2ΔN derivatives.
Remarkably, the phenotype of the ulp2ΔN-NLSSV40 mutant approached that of the congenic WT strain (Figure 8). The buildup of high-molecular-mass poly-SUMO conjugates seen in ulp2ΔN cells was strongly reduced (bracket in Figure 8A), as was the cellular sensitivity to hydroxyurea and benomyl (Figure 8B). The ulp2ΔN-NLSSV40 strain was still slightly temperature sensitive for growth, possibly because of reduced protein stability caused by the large N-terminal truncation. Significantly, the ulp2ΔN-*NLSSV40 mutant strain, which has a single missense mutation in the SV40 NLS, could no longer suppress any of the phenotypic defects associated with the ulp2Δ mutant. [Appending the SV40 NLS to a Ulp2 protein lacking both its NTD and CTD also restored a considerable degree of WT function, suggesting that the CTD is not supplying an essential function in the absence of the NTD; growth did not reach WT rates, but the levels of the chimeric protein were very low (data not shown).] We conclude that the primary function of the Ulp2 NTD is to direct the Ulp2 SUMO protease to the nucleus.
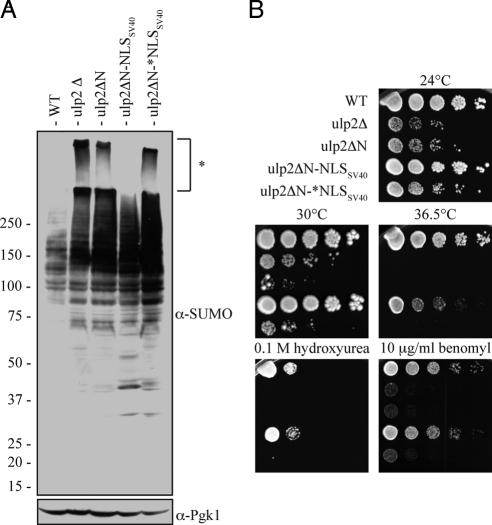
Nuclear targeting is the major function of the Ulp2 NTD. (A) Anti-SUMO immunoblot from whole cell lysates. (The left portion of the blot appears to have incomplete ECL exposure; compare to Figure 1C.) For the bottom panel, the membrane was reprobed with anti-Pgk1. Strains were harvested during logarithmic growth at 24°C. * High-molecular-mass SUMO conjugates that remain largely in the stacking gel. (B) Serial dilutions of yeast cultures spotted onto plates and grown as indicated. Cells were photographed after 4 d of growth, except the 36.5°C and benomyl-containing plates, which were photographed after 2 d.
DISCUSSION
In this study, we have demonstrated that both the N- and C-terminal noncatalytic domains of Ulp2 are required for full enzyme function in vivo. Loss of either domain results in the accumulation of high-molecular-mass poly-SUMO conjugates. Despite its very low sequence conservation, the NTD of Ulp2 is essential for Ulp2 function. This critical activity of the large NTD could be traced to two short sequences that target Ulp2 to the nucleus via the classical import pathway, and the entire NTD could be replaced with a heterologous NLS to yield nearly complete restoration of WT function. We discuss these findings in the context of the potential functions of yeast Ulp2 and poly-SUMO chains in the nucleus and the evolution of the Ulp2 subfamily of SUMO proteases.
Poly-SUMO chains form in both yeast and mammals, but they are not required for any essential function, at least in yeast (Tatham et al., 2001; Bylebyl et al., 2003). In vitro, Ulp2 can cleave the SUMO–SUMO linkages in poly-SUMO chains (Bylebyl et al., 2003). Furthermore, a ulp2Δ strain accumulates large poly-SUMO conjugates. When this strain was engineered to express a mutant version of SUMO that could no longer form poly-SUMO chains, the growth defects were strongly suppressed (Bylebyl et al., 2003). These observations led to the suggestion that the accumulation of large poly-SUMO chains in cells lacking Ulp2 was responsible for the observed growth defects. We find that deletion of the C-terminal extension of Ulp2 causes only a minor growth defect at lower temperatures, although it did lead to mild temperature and hydroxyurea sensitivities. Nevertheless, the ulp2ΔC mutant accumulates significant amounts of high-molecular-mass poly-SUMO conjugates. This suggests that high levels of poly-SUMO conjugates are not solely responsible for the growth defects of cells lacking Ulp2 or that moderate levels of such conjugates can be reasonably well tolerated.
