Abstract
Free full text
Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans
Abstract
Dietary restriction (DR) has the remarkable ability to extend lifespan and healthspan. A variety of DR regimens have been described in species ranging from yeast to mammals. However, whether different DR regimens extend lifespan via universal, distinct, or overlapping pathways is still an open question. Here we examine the genetic pathways that mediate longevity by different DR regimens in Caenorhabditis elegans. We have previously shown that the low-energy sensing AMP-activated protein kinase AMPK/aak-2 and the Forkhead transcription factor FoxO/daf-16 are necessary for longevity induced by a DR regimen that we developed (sDR). Here we find that AMPK and FoxO are necessary for longevity induced by another DR regimen, but are dispensable for the lifespan extension induced by two different DR methods. Intriguingly, AMPK is also necessary for the lifespan extension elicited by resveratrol, a natural polyphenol that mimics some aspects of DR. Conversely, we test if genes previously reported to mediate longevity by a variety of DR methods are necessary for sDR-induced longevity. Although clk-1, a gene involved in ubiquinone biosynthesis, is also required for sDR-induced lifespan extension, we find that four other genes (sir-2.1, FoxA/pha-4, skn-1, and hsf-1) are all dispensable for longevity induced by sDR. Consistent with the observation that different DR methods extend lifespan by mostly independent genetic mechanisms, we find that the effects on lifespan of two different DR regimens are additive. Understanding the genetic network by which different DR regimens extend lifespan has important implications for harnessing the full benefits of DR on lifespan and healthspan.
Introduction
Restricting nutrients without malnutrition extends lifespan and reduces age-dependent decline and diseases in virtually all species (Masoro, 2005). Although a single term [dietary restriction (DR)] is often used to refer to this intervention, there exist a number of methods of restricting nutrients that all result in lifespan extension in species ranging from yeast to mice (Goodrick et al., 1990; Mair et al., 2005; Masoro, 2005; Dilova et al., 2007; Mair & Dillin, 2008; Piper & Bartke, 2008; Skorupa et al., 2008). Whether the different methods of restricting nutrients converge on a common pathway to extend lifespan or whether independent mechanisms are elicited depending on how DR is achieved is still unclear.
The nematode Caenorhabditis elegans provides a good model to study the genetics of lifespan in response to different DR regimens. There exist eight distinct methods of manipulating the diet that all extend lifespan to varying degrees in C. elegans, allowing comparison across DR regimens (Table 1). The standard worm diet consists of attenuated E. coli bacteria (OP50) placed on agarose plates. The DR methods in worms are: (i) a genetic mutation (eat-2) that reduces the pharyngeal pumping rate of the worms, thereby decreasing food intake (Avery, 1993; Lakowski & Hekimi, 1998); (ii and iii) two different methods of diluting the bacteria in liquid cultures (bacterial DR: bDR and liquid DR: lDR) (Klass, 1977; Houthoofd et al., 2003; Bishop & Guarente, 2007; Panowski et al., 2007); (iv and v) two chemically defined liquid medias that induce DR-like phenotype in C. elegans (axenic medium and chemically defined liquid medium: CDLM) (Houthoofd et al., 2002a; Szewczyk et al., 2006); (vi) the dilution of peptone in the agarose plates, which reduces the growth of bacteria (DP: dilution of peptone) (Hosono et al., 1989); (vii) the total absence of bacteria on plates (dietary deprivation: DD) (Kaeberlein et al., 2006; Lee et al., 2006); and (viii) a method that we recently described where bacteria are serially diluted on plates (solid DR: sDR) (Greer et al., 2007).
Table 1
Eight methods of dietary restriction in C. elegans
| Method | Normal conditions | eat-2 | bDR | lDR | Axenic | CDLM | DP | DD | sDR | Resveratrol |
|---|---|---|---|---|---|---|---|---|---|---|
| Medium | Solid | Solid | Liquid | Liquid + Solid | Liquid | Liquid | Solid | Solid | Solid | Solid |
| Source of food | Live E. coli | Live E. coli | Live E. coli (antibiotics) | Live E. coli (antibiotics) | Defined chemical broth | Defined chemical broth | Live E. coli | No E. coli | Live or dead E. coli† | Live or dead E. coli† |
| Genetic mutation | Mutation in the non-α-nicotinic acetylcholine receptor subunit | No | No | No | No | No | No | No | No | |
| Temporal | Birth | Day 2 of adulthood | L4/young adult | Larval day 4 (L4) | Birth | Birth | Day 2 of adulthood | Day 4 of adulthood | Birth | |
| Percentage of Lifespan extension* (%) | 0–57 | 60–73 | 28 | 80–150 | 88 | 33 | 42–50 | 18–35 | 6–14 | |
| Effect on fertility | Decrease | Decrease | Decrease | Decrease | Decrease | Increase | ND | Decrease‡ | No effect | |
| Used by | [1–6] | [2,7,18] | [8] | [6,7,9] | [10] | [11] | [3,4,12] | [13] | [14–17] |
ND, not determined.
[1]: (Lakowski & Hekimi, 1998), [2]: (Panowski et al., 2007), [3]: (Kaeberlein et al., 2006), [4]: (Lee et al., 2006), [5]: (Hansen et al., 2007), [6]: (Houthoofd et al., 2002b), [7]: (Houthoofd et al., 2003), [8]: (Bishop & Guarente, 2007), [9]: (Houthoofd et al., 2002a), [10]: (Szewczyk et al., 2006), [11]: (Hosono et al., 1989), [12]: (Steinkraus et al., 2008), [13]: (Greer et al., 2007), [14]: (Wood et al., 2004), [15]: (Viswanathan et al., 2005), [16]: (Bass et al., 2007), [17]: (Gruber et al., 2007), [18]: (Klass, 1977).
In addition to methods that restrict the diet, a number of chemical compounds have been proposed to act as ‘DR mimetics’, which extend lifespan without inducing the detrimental effects of restricting food (Ingram et al., 2006). For example, the natural polyphenol compound resveratrol has been proposed to act as a DR mimetic in yeast (Howitz et al., 2003), worms (Wood et al., 2004; Viswanathan et al., 2005; Gruber et al., 2007), flies (Wood et al., 2004), fish (Valenzano et al., 2006), and mice on a high fat diet (Baur et al., 2006), although resveratrol did not extend lifespan in flies in one study (Bass et al., 2007) and in mice on a normal diet (Pearson et al., 2008). While resveratrol and various DR regimens can significantly extend lifespan, whether they do so by universal, independent, or overlapping mechanisms is unknown.
A number of genes mediating longevity by different DR methods or by resveratrol have recently been uncovered in invertebrates (Lakowski & Hekimi, 1998; Wood et al., 2004; Hansen et al., 2005; Viswanathan et al., 2005; Wang & Tissenbaum, 2006; Bishop & Guarente, 2007; Greer et al., 2007; Hansen et al., 2007; Panowski et al., 2007; Steinkraus et al., 2008). The NAD-dependent deacetylase of the Sir2 family was one of the first genes identified to be necessary for longevity induced by various DR regimens and by resveratrol in yeast, worms, flies, and possibly mice (Lin et al., 2000; Rogina & Helfand, 2004; Wood et al., 2004; Chen et al., 2005; Viswanathan et al., 2005; Wang & Tissenbaum, 2006), although the importance of Sir2 in DR- or resveratrol-induced longevity is not always observed in these organisms (Kaeberlein et al., 2004; Kaeberlein et al., 2006; Lee et al., 2006; Bass et al., 2007; Hansen et al., 2007). Another pathway that was identified in yeast, worms, and flies, to mediate DR induced longevity is the amino-acid sensing TOR pathway (Kapahi et al., 2004; Kaeberlein et al., 2005; Hansen et al., 2007), although TOR is not always necessary for DR induced longevity in C. elegans (Henderson et al., 2006). In addition, a series of transcriptional regulators involved in the response to oxidative stress have recently been implicated in DR in C. elegans: the Forkhead transcription factor FoxA/pha-4 is necessary for longevity induced by the eat-2 mutation and by bDR (Panowski et al., 2007); the Nrf2 transcription factor skn-1 is necessary for lifespan extension by lDR (Bishop & Guarente, 2007); and the heat-shock transcription factor hsf-1 plays an important role in longevity triggered by DD (Steinkraus et al., 2008), though hsf-1 is dispensable for eat-2 induced lifespan extension (Hsu et al., 2003). Finally, clk-1, a gene encoding a mitochondrial protein involved in ubiquinone synthesis, also appears to be required for longevity induced by the eat-2 mutation in worms (Lakowski & Hekimi, 1998).
