Abstract
Free full text

Cellular response to low adhesion nanotopographies
Abstract
This review focuses on how cells respond to low-adhesion nanotopographies. In order to do this, fabrication techniques, how cells may locate nanofeatures through the use of filopodia and possible mechanotransductive mechanisms are discussed. From this, examples of low-adhesion topographies and sizes and arrangements that may lead to low-adhesion are discussed. Finally, it is hypothesized as to how specifically low-adhesion materials may fit into the outlined mechanotransductive mechanisms.
Introduction
There are many chemical, physical and geometric cues within the extracellular environment in vivo that will provide signals to cells. It has been know for almost one hundred years that cells will react to the shape of their environment, and in 1952 Weiss first used the term contact guidance to describe cell alignment to topography (Weiss and Garber 1952; Curtis 2004). Due to advances in micro-fabrication, researchers have been able to produce accurate micro-scale substrates upon which to study cells. Thus, the effects of micro-topography on cells are becoming well known and include changes in cell adhesion, contact guidance, cytoskeletal organization, apoptosis, macrophage activation and gene expression (Clark et al 1987, 1990, 1991; Wojciak-Stothard, Madeja, et al 1995; Britland et al 1996; Dalby, Riehle, Yarwood, et al 2003). It is further becoming clear that cells will react to their nanoenvironment. In vivo, the nanoenvironment may be provided by protein folding and banding. In vitro testing of the cells response to defined nanotopographies has drawn on a number of fabrication techniques.
The borrowing of lithographical techniques from the microelectronics industry facilitated research into cell response to the topography of their environment. Initially, photolithography was used, which allowed fabrication of micron scale (width, diameter) features such as grooves and pits with sub-micron depths (Wilkinson et al 2002). Every cell type tested responded to the features by contact guidance (Clark et al 1987, 1991; Wojciak-Stothard, Curtis, et al 1995; Wojciak-Stothard, Madeja, et al 1995; Britland et al 1996). Further examination revealed that contact guidance lead to changes in cell adhesion, migration, cytoskeletal organization and genomic regulation (Dalby, Riehle, Yarwood, et al 2003).
It has now been shown that many kinds of cells also respond strongly to nanoscale topographies (Dalby, Riehle, et al 2002a, 2002b; Dalby, Gadegaard, et al 2004; Price et al 2003; Thapa et al 2003; Ward and Webster 2006). The scope of response, as with μm scale topographies ranges from adhesion (Biggs et al 2007a, 2007b) to genomic regulation (Dalby, Yarwood, et al 2002). This is really very interesting as cells can be influenced in similar ways by their μm and nm scale environments, but the mode of action is probably rather different. At the microscale, cells are clearly influenced by features in the same magnitude of size as themselves, ie, encounter mechanical confinement and robust obstacles. At the nanoscale, however, the features are far smaller than the cells themselves and thus must alter cell response through more subtle mechanisms.
So, if nanotopography is more expensive to fabricate and cells respond to microtopography with a large range of responses, why bother pursuing nanotopography as a clinical solution? The answer is that the range of response we can elicit with nanotopography is, in fact, greater and the effects we can produce in cells can range from subtle to strong. A great example of the strength of nanotopographical effect is low-adhesion, as shall be discussed here.
This review will start with a very brief overview of some nanofabrication methods, although it is to be noted that the actual fabrication is not the motivation for the report. The main aim of this report is, however, to present some current, if contentious, thinking on mechanisms of cell response to low-adhesion topographies.
Fabrication
Nanofabrication can be either top-down, starting with a large ‘block’ and creating small features (eg, lithography) or it can be bottom-up, starting with atoms, molecules or polymers and building into regular features (eg, tunneling fabrication or phase separation). Here, just a few techniques will be described that have been routinely used for cell testing.
Starting with top-down, the most high-resolution top-down fabrication technique is that of electron beam lithography (EBL) where a pattern is generated in a radiation sensitive polymer (resist) by a focused beam of electrons. The achievable resolution for this technique is about 3–5 nm (Vieu et al 2000). The area that can be exposed by an EBL tool without movement of the stage (the field size) is limited by the resolution of feature required. For well-defined 35 nm dots, Gadegaard, Thoms, et al (2003) used a field size of 200 × 200 μm. However, larger areas than this are required for cell culture experiments. To make larger areas of pattern, the sample is moved with high precision under interferometric control to allow tiling of the small pattern to fill a larger area (Wilkinson et al 2002; Wilkinson 2004).
EBL is, however, a rather expensive and time-consuming technique. Another top-down fabrication technique is colloidal lithography, where monodispearsed nanocolloids are used as an etch mask from which to create nanofeatures with controlled height and diameter, but random placement (Denis et al 2002a, 2002b; Agheli and Sutherland, submitted). Colloidal lithography allows the rapid production of large areas of nanotopography, but has the pay-off of less controllable feature distribution.
We note at this point that it is envisaged that bottom-up fabrication may play a critical role in production of topographies for cells to react to. An example of currently used bottom up techniques are and polymer demixing where spontaneous phase separation of polymers in a solvent creates nanotopogaphy with reproducible height, but random distribution and diameters (Affrossman et al 1996, 1998, 2000; Affrossman and Stamm 2000). Recently, the Liley group has produced other co-polymer systems that allow better order to be achieved (Minelli et al 2006; Blondiaux et al 2007).
