Abstract
Free full text

Autophagy regulates lipid metabolism
Abstract
The intracellular storage and utilization of lipids are critical to maintain cellular energy homeostasis. During nutrient deprivation, cellular lipids stored as triglycerides in lipid droplets are hydrolysed into fatty acids for energy. A second cellular response to starvation is the induction of autophagy, which delivers intracellular proteins and organelles sequestered in double-membrane vesicles (autophagosomes) to lysosomes for degradation and use as an energy source. Lipolysis and autophagy share similarities in regulation and function but are not known to be interrelated. Here we show a previously unknown function for autophagy in regulating intracellular lipid stores (macrolipophagy). Lipid droplets and autophagic components associated during nutrient deprivation, and inhibition of autophagy in cultured hepatocytes and mouse liver increased triglyceride storage in lipid droplets. This study identifies a critical function for autophagy in lipid metabolism that could have important implications for human diseases with lipid over-accumulation such as those that comprise the metabolic syndrome.
Free fatty acids (FFAs) are taken up by hepatocytes and converted into triglycerides (TGs) for storage with cholesterol in lipid droplets (LDs)1. LD-sequestered TGs continually undergo hydrolysis, generating FFAs that are predominantly re-esterified back into TGs for storage1,2. Nutrient deprivation upregulates TG hydrolysis to supply FFAs for oxidation to meet cellular energy demands3. An alternative energy source in times of nutrient scarcity is provided by the breakdown of cellular components by autophagy4,5. Both macroautophagy (the type of autophagy quantitatively more important and subsequently referred to as autophagy) and lipolysis are regulated hormonally by insulin and glucagon6 and are increased during starvation. Except for the processing of endocytosed lipoproteins, no direct involvement of the lysosomal degradation pathway in lipid metabolism has been established. The regulatory and functional similarities between autophagy and lipolysis, along with the capability of lysosomes to degrade lipids, indicated that autophagy may contribute to LD and TG breakdown (Supplementary Fig. 1).
Inhibition of autophagy increases lipid storage
Pharmacological inhibition of autophagy with 3-methyladenine (3MA)7 significantly increased hepatocyte TG content in the absence or presence of exogenous lipid supplementation with oleate (Fig. 1a). A knockdown of the autophagy gene Atg5 in hepatocytes (siAtg5 cells; Supplementary Fig. 2a) also increased TG levels with oleate or a second endogenous stimulus for TG formation—culture in methionine- and choline-deficient medium (MCDM)8,9 (Fig. 1b and Supplementary Fig. 2b with a second short hairpin RNA, shRNA). TG levels were also increased in embryonic fibroblasts from Atg5 knockout mice (Fig. 1c). Although cellular cholesterol content was unaffected in siAtg5 cells by oleate alone or culture in MCDM, oleate and cholesterol co-treatment led to significantly greater cholesterol content (Supplementary Fig. 2c, d).
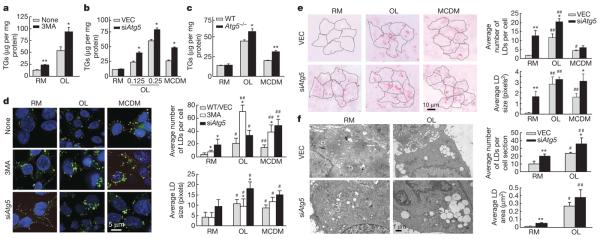
a, TG levels in hepatocytes untreated (None) or treated with 3-methyladenine (3MA) and cultured in regular medium (RM) or oleate (OL) (*P < 0.02, **P < 0.002, n = 3). b, TG levels in vector-infected (VEC) and siAtg5 cells in RM, OL or in MCDM (*P < 0.001, n = 5). OL values are in mM. c, TG levels in wild-type (WT) or Atg5 knockout mice embryonic fibroblasts (Atg5−/−)(*P < 0.01 or **P < 0.0001, n = 5). d–f, Cells from a and b stained with BODIPY 493/503 (d), oil red O (e) or visualized by electron microscopy (f). Right: quantifications of LD number and size (*P < 0.01, **P < 0.001 with untreated wild-type cells; #P < 0.01, ##P < 0.001 with cells in RM; n = 4–6). Nuclei are highlighted with 4,6-diamidino-2-phenylindole (DAPI; d). Error bars, s.e.m.
Consistent with the increased TG levels, lipid staining with BODIPY 493/503 or oil red O revealed increased LD number and size in hepatocytes with oleate or MCDM that were further increased by addition of 3MA or Atg5 knockdown (Fig. 1d, e). These increases were greater than for TG levels because the lipid stains detect all neutral lipids in LDs (TGs and cholesterol). Similar results were obtained with palmitate (Supplementary Fig. 3). An increase in the number and size of LDs in oleate-treated siAtg5 cells was also confirmed by electron microscopy (Fig. 1f). The absence of co-localization of BODIPY 493/503 and an endoplasmic reticulum marker (Supplementary Fig. 4), and the co-localization of lipid with the LD-associated protein TIP47 (also known as M6PRBP1; ref. 1, Supplementary Fig. 5), demonstrated that lipid accumulation occurred preferentially in cytosolic LDs. Thus, inhibition of autophagy triggered increased TG and LD accumulation in hepatocytes challenged with a lipid stimulus.