We have recently discovered that the low energy-sensing kinase AMPK/aak-2 is necessary for longevity induced by sDR in worms (Greer et al., 2007). AMPK can act upstream of the Forkhead transcription factor FoxO/daf-16 to extend lifespan, perhaps via direct phosphorylation (Greer et al., 2007). Like AMPK, FoxO is necessary for longevity induced by sDR (Greer et al., 2007). In contrast, neither AMPK nor FoxO are necessary for the longevity induced by eat-2 (Lakowski & Hekimi, 1998; Curtis et al., 2006). In addition, FoxO is not necessary for longevity induced by other DR methods (bDR, lDR, axenic medium, and DD) (Houthoofd et al., 2003; Kaeberlein et al., 2006; Lee et al., 2006; Bishop & Guarente, 2007; Panowski et al., 2007). Similarly, in Drosophila, FoxO is not absolutely necessary for DR-induced longevity (Giannakou et al., 2008; Min et al., 2008), although FoxO alters the optimal food concentration required for longevity (Clancy et al., 2002; Giannakou et al., 2008; Min et al., 2008). In mammals, the role of AMPK and FoxO in DR-induced longevity has not been examined yet. While a series of genes have been identified as playing important roles in longevity induced by different DR methods, the comparison of the importance of these genes in diverse DR regimens has not been performed. Identifying the different genetic pathways by which the various methods of restricting nutrients promote longevity is important for harnessing the full benefits of DR on lifespan.
Here we test whether distinct DR methods are mediated by specific or common genetic pathways. We find that while AMPK and FoxO are necessary for longevity induced by sDR and by peptone dilution in plates, these genes are not absolutely required for eat-2 and bDR to extend lifespan. Intriguingly, AMPK, but not FoxO, is necessary for the DR mimetic resveratrol to extend lifespan in worms. We then test whether sDR is mediated by genes that were previously found to mediate longevity by other DR methods or DR mimetics. We find that sir-2.1, pha-4, skn-1, and hsf-1 are all dispensable for sDR-induced lifespan extension, but that clk-1 is necessary for this regimen to extend lifespan. Finally, we show that sDR further enhances the lifespan of eat-2 mutant worms, indicating that these two DR methods act additively to promote longevity. Our results are compatible with a model in which sDR induces lifespan extension by a mechanism that is different from, but overlapping with, that of other DR methods. Understanding how different methods of DR induce lifespan extension is pivotal for the identification of all the components of the gene network that orchestrates maximal longevity extension in response to nutrient deprivation.
Results
AMPK/aak-2 and FoxO/daf-16 are necessary for sDR-induced lifespan extension across a gradient of bacteria on plates
Assessing lifespan at only two concentrations of food (ad libitum vs. DR) does not distinguish genes that are truly involved in mediating DR from genes that affect the optimal response to food concentrations required to elicit the DR effect (e.g. chico in flies) (Clancy et al., 2002). Therefore, to test if AMPK/aak-2 and FoxO/daf-16 mediated lifespan extension in response to DR or affected the optimal response to food concentration, we performed lifespan experiments using a gradient of bacterial concentrations. The lifespan extension of WT worms as a function of bacteria concentration follows a parabola-shaped curve (Fig. 1), with moderate reduction of food intake leading to beneficial effects on lifespan (DR) and severe reduction of food intake having detrimental effects leading to death (starvation) (Piper & Partridge, 2007; Mair & Dillin, 2008). In contrast, the lifespan of worms carrying a null mutation in the aak-2 or in the daf-16 gene (aak-2(ok524) and daf-16(mu86)) was never extended, regardless of the concentration of bacteria (Fig. 1A,B; Tables S1A and S1B in Supporting Information). These results indicate that AMPK/aak-2 and FoxO/daf-16 are necessary for lifespan extension in response to sDR and that mutations in these genes do not lead to a shift of the overall organismal dependence on food.
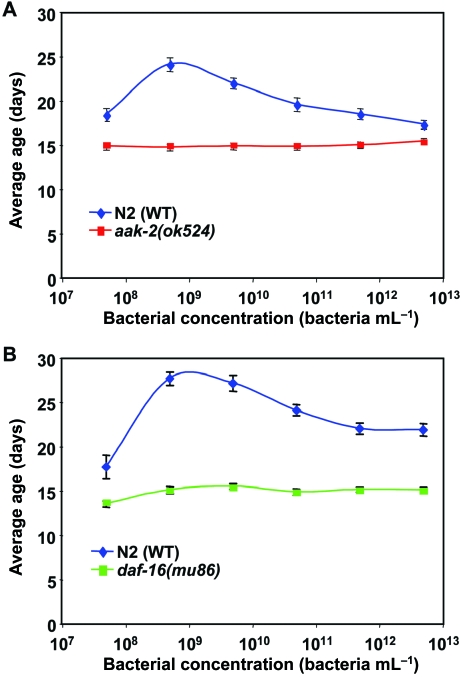
AMPK/aak-2 and FoxO/daf-16 are necessary for lifespan extension by sDR across a gradient of bacteria. (A) A serial dilution of bacteria on plates (5 × 1012 to 5 × 107 bacteria mL−1) extends WT (N2) worm lifespan but does not extend aak-2(ok524) mutant worm lifespan. Two-way anova revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was significantly different from that of aak-2(ok524) mutant worms (P < 0.0001). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S1A. (B) A serial dilution of bacteria on plates extends WT (N2) worm lifespan but does not extend daf-16(mu86) mutant worm lifespan. Two-way anova revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was significantly different from that of daf-16 (mu86) mutant worms (P < 0.0001). Mean, standard errors, and statistical analysis for one experiment performed in triplicate are presented in Table S1B.
The dependency of sDR on AMPK and FoxO was also observed with independent backcrossed worm strains (aak-2(rr48) for AMPK and daf-16(m26) or daf-16(mgDf50) for FoxO) (Fig. 5, data not shown), with bacterial strains from independent laboratories (data not shown), and in the presence or absence of FUdR, an inhibitor of worm reproduction (Figure S1; Table S1C). Together with our previous findings (Greer et al., 2007), these results indicate that AMPK and FoxO are necessary to mediate longevity induced by sDR, a regimen that restricts food intake in worms.