An important step in research for cellular response to nanotopographies (as well as a step closer to mass production of all nanotopographies) and the possibility of improved implant design is the process of producing shims. This technique is borrowed from the DVD manufacture process and involved the plating of Ni onto the master substrates. Firstly, this is done by sputter coating to provide an initial key, next, electroplating is used. Once the master is removed, a negative replica in Ni is left. Polymers can then be injection molded or embossed against the substrate and depending on the design and the polymer used, down to 5 nm fidelity can be achieved. With injection molding, many hundreds of replicas can be produced in a day (Gadegaard, Mosler, et al 2003; Gadegaard, Thoms, et al 2003). The polymers used can either be biodegradable (PCL, PLA) or permanent (eg, polymethylmethacrylate, PMMA or polycarbonate, PC). It is envisaged that in the near future, embossing into metals will be a possibility.
For the described approaches over the past few years, however, there has been considerable research effort into ascertaining the breadth of cellular response to nanoscale features, and again, contact guidance has been observed in many cell types. The physical contact guidance, ie, alignment, however, is on a different scale; that of the filopodia.
Cell filopodia
It was mentioned in the introduction that cells must respond to nanoscale topography on a more subtle scale than the whole cell response to microtopography, it is very likely that filopodia, with tips of approximately 100 nm, are one of the cells’ main nanosensory tools. Gustafson and Wolpert (1961) first described filopodia in living cells in 1961. They observed mesenchymal cells migrating up the interior wall of the blastocoelic cavity in sea urchins and noted that the filopodia produced appeared to explore the substrate. This led them to speculate that they were being used to gather spatial information by the cells. When considering filopodial sensing of topography, fibroblasts have been described as using filopodia to sense and align the cells to microgrooves (Clark et al 1991) and epithelia have been shown to be of importance for alignment to nanogrooves (Teixeira et al 2003). Also, macrophages have been reported to sense grooves down to a depth of 71 nm by actively producing many filopodia and elongating in response to the shallow topography (Wojciak-Stothard, Madeja, et al 1995); in other words, monocyte (rounded) to macrophage (spread) activation occurs.
Cytoskeletal actin bundles drive the filopodia. As the filopodia encounter a favorable guidance cue, they become stabilized following the recruitment of microtubules and accumulation of actin. There is further evidence that nascent focal adhesions then form at the filopodial tips (Dalby, Riehle, Johnstone, et al 2004).
Once cells locate a suitable feature using the filopodia presented on the cells leading edge, lamellipodium are formed which move the cell to the desired site (Dalby, Riehle, Johnstone, et al 2004). These actions require G-protein signaling and actin cytoskeleton. Specifically of interest are Rho, Rac and Cdc42. Rho induces actin contractile stress fiber assembly to allow the cell to pull against the substrate, Rac induces lamellipodium formation, and Cdc42 activation is required for filopodial assembly (Schmitz et al 2000). Rho and Rac are both required for cell locomotion, but cells can translocate when Cdc42 is knocked out. Cells lacking Cdc42 cannot, however, sense chemotactic gradients and simply migrate in a random manner (Jones et al 1998). This, again, presents compelling evidence for filopodial involvement in cell sensing.
There is recent evidence that many types of cells, including mesenchymal stem cells, use filopodia to probe their nanoenvironment (Figure 1) (Dalby, Riehle, et al 2002a, 2002b; Dalby, Childs, et al 2003; Dalby, Gadegaard, et al 2004; Dalby, Riehle, Sutherland, et al 2004a; Hart et al, in press). The smallest feature (thus far) that cells (fibroblasts) have been observed to respond to are 10 nm high polymer demixed islands (Dalby, Riehle, Johnstone, et al 2004).
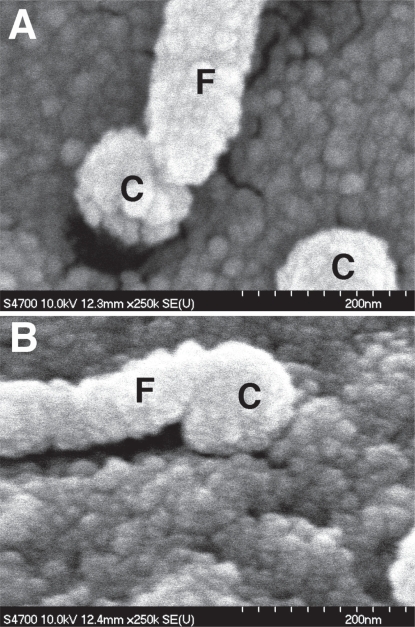
Filopodial sensing of nanocolumns. (A & B) 160 nm high, 100 nm diameter columns originally fabricated by colloidal lithography. Note how the cell filopodia (F) have similar tip dimensions to the columns (C) they interact with.
For low-adhesion materials, however, it is rather the case that filopodia cannot find sites suitable for adhesion. This is likely to be for two reasons. The first and main reason is the surface properties of the materials. Low-adhesion topographies tend to give extremes of environment. If treated with chemicals to normally produce hydrophobicity or hydrophilicity, nanostructures can become super-hydrophobic or super-hydrophilic (Martines et al 2005). This will effect protein interaction with the surfaces and hence cell adhesion; if a surface does not recruit proteins, cells cannot adhere, if a surface recruits proteins so strongly that conformational shape is altered, cells cannot adhere (Kasemo and Lausmaa 1988, 1994).
Secondly is that of surface area. Transmission electron microscopy of EBL pits in highly ordered arrangement has shown that adhesions cannot form across the pits and that filopodia can only start to form adhesions on the raised, interpit, areas (Dalby, Biggs, et al 2007).