Autophagy is required for LD breakdown
To determine how autophagy regulates TG levels, rates of TG synthesis and FFA β-oxidation were examined in siAtg5 cells. Equivalent increases in TG synthesis occurred in control and siAtg5 cells in response to oleate or culture in MCDM (Fig. 2a). Rates of β-oxidation, indicative of the levels of FFA generated by TG hydrolysis10, increased during lipid loading, but to a much lesser extent in cells with inhibited autophagy (Fig. 2b), consistent with reduced lipolysis. TG breakdown in cells cultured in oleate or MCDM was significantly decreased in siAtg5 cells (Fig. 2c, d). TG content after treatment with the lipolysis inhibitor diethylumbelliferyl phosphate (DEUP)11 was higher than after inhibition of autophagy with 3MA (Fig. 2e), consistent with a blockage of all lipolysis by DEUP (Supplementary Fig. 6a)12,13, but a partial inhibition by a loss of autophagy. Co-treatment with 3MA did not have an additive effect on the DEUP-mediated increase (Fig. 2e), supporting the theory that lipid accumulation during autophagy inhibition resulted from blocked lipolysis. DEUP did not affect autophagy, because autophagic flow (determined by changes in levels of LC3-II (also known as MAP1LC3B) in the absence and presence of lysosomal inhibitors) was unaffected by DEUP (Supplementary Fig. 6b).
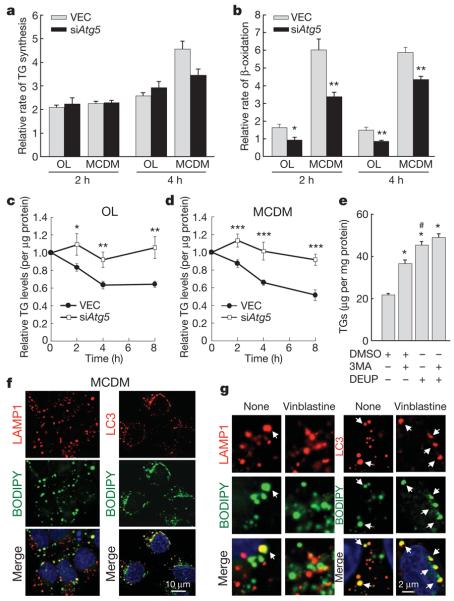
a, b, VEC and siAtg5 cells cultured with oleate (OL) or in MCDM were examined for their rates of TG synthesis (a) and β-oxidation (b) as compared to cells in regular medium alone (*P < 0.03, **P < 0.004 with VEC cells in the same medium, n = 3–4). c, d, Rates of TG decay in OL (c) and MCDM (d) (*P < 0.05, **P < 0.01, ***P < 0.001 as compared to VEC cells, n = 3–7). e, TG levels in wild-type cells treated with dimethyl sulphoxide vehicle (DMSO), 3-methyladenine (3MA) or diethylumbelliferyl phosphate (DEUP) (*P < 0.00001 with DMSO-treated cells, #P < 0.003 with 3MA-treated cells, n = 6). Error bars, s.e.m. f, Co-localization of BODIPY 493/503 (green) with LAMP1 (red) or LC3 (red) in hepatocytes in MCDM. g, High-magnification regions of hepatocytes in MCDM alone (none) or treated with vinblastine and stained as labelled. Arrows indicate co-localization.
Autophagic pathway components associate with LDs
To confirm that lysosomes regulate intracellular lipid, the effect of inhibiting lysosomal hydrolysis on lipid stores was examined. Lysosomal inhibition increased cellular TG and cholesterol content (Supplementary Fig. 7a, b) as well as LD accumulation (Supplementary Fig. 7c) in the absence or presence of a lipid stimulus. Double immunofluorescence studies revealed increased co-localization of LDs with the lysosome-associated membrane protein type 1 (LAMP1) in oleate or MCDM (Fig. 2f and Supplementary Fig. 8). In the absence of a stimulus for TG formation, LD/LAMP1 co-localization was observed only when lysosomal hydrolysis was inhibited, indicating that rapid LD turnover occurred in lysosomes under these conditions (Supplementary Fig. 8). In contrast to MCDM, where lysosomal inhibition increased LD/LAMP1 co-localization to some extent, this treatment did not modify LD association to lysosomes with oleate supplementation. This finding could represent an inability of the autophagic system to accommodate to the sudden FFA increase. Also supporting this possibility, autophagic flow (measured as LC3-II degradation or increased LC3-positive puncta by immunofluorescence; Supplementary Fig. 9a–c) was twofold greater in cells in MCDM compared to regular medium, but did not change with oleate. Despite increased autophagic flow, only a moderate decrease in the activity (20–30%) of the autophagy repressor mTOR (also known as FRAP1) as determined by autophosphorylation and phosphorylation of its downstream substrate p70S6K (also known as RPS6KB1) was observed with MCDM (Supplementary Fig. 9d). In addition, the absence of change in the autophagy activator beclin 1 indicates that autophagy was not induced in response to lipid stimuli. Instead, basal autophagy may be primarily responsible for regulating cellular lipid storage. Consistent with this conclusion, induction of autophagy further decreased lipid stores. Treatment with rapamycin or lithium chloride, activators of autophagy, significantly decreased LD number and TGs, and increased LD/LAMP1 co-localization (more evident with lysosomal inhibition) with lipid stimuli (Supplementary Figs 8 and 10a). As for basal autophagy, the flow of rapamycin-induced autophagy (measured as the increase in LD/LAMP1 co-localization with lysosomal proteolysis inhibition) was considerably reduced in cells exposed to oleate (Supplementary Fig. 8). The different autophagy effectors were unaffected by oleate (Supplementary Fig. 9d), indicating that autophagosome formation is preserved but their clearance is compromised to some extent in these cells and suggesting that increased LDs result from both augmented LD formation and diminished lysosomal breakdown.