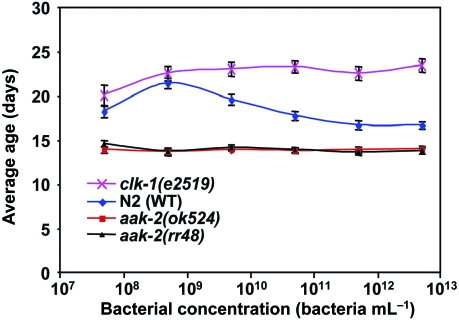
clk-1 is necessary for sDR to extend lifespan. A serial dilution of bacteria on plates extended WT (N2) worm lifespan (28.5%, P < 0.0001) but did not extend two independent aak-2 mutant strains, aak-2(rr48) (1.0%P = 0.5787) and aak-2(ok524) (–1.4%P = 0.7804), or clk-1(e2519) mutant worm lifespans (0%P = 0.6921). Two-way anova revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was significantly different from that of aak-2(ok524) mutant worms (P < 0.0001), aak-2(rr48) mutant worms (P < 0.0001), or clk-1(e2519) mutant worms (P < 0.0001). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S9.
AMPK and FoxO are necessary for the lifespan extension due to peptone dilution
We next asked how general the role of AMPK and FoxO was in longevity induced by different methods of restricting nutrients. Dilution of peptone in bacterial plates extends worm lifespan because it is considered to reduce bacterial growth on the plates (Hosono et al., 1989). We find that the dilution of peptone in bacterial plates extends lifespan in an AMPK/aak-2 and FoxO/daf-16 dependent manner (P < 0.0001 by two-way anova) (Fig. 2A; Table S2). This result indicates that AMPK and FoxO are also necessary for lifespan extension induced by another method of restricting nutrients.
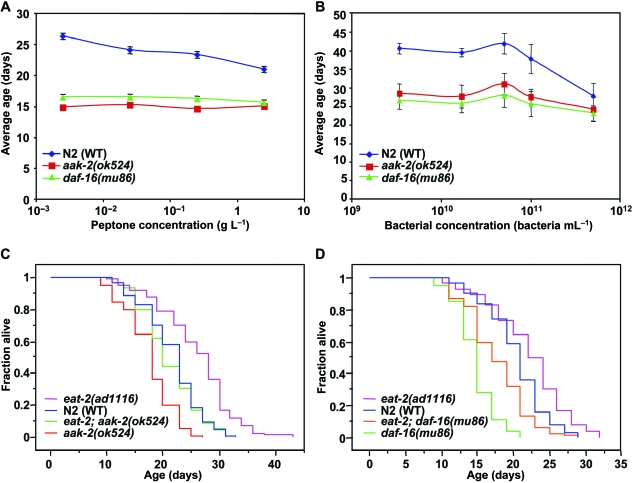
AMPK and FoxO mediate longevity induced by some but not all DR methods. (A) Longevity induced by dilution of peptone (DP) is dependent on AMPK/aak-2 and FoxO/daf-16. Dilution of peptone in the plates extends WT (N2) worm lifespan (25.4%, P < 0.0001) but does not extend aak-2(ok524) mutant lifespan (–1.0%, P = 0.6371) or daf-16(mu86) mutant lifespan (5.7%, P = 0.1402). Two-way anova revealed that the lifespan extension of WT (N2) worms across a peptone gradient was significantly different from that of aak-2(ok524) mutant worms (P < 0.0001) or daf-16(mu86) mutant worms (P < 0.0001). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S2. (B) AMPK/aak-2 and FoxO/daf-16 are not completely necessary for bDR lifespan extension. The average and SEM of three independent experiments indicates that bDR increases WT (N2), aak-2(ok524), and daf-16(mu86) mutant lifespan but appears to increase WT (N2) worm lifespan to a greater extent than aak-2(ok524) or daf-16(mu86) mutant worm lifespans. For each individual experiments, see Figure S2 and Table S3. (C) As previously reported (Curtis et al., 2006), the eat-2(ad1116) mutation extends WT (N2) worm lifespan (19.8%, P < 0.0001) and aak-2(ok524) mutant lifespan (19.4%, P < 0.0001). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S4A. (D) As previously reported (Lakowski & Hekimi, 1998), the eat-2(ad1116) mutation extends WT (N2) worm lifespan (12.9%, P < 0.0001) and daf-16(mu86) mutant lifespan (32.3%P < 0.0001). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S4B.
AMPK and FoxO are not completely necessary for bDR and eat-2 induced longevity
FoxO/daf-16 was shown not to be entirely necessary for longevity induced by the dilution of bacteria in liquid cultures (bDR) (Houthoofd et al., 2003; Panowski et al., 2007). However, the importance of AMPK in bDR-induced lifespan extension has never been examined. We tested whether AMPK and FoxO were required for lifespan extension in response to a gradient of bacteria concentrations in liquid culture. Combining the results of three independent experiments (Fig. 2B), we found that bDR significantly extended the lifespan of WT (N2), aak-2(ok524), and daf-16(mu86) mutant worms (P < 0.0001), indicating that AMPK and FoxO are dispensable for the entire lifespan extension induced by bDR. In two of these experiments, bDR extended WT (N2) lifespan to a larger extent than aak-2 and daf-16 mutant worm lifespan (P < 0.0001 by two-way anova) (Figure S2A and S2B; Table S3). In one experiment however, bDR extended the lifespan of WT (N2) worms to a similar extent as that of aak-2 and daf-16 mutant worms (P = 0.1528 and 0.6643 respectively by two-way anova) (Figure S2C; Table S3). Note that in one of the assays (Figure S2A), starvation was not reached, which may alter the interpretation of the experiment. However, when this assay was omitted for statistical analysis, we still found that bDR extended WT (N2) lifespan to a larger extent than aak-2 and daf-16 mutant worm lifespan (P < 0.001 by two-way anova). Although the parameters for these variations are unknown, these results suggest that AMPK and FoxO are dispensable for bDR to extend lifespan, but that they play a modulatory role in the extension of lifespan by this regimen.
Finally, as previously reported, we confirmed that AMPK/aak-2 and FoxO/daf-16 were completely dispensable for the lifespan extension of eat-2(ad1116) mutant worms, a genetic way to mimic DR (Lakowski & Hekimi, 1998; Curtis et al., 2006) (Fig. 2C,D; Table S4). Together, these results indicate that AMPK and FoxO are required for longevity induced by some (sDR and DP) but not all (bDR and eat-2) DR methods.
Resveratrol extends lifespan in an AMPK-dependent, but FoxO-independent, manner
Resveratrol extends lifespan in many species and has been suggested to act as a DR mimetic (Sinclair, 2005). Resveratrol has recently been found to activate AMPK in cultured cells and in mice (Baur et al., 2006; Zang et al., 2006; Dasgupta & Milbrandt, 2007; Hwang et al., 2007). We thus asked whether AMPK was necessary for increased longevity induced by resveratrol in C. elegans. Resveratrol induced a modest, but statistically significant, increase in WT (N2) worm lifespan. In contrast, resveratrol did not increase the lifespan of aak-2(ok524) mutant worms, suggesting that AMPK is necessary for the beneficial effects of resveratrol on lifespan (Fig. 3A; Table S5), as was proposed by (Bass et al., 2007). Similar to what was previously reported (Viswanathan et al., 2005), we found that resveratrol still extended the lifespan of daf-16(mu86) mutant worms (Fig. 3B; Table S5), indicating that resveratrol extends lifespan by eliciting an AMPK-dependent, FoxO-independent pathway. Therefore, activation of AMPK is not always coupled to that of FoxO, even though AMPK's ability to extend lifespan is dependent on the presence of FoxO (Greer et al., 2007). This result suggests that AMPK also regulates other substrates to extend lifespan, which is consistent with published findings (Apfeld et al., 2004; Narbonne & Roy, 2006). Therefore, these results suggest that different ways of manipulating nutrients and chemical compounds extend lifespan via independent and non-linear genetic pathways.