Mechanotransduction
Filopodial sensing of the nanoenvironment and subsequent changes to the cellular cytoskeleton will almost certainly confer mechanotransductive changes to the nucleus and from here alter gene transcription and protein output. Hence microarray studies have regularly shown changes in genome regulation on both microtopographies and nanotopographies (Dalby, Yarwood, et al 2002; Dalby, Riehle, Yarwood, et al 2003). These mechanotransductive signaling events may be chemical eg, kinase based linked to focal adhesions influenced by cytoskeletal contraction. An example would be intergrin gathering as an adhesion is formed will activate myosin light chain kinase (MCLK) which will generate actin – myosin sliding (the key event in stress fiber contraction) and in turn will change focal adhesion kinase (FAK) activity. Cytoskeletal involvement in contraction against adhesions will also alter calcium influx and G-protein events. These chemical signaling events are collectively known as indirect mechanotransduction (Burridge and Chrzanowska-Wodnicka 1996).
Another form of mechanotransduction, direct mecha-notransduction, is considered to be transduced by the cytoskeletons as an integrated unit. An interesting (although controversial) theory is that of cellular tensegrity, whereby the cells mechanical structure is explained via tensional integrity (Ingber 1993, 2003a, 2003b, 2003c; Maniotis, Bojanowski, et al 1997; Maniotis, Chen, et al 1997; Charras and Horton 2002). Through this tensegrity structure, tensional forces from the extracellular environment (eg, from tissue loading or changes in cell spreading) are possibly conferred to the nucleus and alter genome regulation (Mosgoller et al 1991; Heslop-Harrison et al 1993; Heslop-Harrison 2000; Dalby, Riehle, Sutherland, et al 2004c; Dalby 2005).
We currently support the idea of an integrated cytoskeleton – not necessarily tensegrity, but along those lines – is directly linked to the extracellular matrix through actin anchoring to adhesions and then linked to the nucleus via the intermediate filaments of the cytoskeleton associating with lamin intermediate filaments of the nucleoskeleton. It is further known that the telomeric ends of the interphase chromosomes are intimately linked to the lamins (Foster and Bridger 2005). Thus, tension directed through the cytoskeleton may be passed directly to the chromosomes during gene transcription. Changes in chromosomal three-dimensional arrangement (considering the chromosomes in the interphase nuclei to have relative consistency of position (Heslop-Harrison and Bennett 1990) may effect transcriptional events such as access to the genes by transcription factors and polymerases. Also, changes in DNA tension can also cause polymerase enzymes to slow down, speed up or even stall completely (Bustamante et al 2003). These may be mechanisms by which changes in cell spreading on nanotopographies can change events such as adhesion, proliferation and even differentiation (Dalby, Biggs, et al 2006; Dalby, McCloy, et al 2006a, 2006b) (Figure 2).
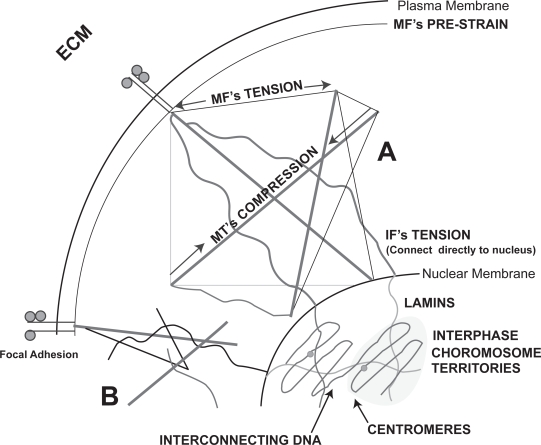
Suggested possible mechanisms for direct mechanotransduction incuding theories of cytoskeletal tensegrity which relys on the interaction of tensile and compressive elements (A) and percolation (B). Cytoskeletal percolation is a simplified form of tensegrity whereby simple integration is required without the rules of tensegrity. The figure shows continuum from the cellular adhesions to the chromosome territories within the nucleus.
Another, possibly connected, theory is that of nano-imprinting into cells by nanofeatures (Curtis et al 2006a, 2006b). This describes a phenomenon that has been clearly seen in platelets and for which some evidence has been provided in more complex cell types (fibroblasts). For nanoimprinting to occur, the pattern of the topography must be transferred to the cytoskeletal filaments, ie, the topographies produce a template that is favorable/unfavorable to condensation of cytoskeletal polymer chains through invagination (or embossing) of the basal membrane against the topography. There is evidence that this leads to increased ‘attempted’ endocytosis, ie, the cells recognize the features (in the referenced case, nanocolumns – 160 nm high) as being in the correct size range to try to endocytose and to form claterin coated pits (Dalby, Berry, et al 2004). Such endocytotic vesicles are moved by actin cables; perhaps it is this mechanism that is causing the topography mimicking actin patterning described by Curtis et al (2006b).
Low adhesion materials
It is fair to say that in biomaterials research there has been major drive to consider how to increase cellular adhesion and proliferation on materials surfaces, however, this is not always the effect desired. There are many applications where low-adhesion is required such as in stents, catheters, heart valves, acetabulum etc where cell migration and tissue formation are problematic. Whilst it is presently hard to see if topography can have roles where tribological issues are involved, it is possible to foresee that in areas such as catheters and valves topography could play a key role.
Topographies that produce low adhesion in fibroblasts and mesenchymal stem cells have already been patterned inside tubes using polymer demixing to form nanoislands (Gadegaard et al 2004; Berry et al 2005, 2006). The next step has to be to produce the islands on the internal surface in the bulk of the tube polymer. This could perhaps be achieved by embossing the low-adhesion shapes and then precision rolling – in a similar manner to that outlined by Seunarine et al (2006). In a similar way, curved surfaces could be achieved.