In support of autophagy mediating delivery of LD content to lysosomes, LD/LAMP1 co-localization was markedly reduced by inhibitors of autophagosome formation (3MA) or autophagosome–lysosome fusion (vinblastine; Fig. 2g and Supplementary Fig. 8). Similarly, LD/LAMP1 co-localization was lower in siAtg5 cells and did not increase when lysosomal proteolysis was blocked (Supplementary Fig. 11a). Furthermore, LD co-localization with the autophagosome marker LC3 demonstrated a direct association between LDs and autophagosomes (Fig. 2f and Supplementary Fig. 12). In the absence of a stimulus for lipid accumulation, LD/LC3 co-localization was more prominent with lysosomal inhibition (Supplementary Fig. 10b), supporting a constitutive function for autophagy in LD regulation. Induction of autophagy by rapamycin or lithium chloride also increased LD/LC3 co-localization in untreated and oleate-treated cells (Supplementary Figs 10b, c and 12). The lack of a significant increase in LD/LC3 co-localization in cells in MCDM during autophagic induction is probably the consequence of their increased autophagic flux ensuring efficient lysosomal clearance of newly formed LD-containing autophagosomes (Supplementary Fig. 12). In contrast to the blocking effect of vinblastine on LD delivery to lysosomes, this drug did not decrease LD/LC3 co-localization (Fig. 2g and Supplementary Fig. 12). This result indicates that LD engulfment by LC3-positive membranes does not require microtubules, arguing against co-localization representing fusion between LDs and previously formed autophagosomes.
Fluorescence real-time video microscopy revealed that BODIPY 493/503-labelled structures (presumably complete LDs and LD-containing vesicles) and lysosomes associate in a dynamic manner (Supplementary Fig. 13; also see Supplementary Videos 1 and 2). Triple labelling for lysosomes, lipids and TIP47 confirmed that all LD components (lipids and proteins) were delivered to lysosomes (Supplementary Fig. 11b). Electron microscopy was used to elucidate further the mechanism of LD sequestration by autophagic vesicles. LDs were easily identifiable as round light-density structures, not limited by a bilayer lipid membrane (Fig. 3a, inset in the top left panel), with homogenous amorphous content and an average diameter of 0.5 μm that increased 10–15-fold in response to lipid stimuli (Supplementary Fig. 14). Double-membrane structures occupying up to 80% of a single LD were identified (Fig. 3a), along with similar density cytosolic autophagolysosome-like vesicles one-tenth of the size of a LD. These vesicles were surrounded by a double membrane and could have originated from sequestration of a portion of a large LD or an entire small LD (Fig. 3a, top and bottom right panels). Immunogold labelling revealed the presence of LC3 on the LD (often concentrated around membranous structures) and on the smaller lipid-containing double-membrane vesicles (autolipophagosomes; Fig. 3b). In some instances a small dense region heavily labelled for LC3 was present in the proximity of a LD or associated to its surface (Fig. 3b, top left panels). In cells in regular media, only a small percentage of double-membrane vesicles with content of density similar to LDs were detected (<20%), and of these less than half contained only lipid cargo (Fig. 3c). However, both stimuli for lipid accumulation increased the number of lipid-containing vesicles up to 80% with almost half containing only lipid, indicating some level of selectivity towards lipid cargo (Fig. 3c). Prolonged serum removal, a known stimulus of lipolysis, also increased LD/LC3 co-localization in hepatocytes to levels comparable to those from a lipid stimulus (Supplementary Fig. 15a). In contrast, co-localization of LC3 with mitochondria, a common cargo of autophagic vesicles, decreased after prolonged serum removal (Supplementary Fig. 15b). These results indicate that, although basal autophagy of LDs is part of an ‘in bulk’ nonselective process, conditions of increased lipolysis switch selectivity towards macroautophagic engulfment of LDs or macrolipophagy.