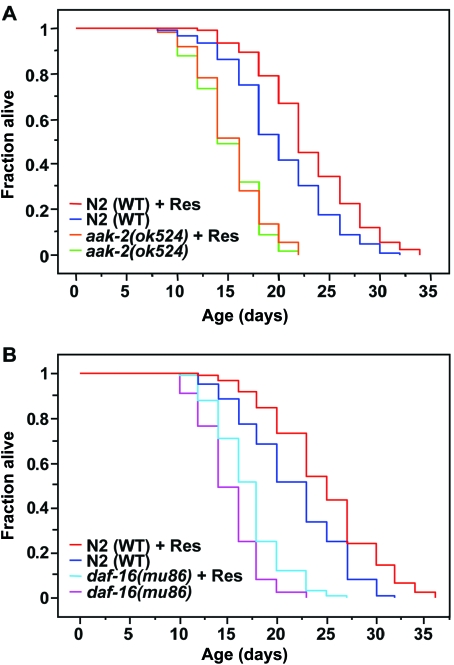
Resveratrol extends lifespan in an AMPK-dependent, but FoxO-independent, manner. (A) Resveratrol (100 µm) extended WT (N2) worm lifespan (14.2%, P = 0.0005), but did not significantly extend aak-2(ok524) mutant worm lifespan (2.2%P = 0.5485). Mean, standard errors, and statistical analysis for three independent experiments performed in triplicate are presented in Table S5. (B) Resveratrol (100 µm) extended both WT (N2) worm lifespan (14.6%P < 0.0001) and daf-16(mu86) mutant worm lifespan (13.7%P < 0.0001). Res, resveratrol. Mean, standard errors, and statistical analysis for one experiment performed in triplicate are presented in Table S5.
sir-2.1, pha-4, skn-1, and hsf-1 are not necessary for sDR-induced lifespan extension
Having shown that FoxO and AMPK were important for some, but not all, DR methods, we next tested if conversely, genes that have been implicated in lifespan extension by resveratrol or by other DR regimens were necessary for sDR to extend lifespan.
We first asked if sir-2.1, which encodes a Sirtuin family protein deacetylase, was required for lifespan extension in response to sDR. sir-2.1 has been found to mediate lifespan extension by resveratrol in some studies (Wood et al., 2004; Viswanathan et al., 2005), but not others (Bass et al., 2007). We used a mutant strain carrying a deletion in the sir-2.1 gene, which is predicted to be a null mutant (sir-2.1(ok434)) (Wang & Tissenbaum, 2006). We found that sDR increased the lifespan of both WT and sir-2.1 mutant worms to the same extent (Fig. 4A; Table S6). There was no statistically significant difference between the effects of sDR on the longevity of WT worms vs. sir-2.1 mutant worms (P = 0.1240 by two-way anova). These results indicate that sir-2.1 is not necessary for lifespan extension by sDR.
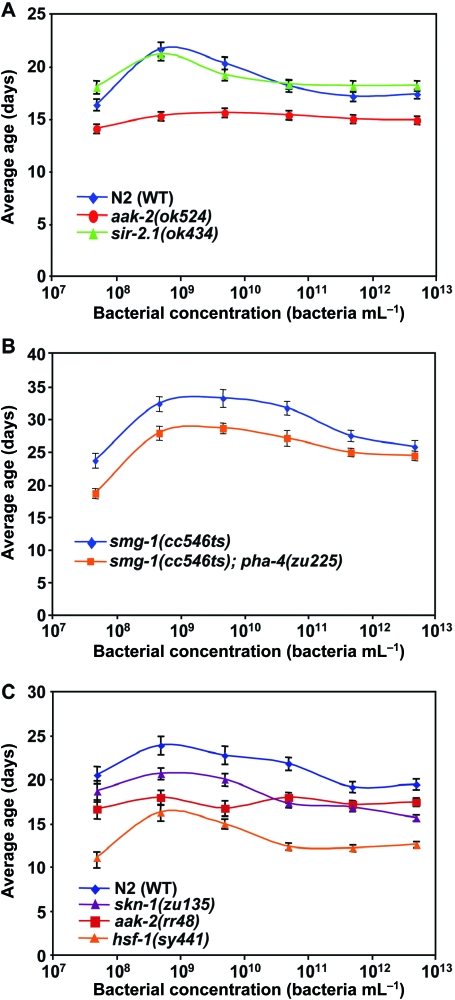
sir-2.1, FoxA/pha-4, skn-1 and hsf-1 are not entirely necessary for sDR to extend lifespan. (A) A serial dilution of bacteria on plates (5 × 1012 to 5 × 107 bacteria mL−1) extends WT (N2) (26.1%, P < 0.0001) and sir-2.1(ok434) (16.6%, P < 0.0001) mutant worm lifespan but does not extend aak-2(ok524) (1.3%, P = 0.6330) mutant worm lifespan. Two-way anova revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was significantly different from that of aak-2(ok524) mutant worms (P < 0.0001) but not statistically different from sir-2.1(ok434) mutant worms (P = 0.1240). Mean, standard errors, and statistical analysis for three independent experiments performed in triplicate are presented in Table S6. (B) A serial dilution of bacteria on plates extended smg-1(cc546ts) and smg-1(cc546ts); pha-4(zu225) mutant worms to a similar extend (P = 0.3724 by two-way anova). Note that this experiment was performed at 15 °C after worms reached adulthood. Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S7A. (C) A serial dilution of bacteria on plates extended WT (N2) (24.9%, P < 0.0001) worm lifespan, skn-1(zu135) (23.1%, P < 0.0001) mutant worm lifespan, and hsf-1(sy441) (33.5%, P < 0.0001) mutant worm lifespan but did not extend aak-2(rr48) (4.6%, P = 0.2566) mutant worm lifespan. Two-way anova revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was significantly different from that of aak-2(ok524) mutant worms (P < 0.0001) but not statistically different from skn-1(zu135) mutant worm lifespan (P = 0.5567) or hsf-1(sy441) mutant worm lifespan (P = 0.2843). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S8 and Table S9.
We then tested the importance of the Forkhead transcription factor FoxA/pha-4, a gene involved in eat-2 and bDR-induced longevity (Panowski et al., 2007), in lifespan extension induced by sDR. We used a temperature sensitive allele of FoxA/pha-4, smg-1(cc546ts); pha-4(zu225), and its control counterpart smg-1(cc546ts) (Gaudet & Mango, 2002). When shifted to the restrictive temperature smg-1(cc546ts); pha-4(zu225) display very little PHA-4 protein and the phenotype of smg-1(cc546ts); pha-4(zu225) is similar to that of the pha-4(q490) null mutant (Kaltenbach et al., 2005; Kiefer et al., 2007). We found that sDR extended lifespan of both the control strain (smg-1(cc546ts)) and the inducible pha-4 mutant strain (smg-1(cc546ts); pha-4(zu225)) (Fig. 4B; Table S7A) (P = 0.3724 by two-way anova). These results indicate that FoxA/pha-4 is not necessary for sDR induced lifespan extension. Consistent with this observation, we found that sDR still increased the lifespan of worms in which FoxA/pha-4 was knocked-down by RNAi, but did not extend the lifespan of worms in which FoxO/daf-16 was knocked-down (Figure S3; Table S7B). These findings suggest that FoxO, but not FoxA, is required for sDR-induced longevity.
We also examined if skn-1, a gene encoding a transcription factor necessary for lDR to extend lifespan (Bishop & Guarente, 2007), was required for longevity in response to sDR. We used a loss of function mutant strain of skn-1, skn-1(zu135), which displays a premature stop codon in all three isoforms of SKN-1 (a, b, and c) (Bishop & Guarente, 2007; Tullet et al., 2008). We showed that sDR extended lifespan of WT worms and skn-1(zu135) mutant worms to a similar extent (P = 0.5567 by two-way anova). Although it is formally possible that some SKN-1 activity remains in the skn-1(zu135) mutant strain, these findings nevertheless suggest that skn-1 is not necessary for sDR-induced longevity (Fig. 4C; Table S8).