Nanoislands produced by polymer demixing are, in fact, tunable to low or high adhesion (Dalby, Pasqui, et al 2004). Very small islands (<20 nm) promote adhesion in most cell types tested (endothelial (Dalby, Riehle, et al 2002a), fibroblastic (Dalby, Riehle, et al 2002b) and mesenchymal stem (Dalby, McCloy, et al 2006a)). As the islands increase in size, they become less adhesive. Islands produced from a blend of polystyrene/ polybutylmethacrylate were adhesive to fibroblasts at 10 nm high, but almost completely non-adhesive at 50 nm high (Dalby, Riehle, Johnstone, et al 2003).
Topographies produced by colloidal lithography have shown similar trend, with very small nanocolumns (11 nm high) producing increased osteoprogenitor response (Dalby, McCloy, et al 2006a), whilst larger nanocolumns (160 nm high) produced a significant (but far from complete) reduction in fibroblast adhesion (Dalby, Riehle, Sutherland, et al 2004a, 2004b). The Sutherland group has also done some interesting work on topographies fabricated through the colloidal route. They firstly showed 100 nm high columns to reduce epithelial cell adhesion (Andersson, Backhed, et al 2003) and then went further to shown, again in line with the polymer demixed nanotopographies, that increasing column size decreased epithelial cell adhesion (Andersson, Brink, et al 2003). In collaboration with Rice and Hunt at Liverpool, they also showed that the 160 nm high nanocolumns imparted properties of low-adhesion on osteoblasts (Rice et al 2003).
Using another fabrication method, that of interference lithography, a recent paper also agreed with these observations of size and adhesion. Choi and others fabricated sharp nanoposts (pointed columns) and nanogratings (very sharp grooves) of different heights. They used 3 size ranges: 50–100 nm high, 200–300 nm high and 500–600 nm high and found decreased adhesion and spreading as size increased (Choi et al 2007).
The notable observation from the above is the theme of tunable height of features, with not much mention about depth. However, there is a very low adhesion pit system that with some cell types can produce almost total non-adhesion. That of nanopits fabricated in a square array by EBL with diameters of approx. 100–150 nm diameters and centre-centre spacing (pitch) in the range of 300 nm. Again, these adhesion effects have been shown to be size dependant. A study that used 3 pit sizes (diameter (nm): pitch (nm) – 35:100, 75:200 and 120:300) and fibroblasts as a cell model showed that as pit diameter increased, filopodial interaction increased, but cell spreading decreased, ie, the interaction of the filopodia with the pits was preventing adhesion (Dalby, Gadegaard, et al 2004). A previous study by Gallagher et al (2002) with pit diameters of 150 nm showed almost no epitenon attachment. However we know that large pits (in the range produced by photolithography) promote increased cellular response in eg, mesenchymal stem cells (Dalby, McCloy, et al 2006b). Thus, there is a large gap in our knowledge of the transition between non-adhesive and adhesive pits (Figure 3).
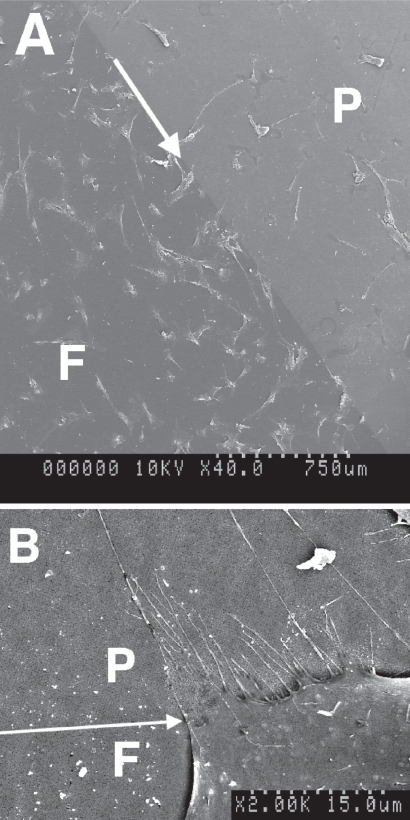
Sensing of low-adhesion borders. In both (A) and (B) there are both flat parts (F) and parts patterned with low-adhesion EBL nanopits (P). (A) Shows that there are many more cells on the planar side of the border, whilst (B) shows a high magnification image of a cell aligning to the border and producing filopodia onto the nanopattern. Arrows show the direction of the planar/topographical borders.
This is further complicated by pit symmetry being implicated as important. This was originally mooted by Curtis and others in 2004 (Curtis et al 2004). Subsequent studies have used 120 nm diameter, 300 nm pitch pits in both square and hexagonal arrangement. Both with mesenchymal stem cells (Hart et al, in press; Dalby et al, submitted) and with fibroblasts (Dalby, Gadegaard, et al 2004; Dalby, Gadegaard, Herzyk, et al 2007; Dalby, Gadegaard, Wilkinson 2007), the hexagonal arrangement has been shown to be the least adhesive.
Mechanisms of low adhesion
It seems likely that the ability of a cell to adhere and spread will alter mechanosensitive pathways in a cell. As adhesions form and act as an anchor for the cytoskeleton and the integrins within the adhesions act as a route for cell/substrate signaling (Burridge and Chrzanowska-Wodnicka 1996), it is probable that low-adhesion topographies will firstly act by changing adhesion formation and morphology and as consequence effect mechanosensitive pathways. Indeed, Biggs et al (2007a, 2007b) have shown the total number of adhesions to reduce significantly on the square and hexagonal low-adhesion EBL arrays. Interestingly, however, they showed the proportion of focal complexes (transient adhesions involved in motility), focal adhesions (mature adhesions to which stress fibers anchor) and fibrilar adhesions (very mature, align to the cells endogenous matrix) to remain constant; thus, whilst a reduction in adhesions was seen, a change in morphology was not.
