Abstract
Free full text

Evaluation of autoSCAN-W/A automated microbiology system for the identification of non-glucose-fermenting gram-negative bacilli.
Abstract
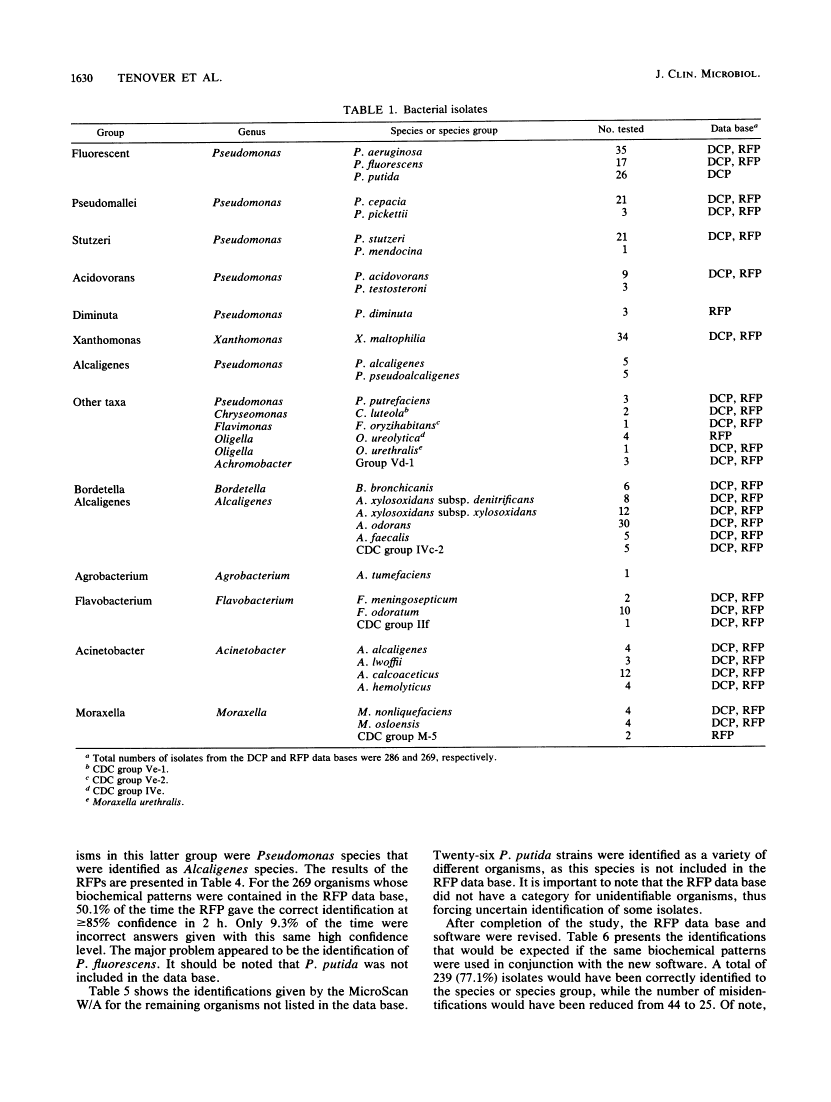
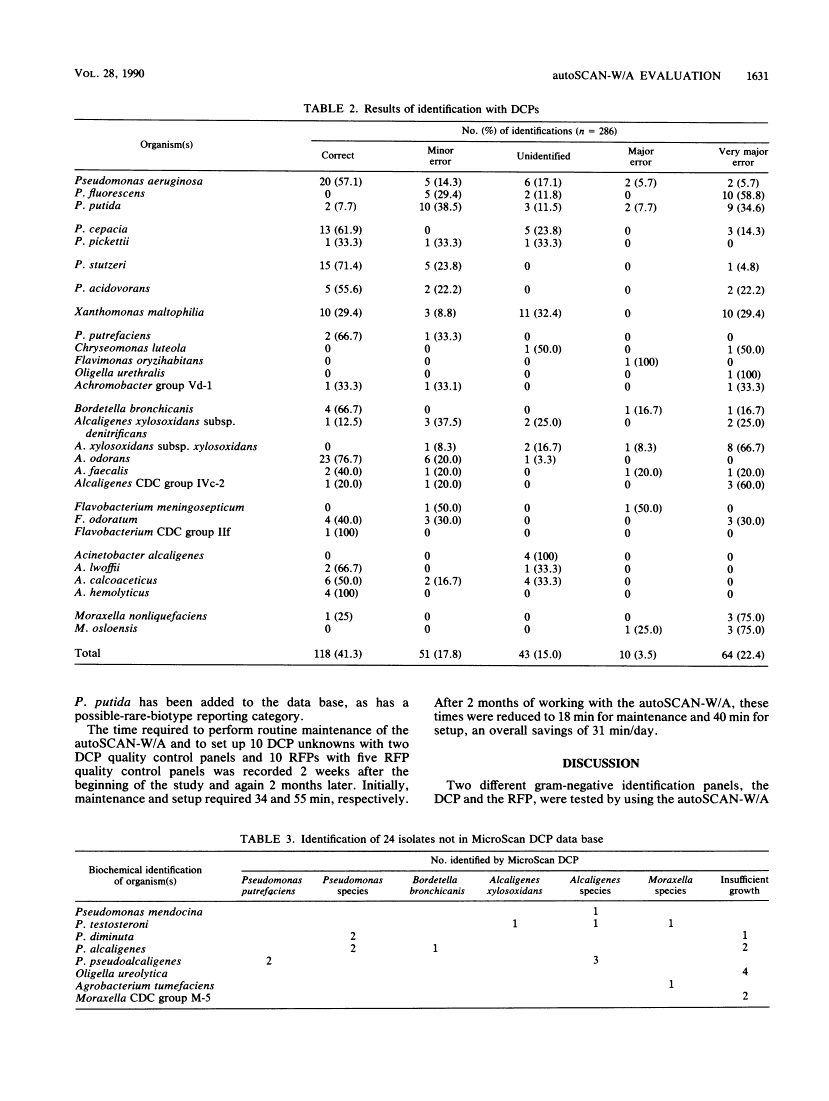
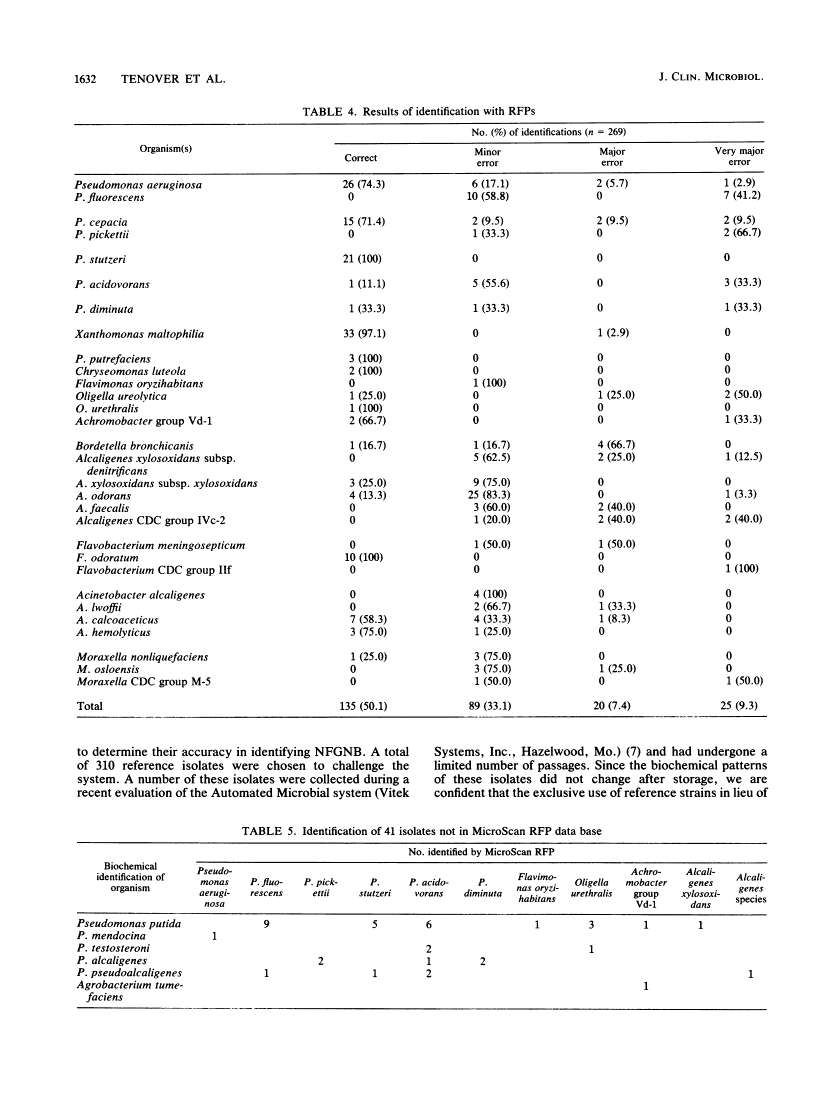
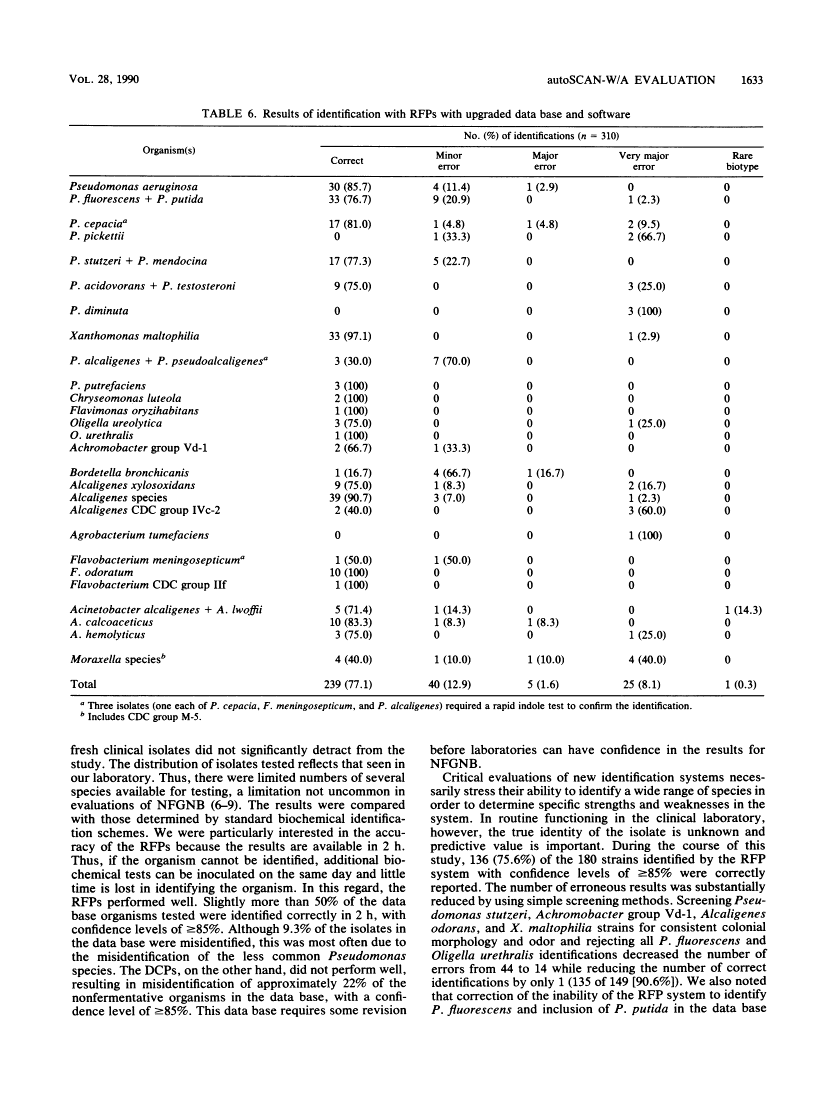
We evaluated the ability of the autoSCAN-W/A (MicroScan Division, Baxter Healthcare Corporation, West Sacramento, Calif.), in conjunction with the dried colorimetric Neg ID type 2 panel (DCP) and new rapid fluorometric Neg ID panel (RFP), to identify non-glucose-fermenting gram-negative bacilli by challenging the system with 310 previously identified reference strains. Of these 310 isolates, 286 organisms were in the DCP data base and 269 were in the RFP data base. Use of the DCP panels resulted in 118 (41.3%) correct and 64 (22.4%) incorrect first choice identifications at greater than or equal to 85% probability, 61 (21.3%) low-probability identifications, and 43 (15.0%) reports of unidentified organisms. The RFP system reported 135 (50.1%) correct and 25 (9.3%) incorrect identifications at greater than or equal to 85% probability and 109 (40.5%) low-probability identifications. Unidentified isolates (DCP system only) and isolates producing low-probability first choice identifications (both systems) required supplementary biochemical testing. Over half (37 of 64 [57.8%]) of the DCP misidentifications were due to four commonly isolated, saccharolytic organisms (Alcaligenes xylosoxidans subsp. xylosoxidans, Pseudomonas putida, Pseudomonas fluorescens, and