Finally, we asked if hsf-1, which encodes a heat-shock responsive transcription factor involved in longevity in response to DD (Steinkraus et al., 2008), was necessary for sDR-induced lifespan extension. We used a mutant strain of hsf-1 (hsf-1(sy441)) that contains a premature stop codon that eliminates the transactivation domain of HSF-1 and is likely to be a null mutant (Hajdu-Cronin et al., 2004). We found that sDR still extended the lifespan in hsf-1(sy441) mutant worms similarly to WT worms (P = 0.2843 by two-way anova), indicating that hsf-1 is not necessary for sDR-induced longevity (Fig. 4C; Table S9).
Together, these data indicate that four genes (sir-2.1, pha-4, skn-1, and hsf-1) that have been previously implicated in longevity in response to a variety of DR methods and DR mimetics do not mediate lifespan extension by sDR. These findings further corroborate the observation that different DR regimens evoke independent pathways.
clk-1 is necessary for sDR-induced lifespan extension
The clk-1 gene encodes a demethoxyubiquinone hydroxylase that is necessary for the biosynthesis of ubiquinone, a component of the electron transport chain (Ewbank et al., 1997; Miyadera et al., 2001). clk-1 mutant worms live longer than their WT counterparts (Lakowski & Hekimi, 1996) and their long lifespan is not further extended by the eat-2 mutation (Lakowski & Hekimi, 1998), suggesting that clk-1 is necessary for eat-2 induced lifespan extension. Although the clk-1 allele, clk-1(e2519), is unlikely to be a null mutant (Lakowski & Hekimi, 1996), we tested if clk-1 was important for sDR-induced lifespan extension. We found that clk-1(e2519) mutant worms, similarly to aak-2(ok524) and aak-2(rr48) mutant worms, no longer responded to sDR (Fig. 5; Table S9). These results suggest that clk-1 is necessary for sDR-induced longevity and are compatible with the observation that clk-1 longevity like sDR-induced lifespan is dependent on daf-16. Although the interpretation of these results is difficult because of the lack of a null allele for clk-1 (Gems et al., 2002), clk-1 may mediate two independent methods of DR, eat-2 and sDR. Thus, in addition to the genes that are specific to DR methods, there may also exist overlapping mechanisms underlying DR-induced longevity.
The effects of sDR and eat-2 on lifespan are additive
The observation that sDR is mediated by AMPK, FoxO, and clk-1 whereas eat-2 is mediated by FoxA and clk-1, raised two possibilities: (i) clk-1 is a common mechanism between both methods of DR but each method also triggers specific pathways in parallel; and (ii) each DR regimen is sensed by different pathways (e.g. by FoxO vs. FoxA), which both converge on clk-1. To distinguish between these two possibilities and to test whether sDR and eat-2 had additive effects on longevity, we tested the combined effect of sDR and eat-2 on lifespan. We found that sDR further extended the long lifespan of eat-2 mutant worms (Fig. 6, Table S4). Thus, both DR regimens are additive and can extend lifespan by up to 57% when combined. Although the eat-2 mutation is not a null mutation, which renders the interpretation of these experiments more difficult, these findings also suggest that eat-2 and sDR evoke mostly independent, though overlapping, pathways to extend lifespan.
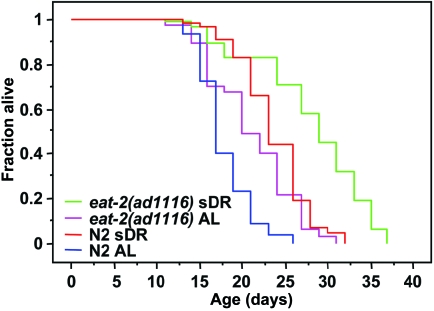
sDR and eat-2 have an additive effect on lifespan. sDR extended WT (N2) worm lifespan (18.2%P < 0.0001) and eat-2(ad1116) mutant worm lifespan (18.0%P < 0.0001) to the same extent. sDR: 5 × 108 bacteria mL−1 and AL (ad libitum): 5 × 1011 bacteria mL−1. Mean, standard errors, and statistical analysis for four independent experiments performed in triplicate are presented in Table S4.
Discussion
In this study, we performed a side-by-side comparison of the role of different genes in lifespan extension elicited by a variety of DR regimens. Our results uncover the importance of the low energy-sensing protein kinase, AMPK, in longevity due to some forms of DR and to resveratrol, a compound that extends lifespan and mimics some aspects of DR in many species. In addition, our findings provide further evidence that, contrary to the assumption that DR is independent of the insulin–FoxO pathway in invertebrates, the FoxO transcription factor daf-16 actually plays a role in DR-induced longevity, depending on how DR is elicited (Fig. 7A). Importantly, our results show that DR is not a uniform condition that triggers a universal and linear genetic pathway. Rather, diverse DR regimens evoke mostly independent genetic pathways, depending on how DR is achieved (Fig. 7B). Identifying the specific genes that mediate DR-induced longevity in C. elegans is likely to have important implications for other species, including yeast, flies, and mammals, because of the conservation of the genes studied and because of the existence of different DR regimens in these other species as well. Understanding the genetic network by which different DR methods extend lifespan should also help harness the effects of this environmental intervention on lifespan and healthspan and will be particularly important to achieve the full benefits of DR.
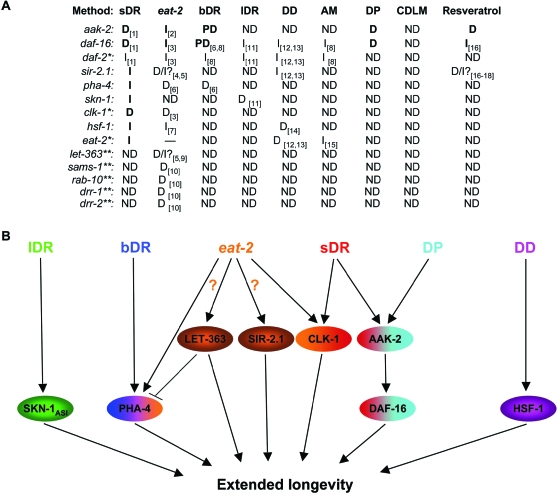
Different DR methods. (A) Table summarizing genes that have been tested for specific DR methods. Question marks indicate conflicting reports in the literature. sDR, ‘solid DR’ (Greer et al., 2007); eat-2 (Lakowski & Hekimi, 1998); bDR, liquid DR (Klass, 1977; Panowski et al., 2007); lDR, Liquid DR (Bishop & Guarente, 2007); DD, dietary deprivation (Kaeberlein et al., 2006; Lee et al., 2006); AM, axenic medium (Houthoofd et al., 2002a; Houthoofd et al., 2002b); DP, dilution of peptone in plates (Hosono et al., 1989); CDLM, chemically defined liquid media (Szewczyk et al., 2006). D, dependent; PD, partially dependent; I, independent; ND, not determined. *Not a null mutant, making results more difficult to interpret. **Experiments were performed with RNAi, making results more difficult to interpret. let-363: C. elegans TOR gene mutant (Henderson et al., 2006; Hansen et al., 2007). [1]: (Greer et al., 2007), [2]: (Curtis et al., 2006), [3]: (Lakowski & Hekimi, 1998), [4]: (Wang & Tissenbaum, 2006), [5]: (Hansen et al., 2007), [6]: (Panowski et al., 2007), [7]: (Hsu et al., 2003), [8]: (Houthoofd et al., 2003) [9]: (Henderson et al., 2006), [10]: (Hansen et al., 2005), [11]: (Bishop & Guarente, 2007), [12]: (Kaeberlein et al., 2006), [13]: (Lee et al., 2006), [14]: (Steinkraus et al., 2008), [15]: (Houthoofd et al., 2002b), [16]: (Viswanathan et al., 2005), [17]: (Wood et al., 2004), [18]: (Bass et al., 2007).(B) Different methods of DR activate distinct signaling pathways. Displayed are molecules that have been shown to play a role in mediating the longevity extension effects of DR methods. Question marks indicate conflicting reports (Henderson et al., 2006; Wang & Tissenbaum, 2006; Hansen et al., 2007). LET-363: C. elegans TOR protein.