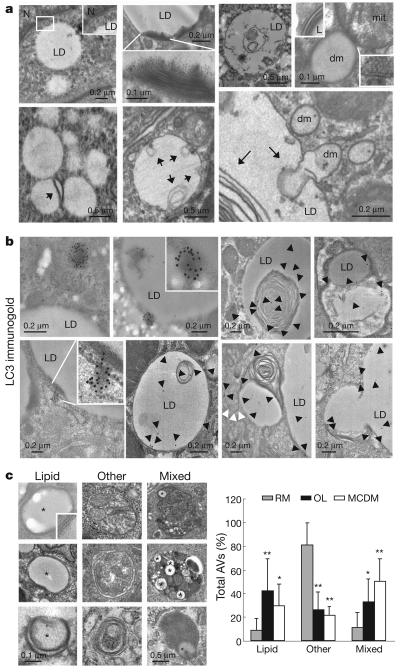
a, Electron micrographs of cultured hepatocytes. dm, double-membrane cytosolic vesicles. Arrows indicate membranes in LDs. Insets show the double membrane in nucleus (N), lipid-containing vesicles (L) and mitochondria (mit), but not in LDs. b, Mouse liver LC3 immunogold. Insets show higher magnification. Arrowheads indicate gold particles (black) and LC3-labelled bilayer membranes (white). c, Percentages of autophagic vacuoles (AVs) containing only lipid (Lipids, *), other cargo (Other) or mixed cargo (Mixed) in cells treated as in b (*P < 0.01, **P < 0.001 with cells in RM, n = 4–6). Left: representative examples. Error bars, s.e.m.
Autophagy regulates hepatic lipid stores in vivo
Starvation induces hepatic autophagy and increases delivery to the liver of FFAs from adipose tissue lipolysis. Electron microscopy revealed that starvation increased the frequency of LDs with areas of increased density and asymmetrically localized multi-membrane structures (Supplementary Fig. 16a) and the number of lipid-containing autolipophagosomes, autophagolysosomes and lysosomes (Supplementary Fig. 16b). LC3-II was undetectable in LDs isolated from the livers of fed mice, but co-purified with LDs from mice starved for 24 h (Fig. 4a). Relative LD purity was demonstrated by enrichment in LD-associated protein adipocyte differentiation-related protein (ADRP, also known as ADFP) and the absence of cytosolic proteins (Fig. 4a). Concomitantly, the percentage of lipid-containing autophagosomes was higher with starvation in both whole liver sections (Fig. 4b) and isolated fractions enriched in autophagosomes/autophagolysosomes (Supplementary Fig. 16b). During the first 24 h of starvation there was a significant increase in autophagic vacuoles with a mixed content (lipids, organelles and cytosol), whereas with progressive starvation autolipophagosomes become more abundant (Fig. 4b), supporting the selectivity of macrolipophagy. In fact, the amount of LD proteins and lipids in autophagic vacuoles and lysosomes increased with starvation (Fig. 4c and Supplementary Fig. 17a, b) whereas lipid content in the endoplasmic reticulum remained unchanged (Supplementary Fig. 17c, d). The increase in lipids in autophagic compartments and the presence of LC3-II in LDs occurred gradually starting within 6 h of fasting (Supplementary Fig. 17e–g). Thus, induction of autophagy and hepatic FFA delivery during starvation increased the association of autophagic components to LDs and the presence of LD components as autophagic cargo.
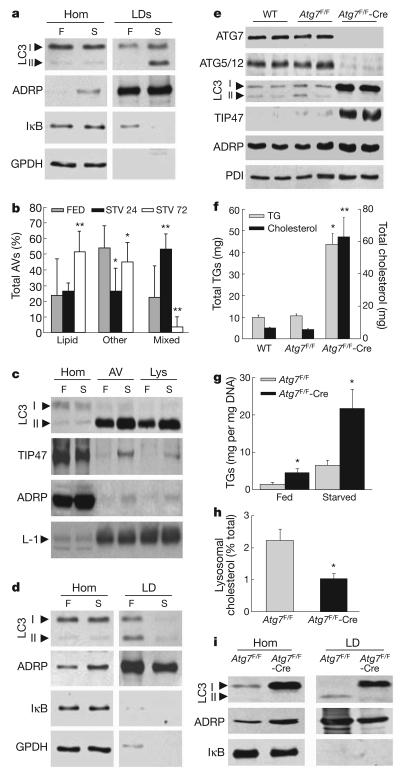
a, Immunoblots of cellular homogenates (Hom) and LDs from fed (F) or 24-h starved (S) mice. IκB, inhibitor of the nuclear factor of kappa light polypeptide gene enhancer; GPDH, glyceraldehyde-3-phosphate dehydrogenase. b, Percentages of autophagic vacuoles (AVs) containing only lipid, other cargo or mixed cargo calculated from samples processed as in Supplementary Fig. 16b (*P < 0.001, **P < 0.0001, n = 4–6). c, Immunoblots of Hom, AV and lysosomes (Lys). L-1, LAMP1. d, Immunoblots of liver Hom and LDs from HFD-fed mice. e, Liver homogenate immunoblots. PDI, protein disulphide isomerase. f, Total hepatic TG and cholesterol content (*P < 0.01, **P < 0.00001, n = 8–17). g, Hepatic TG concentration (*P < 0.05, n = 3). h, Percentage of cellular cholesterol in lysosomes (*P < 0.02, n = 4). i, Immunoblots of homogenates and isolated LDs. Error bars, s.e.m.