Xanthomonas maltophilia), while 7 of 25 (28%) of misidentifications in the RFP system were due to P. fluorescens. Of note, the RFP system identified non-glucose-fermenting gram-negative bacilli within 2 h of panel inoculation, allowing additional conventional biochemical tests to be set up the same day on low-probability isolates, whereas only 13.5% of the DCPs could be read at 18 h, with the remainder requiring 42 h of incubation before reading. When organism identifications were recalculated with the updated RFP data base and revised software, only 8.1% of all 310 isolates were misidentified at greater than or equal to 85% probability while 77.1% of the isolates were now correctly reported at this same high probability.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker CN, Stocker SA, Rhoden DL, Thornsberry C. Evaluation of the MicroScan antimicrobial susceptibility system with the autoSCAN-4 automated reader. J Clin Microbiol. 1986 Jan;23(1):143–148. [Europe PMC free article] [Abstract] [Google Scholar]
- Gavini F, Husson MO, Izard D, Bernigaud A, Quiviger B. Evaluation of autoscan-4 for identification of members of the family Enterobacteriaceae. J Clin Microbiol. 1988 Aug;26(8):1586–1588. [Europe PMC free article] [Abstract] [Google Scholar]
- Kämpfer P, Dott W. Evaluation of the Titertek-NF system for identification of gram-negative nonfermentative and oxidase-positive fermentative bacteria. J Clin Microbiol. 1989 Jun;27(6):1201–1205. [Europe PMC free article] [Abstract] [Google Scholar]