The loss of attachment points for the cytoskeleton will probably result in a reduction of applied tension to the nucleus and a concurrent reduction in integrin related signaling. For the direct mechanosensitive theories (described above), whilst increased cytoskeletal organization, on high-adhesion substrata, would result in increased nuclear tension, reduced organization, on low-adhesion substrata, would result in reduced nuclear tension.
We have investigated this to some extent with low-adhesion topographies. Thus far, it has been seen that reduction of adheion formation potential and cytoskeletal organization changes lamin organization and, via FISH of centromere pairs, relative positions of the chromosomes in the interphase nucleus (Dalby, Riehle, Sutherland, et al 2004c; Dalby, Biggs, et al 2006; Dalby 2005).
Further to this, using human whole genome microarrays and plotting the positions along the chromosome arms of topographically induced gene changes, it has been proposed that the changes in nucleus morphology caused by culture on low-adhesion topographies, leads to spatial changes in genome regulations along the chromosomes. We proposed that rather like a net, opening or closing areas of the DNA via changing morphology alters the ability of the transcription factors to bind the genes (Dalby, Gadegaard, Herzyk, et al 2007; Dalby et al in press). Whilst speculative, these ideas agree with Forgacs theories of cellular percolation and suggest that the extracellular matrix, through fibrilar proteins, the cell, via integrins and the cytoskeleton, the nucleus through the nucleoskeleton and the DNA via the long interphase chromosomes may be in mechanical continuum.
However, as also mentioned in the section on mechanotransduction, indirect mechanisms must not be forgotten and are critically important. A recent microarray study on two low adhesion materials, columns produced by colloidal lithography and EBL pits showed a number of important canonical pathways to be down-regulated by the topographies. These included transforming growth factor, cytokine pathways, G-protein coupled signaling, calcium signaling, integrin signaling, endothelial growth factor, insulin-like growth factor and cyclic AMP signaling amongst others (Dalby, Gadegaard, Herzyk, et al 2007). All of these pathways being reduced will change (decrease) cellular activity such as growth, proliferation and differentiation.
A further array study again demonstrated that culture on nanopits lead to broad gene down-regulation (Dalby, Gadegaard, Wilkinson 2007). Included in the few significant gene up-regulations, and confirmed by clathrin staining, were changes in endocytotic pathways. This shows that the cells were trying to internalize pits as well as the aforementioned nanocolumns and provides further evidence for nanoimprinting.
Summary
It has been shown that nanoscale topography can both increase and reduce adhesion of a broad number of cell types. It has been suggested, in agreement with traditional views, that indirect mechanotransductive pathways play a key role in changes in cell response to low adhesion. Furthermore, preliminary evidence has been presented that more direct mechanisms may be important. It could be speculated that low-adhesion topographies firstly act by reducing adhesion, which impacts not only of integrin related signaling, but also on nanoimprinting through reduction in sites for adhesion and cytoskeletal anchorage. It could be further speculated that low-adhesion topographies reduce tension applied to the cell from a well-organized extracellular matrix via the cytoskeleton to the nucleus effectively shutting the cells down to transcription. However, there is a great deal of work that needs to be done yet in order to prove or disprove these theories of cellular response to nanotopograpies.
Acknowledgments
I am supported by a BBSRC David Phillips Fellowship and an AO Research Fund grant (04-D81) and thank both funders sincerely. I am also thankful to Prof Chris Wilkinson and Dr Nikolaj Gadegaard for EBL fabrication, Dr Duncan Sutherland and Dr Hossein Agheli for colloidal lithography and Dr Stanley Affrossman and Ms Heather Johnstone for polymer demixing. I am especially indebted to Prof Adam Curtis and Drs Mathis Riehle and Catherine Berry for all their support, help and discussion over the years in Glasgow.
References
- Affrossman S, Henn G, O’Neill SA, et al. Surface topography and composition of deuterated polystyrene – poly(bromostyrene) blends. Macromolecules. 1996;29:5010–16. [Google Scholar]
- Affrossman S, Jerome R, O’Neill SA, et al. Surface structure of thin film blends of polystyrene and poly(n-butyl methacrylate) Colloid Polym Sci. 2000;278:993–9. [Google Scholar]
- Affrossman S, O’Neill SA, Stamm M. Topography and surface composition of thin films of blends of polystyrene with bromiated polystyrenes: effects of varying the degree of bromination and annealing. Macromolecules. 1998;31:6280–8. [Google Scholar]
- Affrossman S, Stamm M. The effect of molecular weight on the topography of thin films of blends of poly(4-bromostyrene) and polystyrene. Collid Polymer Science. 2000;278:888–93. [Google Scholar]
- Agheli H, Sutherland DS. Nanofabrication of polymer surfaces utilising colloidal lithography and ion etching. IEEE Trans Nanobioscience (Submitted) [Abstract] [Google Scholar]