Parameters that may affect the mechanisms of DR-induced longevity
Different DR regimens may evoke distinct genetic pathways because some nutrients may be more limiting than others depending on the DR method. For example, sDR might reduce carbohydrates more prominently than amino-acids, which would render it dependent on AMPK and FoxO, while other methods (eat-2, bDR) may reduce amino-acids more readily, which would evoke TOR, a well-known amino-acid responsive pathway (Avruch et al., 2006), and the FoxA/pha-4 transcription factor, which has recently been found to be downstream of TOR (Sheaffer et al., 2008). DR could also trigger different genetic mechanisms depending on the time at which DR is initiated or the tissues/cells by which it is sensed.
Alternatively, different DR regimens may evoke independent pathways because the way in which DR is initiated may trigger other non-DR signaling pathways that in turn have beneficial effects on lifespan. For example, diluting bacteria in sDR might also lead to the reduction in bacterial pathogenicity, which could in turn affect the insulin-FoxO pathway. However, if pathogenicity were the sole cause for premature death, concentrating bacteria above the ad libitum concentration would be detrimental, which we have found not to be the case. We have also shown that sDR is not simply due to a reduction in bacterial pathogenicity because sDR reduces worm fertility (data not shown) and because sDR performed with UV-killed bacteria also extends lifespan (Greer et al., 2007). The way sDR is achieved (by adding diluted amounts of E. coli every 2 days) may induce cycles of feeding and fasting that may cause this method to be AMPK and FoxO dependent whereas other DR methods (eat-2) maintain a more constant bacteria concentration throughout lifespan. sDR may also induce a mild oxidative stress response that could trigger the activation of the AMPK and FoxO pathways, as these pathways are known to transduce stress stimuli (Henderson & Johnson, 2001; Apfeld et al., 2004; Schulz et al., 2007; Lee et al., 2008). Although such stress may be part of DR itself (see the ‘hormesis theory’ of DR (Sinclair, 2005)), it is possible that the other DR methods do not require oxidative stress to extend lifespan.
Conversely, other methods of inducing DR could trigger non-DR signaling pathways. For example, the eat-2 mutation, which affects the acetylcholine receptor, may have other effects in addition to reducing pharyngeal pumping. Liquid methods of DR may also evoke other parameters (e.g. altered oxygen consumption (Honda et al., 1993) or forced exercise (Retzlaff et al., 1966) since worms actively swim in liquid) that may interact with reduction in nutrients to result in lifespan extension in a manner that is dependent on specific genes. Identifying the parameters that are embedded in or interact with DR methods will be important to gain complete insight into the mechanisms by which DR extends lifespan.
Mechanisms of DR in other species
An important question is whether the genetic mechanisms identified to regulate longevity in response to DR in C. elegans are conserved between species. The existence of different DR regimens that extend lifespan in yeast, flies, and mice raises the possibility that some of the mechanisms sensing the restriction of specific nutrients are conserved throughout evolution.
For example, in flies, a number of different methods of restricting diet extend lifespan (for a comprehensive review see (Piper & Partridge, 2007)). Two methods of DR in flies, dilution of yeast (Min et al., 2008) and dilution of both sugar and yeast (Giannakou et al., 2008) have been found to be independent of the Drosophila FoxO transcription factor, dFOXO, although dFOXO modifies the response to DR (Giannakou et al., 2008). While the role of AMPK in DR has not been tested yet, the histone deacetylases Rpd3 and Sir2, and the tumor suppressors Dmp53 and Tsc2, a member of the TOR pathway, have all been shown to be necessary for lifespan extension in different methods of DR in flies (Rogina et al., 2002; Kapahi et al., 2004; Rogina & Helfand, 2004; Bauer et al., 2005). The main method in which DR is initiated in flies, which reduces amino-acids, may primarily evoke the TOR and FoxA pathway rather than the AMPK or FoxO pathway (Kapahi et al., 2004).
In mice, reduction of the total amount of food which mice receive every day (Weindruch et al., 1986) or alternating days of feeding and fasting (EOD) both extend lifespan (Goodrick et al., 1990). The specific genetic components involved in DR in mice have not been as extensively analyzed yet, although the deletion of Sirt1, the mouse Sir2 ortholog, abrogates the beneficial effect of DR on behavioral activity (Chen et al., 2005) and mice expressing additionally copies of the Sirt1 gene have metabolic parameters similar to those induced by DR (Bordone et al., 2007). A reduction of the protein concentration (Goodrick, 1978) or of the amount of methionine in the diet also extend mouse lifespan (Orentreich et al., 1993), raising the possibility that the TOR pathway might be critical in longevity induced by these methods. While the importance of AMPK or FoxO in longevity induced by DR in mammals is still unknown, emerging evidence suggests that these pathways play some role in DR. First, EOD and a 40% restriction of food activate AMPK in the liver of rats (Pallottini et al., 2004). Short term DR (60% reduction of food for 5 days) also activated AMPK in the hippocampus of mice (Dagon et al., 2005). Second, the lifespan of mice that are deficient for the growth hormone receptor is not extended by a 30% reduction of food (Bonkowski et al., 2006). The deficiency in growth hormone receptor is thought to act via a reduction in IGF-1 and insulin levels, which would lead to FoxO activation. These observations raise the possibility that FoxO may mediate longevity in response to DR in mice.
Role of AMPK in longevity induced by ‘DR mimetics’
Chemical compounds that mimic some of the beneficial effects of DR include 2-deoxyglucose (2DG) (Roth et al., 2001), metformin (Dhahbi et al., 2005), and resveratrol (Howitz et al., 2003). Metformin decreases the tumor incidence and extends the lifespan of tumor prone HER-2/neu transgenic mice (Anisimov et al., 2005). Resveratrol extends the lifespan of yeast (Howitz et al., 2003), worms (Wood et al., 2004; Viswanathan et al., 2005; Gruber et al., 2007), flies (Wood et al., 2004), fish (Valenzano et al., 2006) and mice on a high fat diet (Baur et al., 2006), although resveratrol does not extend lifespan under all circumstances (Bass et al., 2007; Pearson et al., 2008). Intriguingly, 2DG, resveratrol, and metformin have all been shown to activate AMPK (Baur et al., 2006; Zang et al., 2006; Dasgupta & Milbrandt, 2007; Hardie, 2007; Hwang et al., 2007). Resveratrol has been proposed to act through sir-2.1 (Wood et al., 2004; Viswanathan et al., 2005), although this was not found in all studies (Bass et al., 2007), and AMPK/aak-2 (this study) to affect lifespan, while 2DG was shown to act through AMPK/aak-2, but not sir-2.1, in C. elegans (Schulz et al., 2007). Thus, AMPK may play a pivotal role in mediating the lifespan extension induced by DR mimetics. However, the ‘DR mimetics’ have less of an impact on lifespan than DR itself. This observation suggests that multiple pathways need to be activated concomitantly to achieve optimal effects on lifespan and healthspan through chemical treatments.