- Plorde JJ, Gates JA, Carlson LG, Tenover FC. Critical evaluation of the AutoMicrobic system gram-negative identification card for identification of glucose-nonfermenting gram-negative rods. J Clin Microbiol. 1986 Feb;23(2):251–257. [Europe PMC free article] [Abstract] [Google Scholar]
- Rhoden DL, Smith PB, Baker CN, Schable B. autoSCAN-4 system for identification of gram-negative bacilli. J Clin Microbiol. 1985 Dec;22(6):915–918. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.28.7.1628-1634.1990
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/28/7/1628.full.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/28/7/1628
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/28/7/1628.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Evaluation of five susceptibility test methods for detection of tobramycin resistance in a cluster of epidemiologically related Acinetobacter baumannii isolates.
J Clin Microbiol, 51(8):2535-2540, 22 May 2013
Cited by: 3 articles | PMID: 23698528 | PMCID: PMC3719664
Evaluation of the BD Phoenix SMIC/ID, a new streptococci identification and antimicrobial susceptibility panel, for potential routine use in a university-based clinical microbiology laboratory.
Diagn Microbiol Infect Dis, 53(2):101-105, 15 Sep 2005
Cited by: 6 articles | PMID: 16168613
Evaluation of the automated phoenix system for potential routine use in the clinical microbiology laboratory.
J Clin Microbiol, 42(4):1542-1546, 01 Apr 2004
Cited by: 56 articles | PMID: 15071001 | PMCID: PMC387561
Evaluation of autoSCAN-W/A and the Vitek GNI+ AutoMicrobic system for identification of non-glucose-fermenting gram-negative bacilli.
J Clin Microbiol, 38(3):1127-1130, 01 Mar 2000