- Andersson AS, Backhed F, Von Euler A, et al. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials. 2003;24:3427–36. [Abstract] [Google Scholar]
- Andersson AS, Brink J, Lidberg U, et al. Influence of Systematically Varied nanoscale Topography on the Morhphology of Epithelial Cells. IEEE Transactions on Nanobioscience. 2003;2:49–57. [Abstract] [Google Scholar]
- Berry CC, Dalby MJ, Mccloy D, et al. The fibroblast response to tubes exhibiting internal nanotopography. Biomaterials. 2005;26:4985–92. [Abstract] [Google Scholar]
- Berry CC, Dalby MJ, Oreffo RO, et al. The interaction of human bone marrow cells with nanotopographical features in three dimensional constructs. J Biomed Mater Res A 2006 [Abstract] [Google Scholar]
- Biggs MJP, Richards RG, Gadegaard N, et al. Characterisation of S-phase primary osteoblast adhesion complexes on nanopitted materials fabricated by electron-beam lithography. Journal of Orthopaedic Research. 2007a;25:273–82. [Abstract] [Google Scholar]
- Biggs MJP, Richards RG, Gadegaard N, et al. The effects of nanoscale pits on primary human osteoblast adhesion and cellular spreading. Journal of Materials Science Materials in Medicine. 2007b;18:399–404. [Abstract] [Google Scholar]
- Blondiaux N, Zurcher S, Liley M, et al. Fabrication of multiscale surface-chemical gradients by means of photocatalytic lithography. Langmuir. 2007;23:3489–94. [Abstract] [Google Scholar]
- Britland S, Morgan H, Wojciak-Stothard B, et al. Synergistic and hierarchical adhesive and topographic guidance of BHK cells. Exp Cell Res. 1996;228:313–25. [Abstract] [Google Scholar]
- Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. [Abstract] [Google Scholar]
- Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–7. [Abstract] [Google Scholar]
- Charras GT, Horton MA. Single cell mechanotransduction and its modulation analyzed by atomic force microscope indentation. Biophys J. 2002;82:2970–81. [Europe PMC free article] [Abstract] [Google Scholar]
- Choi CH, Hagvall SH, Wu BM, et al. Cell interaction with three-dimensional sharp-tip nanotopography. Biomaterials. 2007;28:1672–9. [Abstract] [Google Scholar]
- Clark P, Connolly P, Curtis AS, et al. Topographical control of cell behaviour. I. Simple step cues. Development. 1987;99:439–48. [Abstract] [Google Scholar]
- Clark P, Connolly P, Curtis AS, et al. Topographical control of cell behaviour: II. Multiple grooved substrata. Development. 1990;108:635–44. [Abstract] [Google Scholar]
- Clark P, Connolly P, Curtis AS, et al. 1991Cell guidance by ultrafine topography in vitro J Cell Sci 99 Pt 173–7. [Abstract] [Google Scholar]
- Curtis AS. Small is beautiful but smaller is the aim: review of a life of research. Eur Cell Mater. 2004;8:27–36. [Abstract] [Google Scholar]
- Curtis AS, Gadegaard N, Dalby MJ, et al. Cells react to nanoscale order and symmetry in their surroundings. IEEE Trans Nanobioscience. 2004;3:61–5. [Abstract] [Google Scholar]
- Curtis ASG, Dalby MJ, Gadegaard N. Cell signaling arising from nanotopography: implications for nanomedical devices. Nanomedicine. 2006a;1:67–72. [Abstract] [Google Scholar]
- Curtis ASG, Dalby MJ, Gadegaard N. Nanoimprinting onto Cells. Journal of the Royal Society Interface. 2006b;3:393–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Dalby MJ. Topographically induced direct cell mechanotransduction. Medical Engineering and Physics. 2005;27:730–42. [Abstract] [Google Scholar]
- Dalby MJ, Berry CC, Riehle MO, et al. Attempted endocytosis of nano-environment produced by colloidal lithography by human fibro-blasts. Exp Cell Res. 2004;295:387–94. [Abstract] [Google Scholar]
- Dalby MJ, Biggs MJP, Gadegaard N, et al. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. Journal of Cellular Biochemistry 2006 [Abstract] [Google Scholar]
- Dalby MJ, Biggs MJP, Gadegaard N, et al. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. Journal of Cellular Biochemistry. 2007;100:326–38. [Abstract] [Google Scholar]
- Dalby MJ, Childs S, Riehle MO, et al. Fibroblast reaction to island topography: changes in cytoskeleton and morphology with time. Biomaterials. 2003;24:927–35. [Abstract] [Google Scholar]
- Dalby MJ, Gadegaard N, Andar A, et al. (Submitted) Nanoscale symmetry and disorder are important in human mesenchymal stem cell differentiation [Abstract]
- Dalby MJ, Gadegaard N, Herzyk P, et al. Group analysis of regulation of fibroblast genome on low-adhesion nanostructures. Biomaterials. 2007 In Press. [Abstract] [Google Scholar]
- Dalby MJ, Gadegaard N, Herzyk P, et al. Nanomechanotransduction and interphase nuclear organsiation influence on genomic control. Journal of Cellular Biochemistry (In Press) [Abstract] [Google Scholar]
- Dalby MJ, Gadegaard N, Riehle MO, et al. Investigating filopodia sensing using arrays of defined nano-pits down to 35 nm diameter in size. Int J Biochem Cell Biol. 2004;36:2015–25. [Abstract] [Google Scholar]
- Dalby MJ, Gadegaard N, Wilkinson CDW. The response of fibroblasts to hexagonal nanotopohraphy fabricated by electron beam lithograhy. Journal of Biomedical Materials Research A. 2007 In Press. [Abstract] [Google Scholar]
- Dalby MJ, McCloy D, Robertson M, et al. Osteoprogenitor response to semi-ordered and random nanotopographies. Biomaterials. 2006a;27:2980–7. [Abstract] [Google Scholar]