To confirm the relative inability of autophagy to accommodate to an exogenous lipid load observed in vitro, the association between LC3 and LD was analysed in mice fed regular diet or 16 weeks of a high-fat diet (HFD). Along with the expected increase in LD number and size (Supplementary Fig. 14b), a marked reduction (from 72% to 37%) in the LD-containing membranous structures or autophagosome-like vesicles was seen with starvation in HFD-fed mouse livers by electron microscopy (Supplementary Fig. 18). Autophagic vacuoles were still observed in starved HFD-fed mice, but the percentage of autolipophagosomes was markedly decreased. Surprisingly, immunogold staining (Supplementary Fig. 18) and immunoblot analysis of isolated LDs (Fig. 4d) revealed that LC3-II still associated with LDs of HFD-fed mice, but this association was more prominent under fed rather than starved conditions. Overall, these results are consistent with those in cultured cells that an exogenous lipid challenge reduces the efficiency of the interaction between the autophagic system and LDs.
For direct evidence of the regulation of hepatic lipids by autophagy in vivo, TG content was examined in Atg7F/F-Alb-Cre mice with a hepatocyte-specific knockout of the autophagy gene Atg7. Atg7F/F-Alb-Cre mice had decreased levels of ATG7, conjugated ATG5 and LC3-II in the liver but not in other organs (Fig. 4e and Supplementary Fig. 19a, b), and a marked increase in liver size, as previously reported4. At four months of age, oil red O staining of liver sections (Supplementary Fig. 19c), levels of total TGs (Fig. 4f), TGs normalized to DNA content (Supplementary Fig. 19d), LD-associated proteins TIP47 and ADRP (Fig. 4e) and TG accumulation after starvation (Fig. 4g) were increased in Atg7F/F-Alb-Cre mouse livers compared to control mice. Hepatic total cholesterol content and concentration were also increased in the knockout animals (Fig. 4f and Supplementary Fig. 19e), but the percentage of cholesterol in lysosomes was significantly decreased (Fig. 4h). TGs secreted as VLDL (also known as APOA5) by the liver, derived predominantly from the hydrolysis of TGs in LDs14, was also significantly decreased in Atg7F/F-Alb-Cre mice (Supplementary Fig. 19f). Isolated LDs from Atg7F/F-Alb-Cre mice had no LC3-II but higher levels of LC3-I (Fig. 4i). The fact that LC3-I was still detected in these LDs, as well as in LDs from Atg5 knockout cells (Supplementary Fig. 10d), indicates that conjugation of LC3 is not required before association to LDs, but occurs on the surface of this organelle.
Discussion
These findings demonstrate a previously unknown interrelationship between autophagy and lipid metabolism that we term macrolipophagy (Supplementary Fig. 1). Autophagy regulates lipid content because: inhibition of autophagy increased TGs and LDs in vitro and in vivo; loss of autophagy decreased TG breakdown; TGs and LD structural proteins co-localized with autophagic compartments; and LC3 associated with LDs. Moreover, a reverse relationship exists in which an abnormal increase in intracellular lipid impairs autophagic clearance as shown by decreased LD/LAMP1 co-localization and the absence of autophagic upregulation in hepatocytes cultured with lipids, as well as reduced association of autophagic vacuoles with LDs in response to starvation in HFD-fed mice. This interrelationship may trap hepatocytes in a harmful cycle in which decreased autophagy promotes lipid accumulation that then further suppresses autophagic function, thereby additionally increasing lipid retention (Supplementary Fig. 1).
Although future studies must clarify the mechanism of sequestration/degradation of LDs through autophagy, our data indicate that lysosomes do not fuse directly with LDs but with LD-containing autophagosomes. On the basis of morphometric analysis and LC3 immunogold staining we propose that LC3 is recruited to the LDs where LC3 initiates the formation of a limiting membrane through ATG7-dependent conjugation. Some of the lipid cargo observed within autophagosomes could originate from sequestration of whole small LDs alone or with other cytosolic components that may also become trapped in the sealing vesicle (middle panel, Supplementary Fig. 1). In contrast, for larger LDs, the autophagosome only sequesters a discrete region of the LD, which then pinches off as a double-membrane vesicle enriched in LC3 (autolipophagosome). Such partial sequestration of organelles by autophagy has been described previously for the endoplasmic reticulum and nucleus (piecemeal microautophagy)15-17.
Decreased autophagy in the liver with ageing18 may contribute to hepatic lipid accumulation that occurs along with an increased incidence of the metabolic syndrome in aged humans19. The ability of increased lipid content to impair autophagy also indicates that lipid accumulation could contribute to the decrease in autophagic function with ageing. Therapeutic strategies to increase autophagic function may therefore provide a new approach to prevent the metabolic syndrome and its associated pathologies.