A gene network mediating longevity in response to DR
The pathways that regulate aging in response to DR are unlikely to be linear. Our findings indicate that resveratrol requires AMPK, but not FoxO, to extend lifespan, yet FoxO is necessary to mediate AMPK's effects on lifespan (Greer et al., 2007). Similarly, lifespan extension due to resveratrol is thought to be sir-2.1 dependent (although not in all studies (Bass et al., 2007)) and daf-16 independent (Wood et al., 2004; Viswanathan et al., 2005), yet the lifespan extension in response to sir-2.1 overexpression is daf-16 dependent (Tissenbaum & Guarente, 2001). These observations suggest that genetic pathways branching downstream of Sir-2 and AMPK probably exist and that negative and positive feedback mechanisms may also be involved. For example, AMPK substrates other than FoxO may contribute to AMPK lifespan extension, as proposed (Apfeld et al., 2004; Narbonne & Roy, 2006). The identification of additional AMPK substrates will be important for understanding the mechanisms of AMPK action on longevity. Our study may provide an explanation for the controversies regarding the implication of daf-16 downstream of pro-longevity genes (Lakowski & Hekimi, 1996; Murakami & Johnson, 1996; Braeckman et al., 1999; Henderson et al., 2006; Wang & Tissenbaum, 2006; Hansen et al., 2007). Lifespan assay protocols might inadvertently induce sDR, which would render lifespan daf-16 dependent. Thus, maintaining worms on ad libitum concentrations of food throughout lifespan experiments is likely crucial to prevent confounding variables, which could influence whether longevity is dependent on daf-16.
Finally, it is interesting to note that the genes involved in mediating different DR methods encode proteins from similar families (the Forkhead transcription factors FoxO and FoxA or the nutrient sensing kinases AMPK and TOR). In addition, the pathways mediating DR-induced longevity have already been shown to cross-talk extensively, at least in mammalian systems (e.g. AMPK and mTOR, Sir2 and AMPK) (Greer & Brunet, 2008). These observations underscore that DR-induced longevity is likely to be mediated by a network of genes rather than by linear pathways. Manipulating more than one ‘node’ in this network may allow additive or even synergistic lifespan and healthspan benefits.
Materials and methods
Worm strains and RNA interference
N2 and daf-16(mu86) strains were a kind gift from Dr Man-Wah Tan. The aak-2(ok524), aak-2(rr48), sir-2.1(ok434), skn-1(zu135), hsf-1(sy441), clk-1(e2519), daf-16(mgDf50), daf-16(m26), and daf-16(mu86), strains were provided by the Caenorhabditis Genetics Center. The eat-2(ad1116), daf-16(mu86); eat-2(ad1116), N2, and a three times backcrossed aak-2(ok524) strains were generously provided by Cynthia Kenyon. The smg-1(cc546ts) and smg-1(cc546ts); pha-4(zu225) strains were generously provided by Susan Mango. The aak-2(ok524) strain was crossed to eat-2(ad1116) to generate the double mutant strain aak-2(ok524); eat-2(ad1116). Unless otherwise noted, aak-2(ok524) and daf-16(mu86) strains were used. HT115 (DE3) bacteria transformed with vectors expressing dsRNA of the genes of interest were obtained from the Ahringer library (a gift from Dr M.-W. Tan) and were grown at 37 °C and seeded onto standard nematode growth medium (NGM) plates containing Ampicillin (100 µg mL−1) and IPTG (0.4 mm). Adult worms were placed on standard NGM plates and removed after 4–6 h to obtain synchronized populations of worms. L1 or L4 worms obtained from these synchronized populations were placed on NGM plates containing Ampicillin (100 µg mL−1) and IPTG (0.4 mm) seeded with the respective bacteria. Each vector was sequenced to verify the presence of the appropriate gene of interest.
Lifespan assays
Worm lifespan assays were performed at 20 °C unless noted differently. Worm populations were synchronized by placing young adult worms on NGM plates seeded with the E. coli strain OP50–1 (unless otherwise noted) for 4–6 h and then removed. OP50–1 is a strain derived from OP50 that contains a streptomycine resistance gene (Kawli & Tan, 2008). The hatching day was counted as day 1 for all lifespan measurements. Worms were changed every other day to new plates to eliminate confounding progeny, and were marked as dead or alive. Worms were scored as dead if they did not respond to repeated prods with a platinum pick. Worms were censored if they crawled off the plate or died from vulval bursting. For each lifespan assay, 90 worms per condition were used in three plates (30 worms per plate). The data were plotted in StatView 5.0.1 using the Kaplan–Meier Survival curves and statistical significance was determined by Log-rank (Mantel-Cox) tests. Lifespan assays were repeated at least once unless otherwise mentioned. Representative Kaplan-Meier survival curves are shown in the figures. For lifespan assays performed on a gradient of bacterial concentrations mean and standard error values were taken from Kaplan–Meier Survival curves plotted in StatView 5.0.1 and plotted in Excel. Representative curves are presented unless noted otherwise. For smg-1 and smg-1; pha-4 lifespan assays, worms were grown at permissive temperature 24 °C until the first day of adulthood when they were switched to 15 °C which allows for the degradation of pha-4(zu225) (Gaudet & Mango, 2002).
DR assays
OP50–1 bacteria were serially diluted from 5 × 1012 to 5 × 104 bacteria mL−1. Bacteria were resuspended in S Medium to inhibit bacterial growth. Adult worms were placed on these various concentrations of bacteria starting at day 7 of life (day 4 of adulthood). Worms placed on a gradient of bacterial concentrations ranging from 5 × 104 to 5 × 107 bacteria mL−1 died 2 days after being placed on these extreme DR diets. For specific assays, sDR was considered 5 × 108 bacteria mL−1 and ad libitum was 5 × 1011 bacteria mL−1 (Greer et al., 2007).
Peptone dilution assays
Assays were performed as in (Hosono et al., 1989). Synchronized populations of worms were obtained by placing adult worms on plates with decreasing concentrations of peptone (from 2.5 to .0025 g L−1 with 150 µL of 5 × 1012 bacteria mL−1 seeded on each plate) and removing the worms after 4–6 h. Worms were switched every other day to fresh plates and were scored as alive as described above.
Liquid DR assays
Assays were performed as in (Panowski et al., 2007). Briefly worms were grown until day 1 of adulthood on NGM plates seeded with 150 µL of 5 × 1011 bacteria mL−1. They were then transferred for 1 day to NGM plates with FUdR (100 mg L−1) seeded with 150 µL of 5 × 1011 bacteria mL−1. Approximately 20 worms were then placed in each well of a 12-well plate with 1 mL of S-basal supplemented with cholesterol (5 mg L−1), Ampicillin (50 µg L−1), Kanamycin (10 µg L−1), Tetracycline (1 µg L−1) and FUdR (100 mg L−1) containing OP50–1 bacteria at various concentrations (from 3.33 × 109 to 5 × 1011 bacteria mL−1). Approximately 90 total worms were used for each condition (4 wells containing approximately 22 worms per well). Plates were gently shaken at 20 °C and worms were scored as alive as described before. Live worms were switched to fresh liquid medium containing the appropriate dilution of bacteria every third day.
Statistical analysis
Statistical analysis of lifespan was performed on Kaplan–Meier survival curves in StatView 5.0.1. For statistical comparison of independent replicates, Fischer's combined probability tests were performed. To compare the interaction between genotype and food concentration, two-way anova tests were performed in Prism 4.0 c using the mean and standard error values obtained from the Kaplan–Meier survival curves. To compare the interaction between genotype and food concentration in an independent manner, regression analyses were also performed on raw lifespan data using a Cox proportional hazard analysis in r 2.7.1. The values from the Fisher's combined probability tests, two-way anova, and Cox proportional hazard analysis are included in the supplemental tables.