The most likely inference from the ulp2ΔC analysis is that the growth-regulating roles of Ulp2 involve cleavage of SUMO from specific target proteins in addition to poly-SUMO chain cleavage. Previously, Ulp2 was indicated to be capable of cleaving substrate-SUMO isopeptide bonds in vivo because overexpression of Ulp2 eliminated monosumoylated forms of one of its substrates, Pds5 (Stead et al., 2003). Moreover, data from Bylebyl et al. (2003) suggest that the correlation between poly-SUMO conjugate accumulation and growth defects in ulp2Δ cells does not always hold. Mutation of SUMO Lys54 and Lys58 strongly suppressed the ulp2Δ growth defects, but poly-SUMO conjugates accumulated to the same extent as with WT SUMO. These SUMO mutants may suppress ulp2Δ through a reduced ability to be conjugated to specific substrates. Conversely, deletion of the Siz2 SUMO ligase from ulp2Δ cells virtually eliminated the poly-SUMO conjugates detectable in the stacking gel, but the growth defect was not suppressed (Bylebyl et al., 2003). Thus, although Ulp2 contributes to poly-SUMO chain disassembly, it seems to have critical functions in the cleavage of specific (poly)SUMO-substrate isopeptide linkages as well. This idea fits well with a recent biochemical analysis of the human SENP6 and SENP7 orthologues of Ulp2, which demonstrated that the purified enzymes also efficiently cleaved the isopeptide bond between SUMO2/3 and a model substrate (Lima and Reverter, 2008).
The abundant high-molecular-mass poly-SUMO conjugates observed in ulp2ΔN cells appear to be nuclear, because appending an NLS to the ulp2ΔN protein strongly suppressed their accumulation (Figure 8A). The Ulp2 import defect in the kap95-14 mutant also correlated with an accumulation of poly-SUMO conjugates, consistent with the idea that most of these conjugates are nuclear; this result also suggests that a component of the pleiotropic kap95 phenotype could result from the reduced import of Ulp2. It is interesting in this regard that poly-SUMO–modified proteins might be preferred substrates for the nuclear Slx5-Slx8 ubiquitin ligase and its orthologues (Uzunova et al., 2007; Xie et al., 2007; Mullen and Brill, 2008; Tatham et al., 2008). Whether the conjugates in ulp2Δ cells are mostly free poly-SUMO chains or poly-SUMO–protein conjugates is not yet clear. The critical role for nuclear localization in Ulp2 function is also consistent with the nuclear localization of two substrates, topoisomerase Top2 and the chromatid cohesion factor Pds5.
Ulp2 and all its known orthologues have large but otherwise poorly conserved NTDs. Conservation of size but not sequence implies that short but critical sequence elements are dispersed throughout the large domain, which is true for Ulp2. Within the >400-residue NTD, only two short, noncontiguous sequences appear to be essential for Ulp2 function. Mutation of both sequences cause a phenotype similar to a complete ULP2 knockout, and the entire NTD becomes dispensable if a heterologous NLS is added to the enzyme. In our assays, the two NLSs acted redundantly, but this might not be true under all conditions, which could account for why both are maintained.
Why is the relatively large size of the NTDs of Ulp2 family proteins nevertheless conserved? We note that large fractions of all the Ulp2 family NTD sequences are predicted to be intrinsically disordered (Linding et al., 2003). These are protein regions that lack a stable tertiary structure in the isolated proteins. For yeast Ulp2, ~38% of the NTD is predicted to be intrinsically disordered, whereas for the human Ulp2 orthologues SENP6 and SENP7, this fraction is even higher. More than 60% of the Ulp2 CTD is also predicted to lack a stable tertiary fold. Such protein regions are often involved in low-affinity but high-specificity protein–protein interactions (Dyson and Wright, 2005). The disordered regions in Ulp2 might also facilitate substrate binding, although our results suggest that at least some Ulp2-dependent processes do not absolutely require these regions.
SUMO has been shown to bind noncovalently to SUMO-interacting motifs (SIMs) on interacting proteins. The core consensus SIM has been determined as I/V-X-I/V-I/V or the inverted motif and is usually flanked on one side by a cluster of acidic residues (Song et al., 2004). Ulp2 has two SIMs that fit this consensus, one next to the catalytic site within the UD and the second within the CTD. The SIMs could be binding to poly-SUMO for desumoylation by Ulp2. In accordance with this, mutating the SIM within the CTD causes a slight but reproducible accumulation of poly-SUMO in vivo, but poly-SUMO binding in vitro was not reduced (data not shown). These data suggest that other elements of the CTD contribute to SUMO binding; the identified SIM might help orient the poly-SUMO substrate for optimal cleavage by the enzyme.