METHODS SUMMARY
The hepatocyte cell line RALA255-10G was cultured under nontransformed conditions, as described previously20. Wild-type and Atg5−/− mouse embryonic fibroblasts provided by N. Mizushima21 were cultured as described previously22. TG content was determined by the Trig/GB Kit (Roche Diagnostics), cholesterol content by the Amplex Red Cholesterol Assay (Invitrogen), fatty acid β-oxidation by a modification of a previously used method23, and TG decay in cells radiolabelled with [14C]oleate and TG synthesis by standard methods12. shRNAs were cloned into pSUPER (Ambion) and then pCCL.sin.PPT.hPGK.GFPWpre24. Protein isolation and western blotting were performed as described previously25. Fluorescence microscopy for BODIPY 493/503 (Invitrogen) and immunofluorescence were performed as described previously26. Atg7F/F mice4 were crossed with Alb-Cre mice27 to generate Atg7F/F-Alb-Cre mice. Some animals were fed a high-fat diet (60% kcal in fat; Research Diets, D12492). Electron microscopy and immunogold labelling were performed as described previously26. LDs from mouse livers were isolated by sucrose density gradient centrifugation28 and autophagic vacuoles and lysosomes by centrifugation in metrizamide discontinuous density gradients29.
METHODS
Cells and cell culture
The rat hepatocyte line RALA255-10G was cultured under nontransformed conditions, as described previously20. Some cells were cultured in high glucose or serum-free DMEM lacking methionine and choline (Atlanta Biologicals) or pre-treated with 10 mM 3-methyladenine, 100 μM DEUP, 50 μM chloroquine (Sigma), 20 mM ammonium chloride or 100 μM leupeptin (Fisher). Oleic and palmitic acid were conjugated to albumin, as described previously30, and cells were treated with 0.125 or 0.25 mM oleate or palmitate for 24 h in high glucose, serum-free DMEM. Mouse embryonic fibroblasts were maintained in high glucose DMEM with 10% fetal bovine serum.
Lentiviral Atg5 siRNA construction
The following small hairpin (sense–loop–antisense) RNAs (shRNA) for Atg5 were cloned into lentiviral vectors: 5′-GATCCCCGTCAGGTGATCAACGAAATTTCAAGAGAATTTCGTTGATCACCTGACTTTTTC-3′ (sense), 5′-TCGAGAAAAAGTCAGGTGATCAACGAAATTCTCTTGAAATTTCGTTGATCACCTGACGGG-3′ (antisense) for shAtg5, and 5′-GATCCCCGATGTTAGTGAGATTTGGTTTCAAGAGAACCAAATCTCACTAACATCTTTTTC-3′ (sense), 5′-TCGAGAAAAAGATGTTAGTGAGATTTGGTTCTCTTGAAACCAAATCTCACTAACATCGGG-3′ (antisense) for shAtg5 number 2. The hairpins were cloned between the BglII–XhoI sites of pSUPER (Ambion), and after SmaI–XhoI digestion the fragments, which included the H1 promoter-shRNA cassette, were subcloned into the EcoRV–XhoI sites of the vector pCCL.sin.PPT.hPGK.GFPWpre24. Lentiviral stocks were prepared by calcium phosphate transfection of these vectors and the packaging vectors pMDLg/pRRE, pRSV-Rev and pMD2.VSVG into HEK-293T cells. Supernatants were collected over 36 to 48 h, titred by plaque assay and used at a multiplicity of infection of 5 to infect RALA hepatocytes. The efficiency of infection at 72 h, determined by the number of green-fluorescent-protein-positive cells, exceeded 98% for all constructs.
Western blotting
Cell lysates and liver homogenates were subjected to western blot analysis, as previously described25. Membranes were incubated with the following primary antibodies: rabbit anti-ATG7, rabbit anti-LC3, rabbit anti-total and phospho-(Ser 2448 and Ser 2481) mTOR, rabbit anti-total and phospho-p70SK6, rabbit anti-total and phospho-Akt (Cell Signaling Technology), guinea pig anti-ADRP (Progen Biotechnik), rabbit anti-ATG5 (Novus Biologicals), mouse anti-beclin 1 (BD Biosciences), mouse anti-GPDH (Abcam), rat anti-LAMP1 (Developmental Studies Hybridoma Bank, University of Iowa), rabbit anti-IκB (Santa Cruz Biotechnology) and rabbit anti-TIP47 (ProSci Incorporated). Western blot for β-actin (AbCam) or protein disulphide isomerase (a gift from R. Stockert) was used as loading control.
As a measure of autophagic flow, immunoblots for LC3 were performed in untreated cells and cells treated with the lysosomal inhibitors ammonium chloride and leupeptin. Autophagic flow was determined by the ratio of the densitometric value for LC3-II in the presence of inhibitors to that in the absence of inhibitors, as described previously5.