Cited by: 8 articles | PMID: 10699007 | PMCID: PMC86355
Evaluation of the VITEK 2 system for the identification and susceptibility testing of three species of nonfermenting gram-negative rods frequently isolated from clinical samples.
J Clin Microbiol, 39(9):3247-3253, 01 Sep 2001
Cited by: 58 articles | PMID: 11526158 | PMCID: PMC88326
Go to all (15) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Parallel comparison of accuracy of API 20E, Vitek GNI, MicroScan Walk/Away Rapid ID, and Becton Dickinson Cobas Micro ID-E/NF for identification of members of the family Enterobacteriaceae and common gram-negative, non-glucose-fermenting bacilli.
J Clin Microbiol, 31(12):3165-3169, 01 Dec 1993
Cited by: 25 articles | PMID: 8308108 | PMCID: PMC266369
Comparison of the autoSCAN-W/A rapid bacterial identification system and the Vitek AutoMicrobic system for identification of gram-negative bacilli.
J Clin Microbiol, 29(7):1422-1428, 01 Jul 1991
Cited by: 23 articles | PMID: 1885737 | PMCID: PMC270128
Evaluation of autoSCAN-W/A and the Vitek GNI+ AutoMicrobic system for identification of non-glucose-fermenting gram-negative bacilli.
J Clin Microbiol, 38(3):1127-1130, 01 Mar 2000
Cited by: 8 articles | PMID: 10699007 | PMCID: PMC86355
Evaluation of Vitek GNI+ and Becton Dickinson Microbiology Systems Crystal E/NF identification systems for identification of members of the family Enterobacteriaceae and other gram-negative, glucose-fermenting and non-glucose-fermenting bacilli.
J Clin Microbiol, 35(12):3269-3273, 01 Dec 1997
Cited by: 19 articles | PMID: 9399532 | PMCID: PMC230160