It is likely that the localization and functional data on Ulp2 are relevant to how the mammalian orthologues of Ulp2 function. Previous work determined that a large region of the N-terminal domain of human SENP6, residues 84-448 that contains multiple predicted NLSs, is responsible for the localization of this protein (Mukhopadhyay et al., 2006). Although the NTDs of Ulp2 and SENP6 are not readily aligned, the presence of short redundant elements involved in nuclear import that are dispersed over a relatively large and intrinsically disordered domain appears to be similar. We predict that the NTD of SENP6, and probably that of SENP7, will also be essential for its in vivo function and that a substantial part of this activity is due to the nuclear localization role of the NTD.
ACKNOWLEDGMENTS
We thank Rachael Felberbaum for comments on the manuscript, Bill Hankey for help with yeast strain construction, and Chris Ptak (University of Alberta) and Rick Wozniak (University of Alberta) for the kap95-14 strain. M.B.K. was supported in part by an National Science Foundation predoctoral fellowship. This study was supported by National Institutes of Health Grant GM053756 to M.H.
Abbreviations used:
| CTD | C-terminal domain |
| NLS | nuclear localization signal |
| NTD | N-terminal domain. |
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-10-1090) on February 18, 2009.
REFERENCES
- Abramoff M. D., Magelhaes P. J., Ram S. J. Image processing with Image J. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Amerik A. Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. [Abstract] [Google Scholar]
- Ausubel F., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1989. [Google Scholar]
- Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S. J. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. [Abstract] [Google Scholar]
- Bossis G., Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:13. [Europe PMC free article] [Abstract] [Google Scholar]
- Bylebyl G. R., Belichenko I., Johnson E. S. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 2003;278:44113–44120. [Abstract] [Google Scholar]
- Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. [Abstract] [Google Scholar]
- Darst R. P., Garcia S. N., Koch M. R., Pillus L. Slx5 promotes transcriptional silencing and is required for robust growth in the absence of Sir2. Mol. Cell. Biol. 2008;28:1361–1372. [Europe PMC free article] [Abstract] [Google Scholar]
- Drag M., Salvesen G. S. DeSUMOylating enzymes-SENPs. IUBMB Life. 2008;60:734–742. [Abstract] [Google Scholar]
- Dyson H. J., Wright P. E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. [Abstract] [Google Scholar]
- Ferris H. U., Furukawa Y., Minamino T., Kroetz M. B., Kihara M., Namba K., Macnab R. M. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 2005;280:41236–41242. [Abstract] [Google Scholar]
- Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. [Abstract] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4102–4110. [Abstract] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. [Abstract] [Google Scholar]
- Hodel M. R., Corbett A. H., Hodel A. E. Dissection of a nuclear localization signal. J. Biol. Chem. 2001;276:1317–1325. [Abstract] [Google Scholar]
- Johnson E. S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. [Abstract] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. [Abstract] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. [Abstract] [Google Scholar]
- Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. [Abstract] [Google Scholar]
- Lewis A., Felberbaum R., Hochstrasser M. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 2007;178:813–827. [Europe PMC free article] [Abstract] [Google Scholar]
- Li S. J., Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. [Abstract] [Google Scholar]
- Li S. J., Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–2377. [Europe PMC free article] [Abstract] [Google Scholar]
- Li S. J., Hochstrasser M. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 2003;160:1069–1081. [Europe PMC free article] [Abstract] [Google Scholar]
- Lima C. D., Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J. Biol. Chem. 2008;283:32045–32055. [Europe PMC free article] [Abstract] [Google Scholar]
- Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. [Abstract] [Google Scholar]
- Makhnevych T., Ptak C., Lusk C. P., Aitchison J. D., Wozniak R. W. The role of karyopherins in the regulated sumoylation of septins. J. Cell Biol. 2007;177:39–49. [Europe PMC free article] [Abstract] [Google Scholar]
- Mossessova E., Lima C. D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. [Abstract] [Google Scholar]
- Mukhopadhyay D., Ayaydin F., Kolli N., Tan S. H., Anan T., Kametaka A., Azuma Y., Wilkinson K. D., Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J. Cell Biol. 2006;174:939–949. [Europe PMC free article] [Abstract] [Google Scholar]