Fluorescence microscopy
Cells were fixed with 3% paraformaldehyde, blocked and incubated with the primary and corresponding Cy5- and texas-red-conjugated secondary antibodies. Lipid droplets were stained by incubating cells with BODIPY 493/503 (Invitrogen) for 30 min, fixed and processed for immunofluorescence as described previously26. Lysosomes were highlighted with Lysotracker and mitochondria with Mitotracker (Invitrogen). Mounting medium contained DAPI stain to highlight the cell nucleus. Images were acquired with an Axiovert 200 fluorescence microscope (Carl Zeiss Ltd) with a ×63 objective and 1.4 numerical aperture, subjected to deconvolution with the manufacturer's software and prepared using Adobe Photoshop 6.0 software (Adobe Systems Inc.). Quantification was performed in individual frames after deconvolution and thresholding using ImageJ software (NIH) in a minimum of 20 cells per slide. Particle number was quantified with the ‘analyze particles’ function in thresholded single sections with size (pixel2) settings from 0.1 to 10 and circularity from 0 to 1. Co-localization was calculated by JACoP plugin in single Z-stack sections of deconvoluted images. Real-time video microscopy was performed using 8-chamber slides in medium buffered with HEPES and maintained at 37 °C in a temperature-controlled stage. Images were captured at 30-s intervals with a ×40 objective and ×1.6 Optovar system, and frames in the different fluorescence channels merged using the manufacturer's software.
Oil red O staining
Oil red O staining was performed, as described previously31. Staining was assessed by bright-field microscopy and quantified by the Image J software after appropriate thresholding.
Measurement of TG synthesis
TG synthesis was determined by standard methods12. Cells were cultured with [14C]oleate–BSA complex and the cellular lipids extracted with hexane/isopropanol (3:2, vol:vol). Lipids were then dried with nitrogen gas, redissolved into chloroform and resolved by thin-layer chromatography using successive solvent systems containing chloroform, acetone, methanol, acetic acid and water in volumetric ratios of 10:4:2:2:1, and hexane, methanol and acetic acid in ratios of 80:20:1. Phosphorimages were obtained with a Storm Gel and Blot Imaging System (GE Healthcare).
Fatty acid β-oxidation assay
Rates of fatty acid β-oxidation were determined by a modification of a previously used method23, in which the rate of carbon dioxide production from the oxidation of [14C]oleate was measured. Cells were cultured in the presence of [14C]oleate–BSA complex and the released [14C]carbon dioxide trapped for 1 h at 37 °C onto filter paper soaked in 100 mM sodium hydroxide. The rate of β-oxidation was calculated as the amount of trapped [14C]carbon dioxide in relative units produced per mg protein per hour.
TG decay determination
Cells were cultured in the presence of [14C]oleate–BSA complex for 24 h following which the cells were washed and the medium replaced. At different times TGs were extracted from the cells and quantified by thin-layer chromatography as described previously.
Animals
Atg7F/F mice4 were crossed with Alb-Cre mice27 to generate Atg7F/F-Alb-Cre mice with a hepatocyte-specific knockout of Atg7. Starved mice were allowed free access to water. Some animals were fed a HFD (34 g per 100 g diet fat, 0.03 g per 100 g diet cholesterol with 60% of the total kcal in fat; Research Diets, D12492). The HFD was begun at 3 weeks of age and continued for a total of 16 weeks. All studies were approved by the Animal Care and Use Committee of the Albert Einstein College of Medicine and followed the National Institutes of Health guidelines for animal care.
Electron microscopy and immunogold
Cells cultured in monolayers and liver blocks (1 mm3) were fixed in 2.5% gluteraldehyde in 100 mM sodium cacodylate, pH 7.43, and post-fixed in 1% osmium tetroxide in sodium cacodylate followed by 1% uranyl acetate. After ethanol dehydration and embedding in LX112 resin (LADD Research Industries), ultrathin sections were stained with uranyl acetate followed by lead citrate. Immunogold labelling was performed in ultrathin sections of samples fixed in 4% paraformaldehyde/0.1% gluteraldehyde in sodium cacodylate, dehydrated and embedded in Lowicryl. Grids were washed in 50 mM glycine in phosphate buffered saline, blocked, incubated with LC3 antibody for 2 h, washed extensively and incubated with the gold-conjugated secondary antibody (1:100) for 2 h. Control grids were incubated with the secondary antibody alone or with an irrelevant immunoglobulin G. After extensive washing, samples were fixed a second time for 5 min in 2% gluteraldehyde, washed and negatively stained with 1% uranyl acetate for 15 min. All grids were viewed on a JEOL 100CX II transmission electron microscope at 80 kV. Morphometric analysis was performed using ImageJ in 15–20 different micrographs for each condition after thresholding. Autophagic vacuoles were identified using previously established criteria32,33. Autophagic vacuoles (vesicles <0.5 μm) were classified as autophagosomes when they met two or more of the following criteria: double membranes (complete or at least partially visible), absence of ribosomes attached to the cytosolic side of the membrane, luminal density similar to cytosol, and identifiable organelles or regions of organelles in their lumen. Vesicles of similar size but with a single membrane (or less than 40% of the membrane visible as double), luminal density lower than the surrounding cytosol or multiple single membrane-limited vesicles containing light or dense amorphous material were classified as autophagolysosomes. For the classification of autophagic contents, autophagosomes and autophagolysosomes were grouped under the term autophagic vacuoles. Autophagic vacuoles containing only lipids were those with double membranes, homogenous density comparable to that of LDs and lacking other content.