- Dalby MJ, McCloy D, Robertson M, et al. Osteoprogenitor response to defined topographies with nanoscale depths. Biomaterials. 2006b;27:1306–15. [Abstract] [Google Scholar]
- Dalby MJ, Pasqui D, Affrossman S. Cell response to nano-islands produced by polymer demixing: a brief review. IEE Proc. Nanobiotechnol. 2004;151:53–61. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Johnstone H, et al. In vitro reaction of endothelial cells to polymer demixed nanotopography. Biomaterials. 2002a;23:2945–54. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Johnstone HJ, et al. Polymer-demixed nanotopography: control of fibroblast spreading and proliferation. Tissue Eng. 2002b;8:1099–108. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Johnstone HJ, et al. Nonadhesive nanotopography: fibroblast response to poly(n-butyl methacrylate)-poly(styrene) demixed surface features. J Biomed Mater Res. 2003;67A:1025–32. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Johnstone H, et al. Investigating the limits of filopodial sensing: a brief report using SEM to image the interaction between 10 nm high nano-topography and fibroblast filopodia. Cell Biol Int. 2004;28:229–36. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Sutherland DS, et al. Changes in fibroblast morphology in response to nano-columns produced by colloidal lithography. Biomaterials. 2004a;25:5415–22. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Sutherland DS, et al. Fibroblast response to a controlled nanoenvironment produced by colloidal lithography. J Biomed Mater Res. 2004b;69A:314–22. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Sutherland DS, et al. Use of nanotopography to study mechanotransduction in fibroblasts – methods and perspectives. Eur J Cell Biol. 2004c;83:159–69. [Abstract] [Google Scholar]
- Dalby MJ, Riehle MO, Yarwood SJ, et al. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284:274–82. [Abstract] [Google Scholar]
- Dalby MJ, Yarwood SJ, Riehle MO, et al. Increasing fibroblast response to materials using nanotopography: morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands. Exp Cell Res. 2002;276:1–9. [Abstract] [Google Scholar]
- Denis FA, Hanarp P, Sutherland DS, et al. Fabrication of nanostructured polymer surfaces using colloidal lithography and spin coating. Nanoletters. 2002a;2:1419–25. [Google Scholar]
- Denis FA, Hanarp P, Sutherland DS, et al. Protein adsorption on model surfaces with controlled nanotopography and chemistry. Langmuir. 2002b;18:819–28. [Google Scholar]
- Foster HA, Bridger JM. The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114:212–29. [Abstract] [Google Scholar]
- Gadegaard N, Dalby MJ, Riehle MO, et al. Tubes with controllable internal nanotopography. Advanced Materials. 2004;16:1857–60. [Google Scholar]
- Gadegaard N, Mosler S, Larsen NB. Biomimetic polymer nano-structures by injection moulding. Macromeolecular Materials and Engineering. 2003;288:76–83. [Google Scholar]
- Gadegaard N, Thoms S, Macintyre DS, et al. Arrays of nano-dots for cellular engineering. Microelectronic Engineering. 2003:67–68. 162–8. [Google Scholar]
- Gallagher JO, McGhee KF, Wilkinson CDW, et al. Interaction of Animal Cells with Ordered Nanotopography. IEEE Transactions on Nanobioscience. 2002;1:24–8. [Abstract] [Google Scholar]
- Gustafson T, Wolpert L. Studies on the cellular basis of morphogenesis in the sea urchin embryo. Directed movements of primary mesenchvme cells in normal and vegetalized larvae. Exp Cell Res. 1961;24:64–79. [Abstract] [Google Scholar]
- Hart A, Gadegaard N, Wilkinson CDW, et al. Osteoprogenitor response to low-adhesion nanotopographies originally fabricated by electron beam lithography. Journal Of Materials Science Materials In Medicine In Press. [Abstract] [Google Scholar]
- Heslop-Harrison JS. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell. 2000;12:617–36. [Abstract] [Google Scholar]
- Heslop-Harrison JS, Bennett MD. Nuclear architecture in plants. Trends Genet. 1990;6:401–5. [Abstract] [Google Scholar]
- Heslop-Harrison JS, Leitch AR, Schwarzacher T. The physical organisation of interphase nuclei. In: Heslop-Harrison JS RB F, editor. The chromosome. Oxford: Bios; 1993. [Google Scholar]
- Ingber DE. 1993Cellular tensegrity: defining new rules of biological design that govern the cytoskeleton J Cell Sci 104 (Pt 3)613–27. [Abstract] [Google Scholar]
- Ingber DE. Mechanosensation through integrins: cells act locally but think globally. Proc Natl Acad Sci USA. 2003a;100:1472–4. [Europe PMC free article] [Abstract] [Google Scholar]
- Ingber DE. Tensegrity I. Cell structure and hierarchical systems biology. J Cell Sci. 2003b;116:1157–73. [Abstract] [Google Scholar]
- Ingber DE. Tensegrity II. How structural networks influence cellular information processing networks. J Cell Sci. 2003c;116:1397–408. [Abstract] [Google Scholar]
- Jones GE, Allen WE, Ridley AJ. The Rho GTPases in macrophage motility and chemotaxis. Cell Adhes Commun. 1998;6:237–45. [Abstract] [Google Scholar]
- Kasemo B, Lausmaa J. Biomaterial and implant surfaces: a surface science approach. Int J Oral Maxillofac Implants. 1988;3:247–59. [Abstract] [Google Scholar]