- Mukhopadhyay D., Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 2007;32:286–295. [Abstract] [Google Scholar]
- Mullen J. R., Brill S. J. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 2008;283:19912–19921. [Europe PMC free article] [Abstract] [Google Scholar]
- Nathan D., et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. [Europe PMC free article] [Abstract] [Google Scholar]
- Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. [Abstract] [Google Scholar]
- Panse V. G., Kuster B., Gerstberger T., Hurt E. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 2003;5:21–27. [Abstract] [Google Scholar]
- Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. [Abstract] [Google Scholar]
- Schwartz D. C., Felberbaum R., Hochstrasser M. The Ulp2 SUMO protease is required for cell division following termination of the DNA damage checkpoint. Mol. Cell. Biol. 2007;27:6948–6961. [Europe PMC free article] [Abstract] [Google Scholar]
- Schwienhorst I., Johnson E. S., Dohmen R. J. SUMO conjugation and deconjugation. Mol. Gen. Genet. 2000;263:771–786. [Abstract] [Google Scholar]
- Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA. 2004;101:14373–14378. [Europe PMC free article] [Abstract] [Google Scholar]
- Stead K., Aguilar C., Hartman T., Drexel M., Meluh P., Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 2003;163:729–741. [Europe PMC free article] [Abstract] [Google Scholar]
- Storici F., Lewis L. K., Resnick M. A. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. [Abstract] [Google Scholar]
- Strunnikov A. V., Aravind L., Koonin E. V. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158:95–107. [Europe PMC free article] [Abstract] [Google Scholar]
- Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. [Abstract] [Google Scholar]
- Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. [Abstract] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. [Abstract] [Google Scholar]
- Uzunova K., et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. [Abstract] [Google Scholar]
- Xie Y., Kerscher O., Kroetz M. B., McConchie H. F., Sung P., Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007;282:34176–34184. [Abstract] [Google Scholar]
- Xu P., Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim. Biophys. Acta. 2006;1764:1940–1947. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Molecular Biology of the Cell are provided here courtesy of American Society for Cell Biology
Full text links
Read article at publisher's site: https://doi.org/10.1091/mbc.e08-10-1090
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2669027?pdf=render
Free to read after 2 months at www.molbiolcell.org
http://www.molbiolcell.org/cgi/reprint/20/8/2196.pdf
Free to read after 2 months at www.molbiolcell.org
http://www.molbiolcell.org/cgi/content/full/20/8/2196
Free to read at www.molbiolcell.org
http://www.molbiolcell.org/cgi/content/abstract/20/8/2196
Citations & impact
Impact metrics
Citations of article over time
Article citations
The polySUMOylation axis promotes nucleolar release of Tof2 for mitotic exit.
Cell Rep, 43(7):114492, 13 Jul 2024
Cited by: 1 article | PMID: 39002125 | PMCID: PMC11298248
DeSUMOylation of a Verticillium dahliae enolase facilitates virulence by derepressing the expression of the effector VdSCP8.
Nat Commun, 14(1):4844, 10 Aug 2023
Cited by: 2 articles | PMID: 37563142 | PMCID: PMC10415295
Histone sumoylation promotes Set3 histone-deacetylase complex-mediated transcriptional regulation.
Nucleic Acids Res, 48(21):12151-12168, 01 Dec 2020
Cited by: 28 articles | PMID: 33231641 | PMCID: PMC7708062
The role of SUMOylation during development.
Biochem Soc Trans, 48(2):463-478, 01 Apr 2020
Cited by: 18 articles | PMID: 32311032 | PMCID: PMC7200636
Review Free full text in Europe PMC
Siz2 Prevents Ribosomal DNA Recombination by Modulating Levels of Tof2 in Saccharomyces cerevisiae.
mSphere, 4(6):e00713-19, 27 Nov 2019
Cited by: 4 articles | PMID: 31776241 | PMCID: PMC6881720
Go to all (28) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Binding to small ubiquitin-like modifier and the nucleolar protein Csm1 regulates substrate specificity of the Ulp2 protease.
J Biol Chem, 293(31):12105-12119, 14 Jun 2018
Cited by: 5 articles | PMID: 29903909 | PMCID: PMC6078457
In Vitro Studies Reveal a Sequential Mode of Chain Processing by the Yeast SUMO (Small Ubiquitin-related Modifier)-specific Protease Ulp2.
J Biol Chem, 290(19):12268-12281, 01 Apr 2015
Cited by: 9 articles | PMID: 25833950 | PMCID: PMC4424358
Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity.
Genes Dev, 31(8):802-815, 01 Apr 2017
Cited by: 26 articles | PMID: 28487408 | PMCID: PMC5435892
Cytoplasmic sumoylation by PIAS-type Siz1-SUMO ligase.
Cell Cycle, 7(12):1738-1744, 16 Jun 2008
Cited by: 16 articles | PMID: 18583943
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: GM053756
Grant ID: R01 GM053756
 †
†