Isolation of LDs
LDs were isolated from mouse livers by density gradient centrifugation following a modification of a method described previously28. Livers homogenized in 0.25 M sucrose were centrifuged at 6,800g for 5 min at 4 °C. The supernatant and fatty layer were centrifuged at 17,000g for 10 min at 4 °C to pellet autophagic vacuoles and lysosomes. The supernatant was adjusted to 20% sucrose in hypotonic lysis buffer (20 mM Tris, 1 mM EDTA, pH 7.4), placed in a centrifuge tube and sequentially overlaid with 5 ml each of 5% sucrose (w/v) in hypotonic lysis buffer. After centrifugation at 28,000g for 30 min at 4 °C, the LD fraction was collected from the top of the tube. LD fractions were delipidated with acetone followed by sequential washes in acetone/ether (1:1, v:v) and ether. The protein pellets were solubilized in SDS and analysed by western blotting.
Autophagic vacuole isolation
Autophagic vacuoles and lysosomes were isolated from mouse livers by differential centrifugation and centrifugation in discontinuous density gradients of metrizamide, following a protocol modified from ref. 34, as described previously29. A fraction enriched in endoplasmic reticulum resealed vesicles (microsomes) was prepared by centrifugation of the supernatant of the 17,000g centrifugation at 100,000g for 1 h.
Assay of hepatic TG secretion
TG secretion was determined as described previously35. Mice were fasted overnight and injected with 1 g per kg of P-407 (Sigma). TG levels were determined on serum drawn immediately before and 6 h after injection. TG production was calculated from the difference in serum TG levels over the 6 h period and expressed as mg per kg per h.
Statistical analysis
All numerical results are reported as mean and s.e.m., and represent data from a minimum of three independent experiments unless otherwise stated. We determined the statistical significance of the difference between experimental groups in instances of single comparisons by the two-tailed unpaired Student's t-test of the means with Sigma Plot (Jandel Scientific) software. In instances of multiple means comparisons, we used one-way analysis of variance (ANOVA) followed by the Bonferroni post hoc test to determine statistical significance.
Acknowledgements
We thank D. Silver for his discussions, N. Mizushima for providing the Atg5−/− mouse embryonic fibroblasts, R. Stockert for the protein disulphide isomerase antibody and the personnel at the Analytical Imaging Facility for their technical assistance. This work was supported by National Institutes of Health grants from the National Institute of Diabetes and Digestive and Kidney Diseases and National Institute on Aging, a Glenn Award and an American Liver Foundation Postdoctoral Research Fellowship Award (R.S.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature. A summary figure is also included.
Reprints and permissions information is available at www.nature.com/reprints.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature07976
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2676208?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Lipids associated with autophagy: mechanisms and therapeutic targets.
Cell Death Discov, 10(1):460, 30 Oct 2024
Cited by: 0 articles | PMID: 39477959 | PMCID: PMC11525783
Review Free full text in Europe PMC
Targeting Lipid Metabolism in Cancer Stem Cells for Anticancer Treatment.
Int J Mol Sci, 25(20):11185, 17 Oct 2024
Cited by: 0 articles | PMID: 39456967 | PMCID: PMC11508222
Review Free full text in Europe PMC
Coronavirus M protein promotes mitophagy over virophagy by recruiting PDPK1 to phosphorylate SQSTM1 at T138.
Nat Commun, 15(1):8927, 16 Oct 2024
Cited by: 0 articles | PMID: 39414765 | PMCID: PMC11484861
Thyroid Hormone-Mediated Selective Autophagy and Its Implications in Countering Metabolic Dysfunction-Associated Steatotic Liver Disease.
Endocrinol Metab (Seoul), 39(5):686-692, 14 Oct 2024
Cited by: 0 articles | PMID: 39397515 | PMCID: PMC11525703
Review Free full text in Europe PMC
Identification of KW-2449 as a dual inhibitor of ferroptosis and necroptosis reveals that autophagy is a targetable pathway for necroptosis inhibitors to prevent ferroptosis.
Cell Death Dis, 15(10):764, 21 Oct 2024
Cited by: 0 articles | PMID: 39433736 | PMCID: PMC11493980
Go to all (2,206) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis.
Nat Cell Biol, 17(6):759-770, 11 May 2015
Cited by: 344 articles | PMID: 25961502 | PMCID: PMC4449813
Autophagy-lysosomal pathway is involved in lipid degradation in rat liver.
Physiol Res, 61(3):287-297, 05 Apr 2012
Cited by: 37 articles | PMID: 22480422
Direct lysosome-based autophagy of lipid droplets in hepatocytes.
Proc Natl Acad Sci U S A, 117(51):32443-32452, 07 Dec 2020
Cited by: 87 articles | PMID: 33288726 | PMCID: PMC7768785
Regulation of lipid droplets by autophagy.
Trends Endocrinol Metab, 22(6):234-240, 16 Mar 2011
Cited by: 125 articles | PMID: 21419642 | PMCID: PMC3118855
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIA NIH HHS (4)
Grant ID: P01 AG031782
Grant ID: R01 AG021904
Grant ID: P01 AG031782-01A1
Grant ID: R01 AG021904-07
NIDDK NIH HHS (3)
Grant ID: R01 DK061498-05
Grant ID: K01 DK087776
Grant ID: R01 DK061498