- Kasemo B, Lausmaa J. Material-tissue interfaces: the role of surface properties and processes. Environ Health Perspect. 1994;1025(Suppl):41–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Maniotis AJ, Bojanowski K, Ingber DE. Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J Cell Biochem. 1997;65:114–30. [Abstract] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94:849–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Martines E, Seunarine K, Morgan H, et al. Superhydrophobicity and superhydrophilicity of regular nanopatterns. Nanoletters. 2005;5:2097–103. [Abstract] [Google Scholar]
- Minelli C, Geissbuehler I, Hinderling C, et al. Organization of nanoparticles on hard substrates using block copolymer films as templates. J Nanosci Nanotechnol. 2006;6:1611–19. [Abstract] [Google Scholar]
- Mosgoller W, Leitch AR, Brown JK, et al. Chromosome arrangements in human fibroblasts at mitosis. Hum Genet. 1991;88:27–33. [Abstract] [Google Scholar]
- Price RL, Haberstroh KM, Webster TJ. Enhanced functions of osteoblasts on nanostructured surfaces of carbon and alumina. Med Biol Eng Comput. 2003;41:372–5. [Abstract] [Google Scholar]
- Rice JM, Hunt JA, Gallagher JA, et al. Quantitative assessment of the response of primary derived human osteoblasts and macrophages to a range of nanotopography surfaces in a single culture model in vitro. Biomaterials. 2003;24:4799–818. [Abstract] [Google Scholar]
- Schmitz AA, Govek EE, Bottner B, et al. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261:1–12. [Abstract] [Google Scholar]
- Seunarine K, Gadegaard N, Torman M, et al. 3D Polymer Scaffolds for Tissue Engineering. Nanomedicine. 2006;1:281–96. [Abstract] [Google Scholar]
- Teixeira AI, Abrams GA, Bertics PJ, et al. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J Cell Sci. 2003;116:1881–92. [Europe PMC free article] [Abstract] [Google Scholar]
- Thapa A, Webster TJ, Haberstroh KM. Polymers with nano-dimensional surface features enhance bladder smooth muscle cell adhesion. J Biomed Mater Res. 2003;67A:1374–83. [Abstract] [Google Scholar]
- Vieu C, Carcenac F, Pepin A, et al. Electron beam lithography: resolution limits and applications. Applied Surface Science. 2000;164:111–17. [Google Scholar]
- Ward BC, Webster TJ. The effect of nanotopography on calcium and phosphorus deposition on metallic materials in vitro. Biomaterials. 2006;27:3064–74. [Abstract] [Google Scholar]
- Weiss P, Garber B. Shape and movement of mesenchyme cells as functions of the physical structure of the medium. Proc. Natl. Acad. Sci. USA. 1952;38:264–80. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilkinson CD. Making structures for cell engineering. Eur Cell Mater. 2004;8:21–6. [Abstract] [Google Scholar]
- Wilkinson CDW, Riehle M, Wood M, et al. The use of materials patterned on a nano- and micro-metric scale in cellular engineering. Materials Science and Engineering. 2002;19:263–9. [Google Scholar]
- Wojciak-Stothard B, Curtis ASG, Monaghan W, et al. Role of the cytoskeleton in the reaction of fibroblasts to multiple grooved substrata. Cell Motil Cytoskeleton. 1995;31:147–58. [Abstract] [Google Scholar]
- Wojciak-Stothard B, Madeja Z, Korohoda W, et al. Activation of macrophage-like cells by multiple grooved substrata - topographical control of cell behavior. Cell Biol Int. 1995:485–90. [Abstract] [Google Scholar]
Articles from International Journal of Nanomedicine are provided here courtesy of Dove Press
Citations & impact
Impact metrics
Citations of article over time
Article citations
3D Cell Culture Systems: Tumor Application, Advantages, and Disadvantages.
Int J Mol Sci, 22(22):12200, 11 Nov 2021
Cited by: 133 articles | PMID: 34830082 | PMCID: PMC8618305
Review Free full text in Europe PMC
Micro/nano-textured hierarchical titanium topography promotes exosome biogenesis and secretion to improve osseointegration.
J Nanobiotechnology, 19(1):78, 19 Mar 2021
Cited by: 23 articles | PMID: 33741002 | PMCID: PMC7980346
The Impacts of Crystalline Structure and Different Surface Functional Groups on Drug Release and the Osseointegration Process of Nanostructured TiO2.
Molecules, 26(6):1723, 19 Mar 2021
Cited by: 1 article | PMID: 33808785 | PMCID: PMC8003584
Design, construction and characterisation of a novel nanovibrational bioreactor and cultureware for osteogenesis.
Sci Rep, 9(1):12944, 10 Sep 2019
Cited by: 12 articles | PMID: 31506561 | PMCID: PMC6736847
Formation of nanotubular TiO2 structures with varied surface characteristics for biomaterial applications.
J Biomed Mater Res A, 106(5):1341-1354, 31 Jan 2018
Cited by: 5 articles | PMID: 29316200
Go to all (24) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The effects of nanoscale pits on primary human osteoblast adhesion formation and cellular spreading.
J Mater Sci Mater Med, 18(2):399-404, 01 Feb 2007
Cited by: 55 articles | PMID: 17323174
Decreased fibroblast and increased osteoblast adhesion on nanostructured NaOH-etched PLGA scaffolds.
Int J Nanomedicine, 2(3):383-388, 01 Jan 2007
Cited by: 19 articles | PMID: 18019837 | PMCID: PMC2676656
Neurons on nanotopographies: behavioral responses and biological implications.
J Nanosci Nanotechnol, 14(1):513-521, 01 Jan 2014
Cited by: 6 articles | PMID: 24730279
Review
Nanotopography: cellular responses to nanostructured materials.
J Nanosci Nanotechnol, 6(7):1905-1919, 01 Jul 2006
Cited by: 42 articles | PMID: 17025103
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (1)
BBSRC David Phillips Fellowship Nano-biomimetics for bone tissue engineering: Progenitor cell adhesion and differentiation
Dr Dalby, University of Glasgow
Grant ID: JF20604