Acknowledgments
We thank Andy Dillin, Cynthia Kenyon, Susan Mango, and Man-Wah Tan for generously sharing protocols, reagents, and for helpful discussion. We thank Keith Blackwell for helpful suggestions. We thank Alex McMillan, Art Owen, and Dario Valenzano for help with statistical analysis. We thank Max Banko and Hirokazu Fukui for experimental help. We thank members of the Brunet laboratory as well as Paul Greer, Stuart Kim, Judy Lieberman, and Adolfo Sanchez-Blanco for critically reading the manuscript. We are grateful to Theresa Stiernagle and the Caenorhabditis Genetics Center funded by the National Center for Research Resources of the NIH for providing worm strains. This research was supported by an NIH grant (NIA AG026648–01), an American Institute for Cancer Research grant, and by a gift from the Glenn Foundation to A.B. E.L.G. was supported by a PHS grant (CA 09302) from the NCI and by an NSF graduate fellowship.
Additional Note
Since this manuscript was accepted, another method of DR in C. elegans, termed IF for intermittent fasting, has been reported. This DR method is dependent on RHEB-1 and FoxO/daf-16, but independent of AMPK/aak-2 (Honjoh S, Yamamoto T, Uno M, Nishida E (2009) Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 457, 726–730.)
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1 Addition of FUdR does not alter the dependency of sDR-induced lifespan extension on AMPK/aak-2 or FoxO/daf-16. sDR extended WT (N2) worm lifespan (31.0% P < 0.0001) but did not significantly extend aak-2(ok524) mutant worm lifespan (2.3% P = 0.3295) or daf-16(mu86) mutant worm lifespan (4.3% P = 0.0624) on plates with 100 mg L-1 FUdR. Mean, standard errors, and statistical analysis for one experiment performed in triplicate are presented in Table S1C.
Figure S2 AMPK and FoxO are not completely necessary for bDR lifespan extension. (A) In one experiment, bDR extended WT (N2) worm lifespan (101%, P < 0.0001), aak-2(ok524) mutant lifespan (47.3%, P < 0.0001), and daf-16(mu86) mutant lifespan (39.6%, P < 0.0001). Two-way ANOVA revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was significantly different from that of aak-2(ok524) mutant worms (P < 0.0001) or daf-16(mu86) mutant worms (P < 0.0001). (B) In a second experiment, bDR extended WT (N2) worm lifespan (59.1%, P < 0.0001), aak-2(ok524) mutant lifespan (27.4%, P<0.0001), and daf-16(mu86) mutant lifespan (14.6%, P = 0.0120). Two-way ANOVA revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was significantly different from that of aak-2(ok524) mutant worms (P < 0.0001) or daf-16(mu86) mutant worms (P < 0.0001). (C) In a third experiment, bDR extended WT (N2) worm lifespan (32.3%, P<0.0001), aak-2(ok524) mutant lifespan (20.8%, P<0.0001), and daf-16(mu86) mutant lifespan (26.7%,Two-way ANOVA revealed that the lifespan extension of WT (N2) worms across a bacterial gradient was not significantly different from that of aak-2(ok524) mutant worms (P = 0.1528) and daf-16(mu86) mutant worms (P = 0.6643). Mean, standard errors, and statistical analysis for these experiments performed in quadruplicates are presented in Table S3.
Figure S3 FoxA/pha-4 is not necessary for sDR to extend lifespan. (A) sDR extended the lifespan of WT (N2) worms treated with empty vector RNAi (E.V.) (34.7% P < 0.0001) or pha-4 RNAi (36.7% P < 0.0001) initiated at L1, but not the lifespan of worms treated with daf-16 RNAi (7.0 %P = 0.1562). The efficacy of pha-4 RNAi was confirmed by its ability to decrease worm fertility, consistent with its known role in pharyngeal organogenesis (Mango et al., 1994), and its effects on lifespan (Panowski et al., 2007). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S7B. (B) sDR extended the lifespan of WT (N2) worms treated with empty vector RNAi (E.V.) (30.8% P < 0.0001) or pha-4 RNAi (26.4% P < 0.0001) initiated at L4 but not the lifespan of worms treated with daf-16 RNAi (6.1 P = 0.1801). Mean, standard errors, and statistical analysis for two independent experiments performed in triplicate are presented in Table S7B.
Table S1 AMPK/aak-2 and FoxO/daf-16 are necessary for lifespan extension by sDR across a gradient of bacteria. (A) A serial dilution of bacteria extends WT (N2) worm lifespan but does not extend aak-2(ok524) mutant worm lifespan. Experiment #1 is displayed in Figure 1A. Combined p values were calculated using Fisher's combined probability test. (B) A serial dilution of bacteria extends WT (N2) worm lifespan but does not extend daf-16 (mu86) mutant worm lifespan. This experiment is displayed in Figure 1B. (C) Addition of FUdR does not alter the dependency of sDR-induced lifespan extension on AMPK/aak-2 or FoxO/daf- 16. This experiment is displayed in Figure S1. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from triplicate samples of 30 worms each. n, number of observed dead worms/number of total worms.
Table S2 Dilution of peptone (DP) extends lifespan in an AMPK/aak-2 and FoxO/daf-16 dependent manner. Experiment #2 is displayed in Figure 2A. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from triplicate samples of 30 worms each. n, number of observed dead worms/number of total worms. Combined P-values were calculated using Fisher#x0027;s combined probability test.
Table S3 bDR increase in worm lifespan is partially dependent on AMPK/aak-2 and FoxO/daf-16. The average of these three experiments is displayed in Figure 2B. Each experiment is displayed in Figure S2. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from quadruplicate samples of approximately 22 worms each. n, number of observed dead worms/number of total worms. Combined P-values were calculated using Fisher's combined probability test.
Table S4 (A) eat-2(ad1116) induced lifespan extension is independent of AMPK/aak-2. Experiment #1 is displayed in Figure 2C. Experiment #2 is displayed in Figure 6. (B) eat-2(ad1116) induced lifespan extension is independent of FoxO/daf-16. Experiment #2 is displayed in Figure 2D. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from triplicate samples of 30 worms each. n, number of observed dead worms/number of total worms. Combined Pvalues were calculated using Fisher'427s combined probability test.
Table S5 Resveratrol increases worm lifespan in an AMPK/aak-2 dependent but FoxO/daf-16 independent manner.Experiment #2 is displayed in Figure 3A. Experiment #3 is displayed in Figure 3B. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from triplicate samples of 30 worms each. n, number of observed dead worms/number of total worms. Combined P-values were calculated using Fisher's combined probability test.
Table S6 sDR increases worm lifespan in an AMPK/aak-2 dependent and SIR2/sir-2.1 independent manner. Experiment #3 is displayed in Figure 4A. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from triplicate samples of 30 worms each. n, number of observed dead worms/ number of total worms. Combined P-values were calculated using Fisher's combined probability test.
Table S7 sDR increases worm lifespan in a FoxA/pha-4 independent manner. A) smg-1 and smg-1; pha-4 worms were grown at permissive temperature (24 °C) until the first day of adulthood when they were switched to 15 °C. Experiment #1 is displayed in Figure 4B. B) sDR extends the lifespan of WT(N2) worms treated with empty vector RNAi (E.V.) or pha-4 RNAi bute not daf-16 RNAi initiated at larval stage L1 or larval stage L4. L1 Experiment #1 is displayed in Figure S3A. L4 Experiment #2 is displayed in Figure S3B. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from triplicate samples of 30 worms each. n, number of observed dead worms/number of total worms. Combined P-values were calculated using Fisher's combined probability test.
Table S8 sDR increases worm lifespan in an AMPK/aak-2 dependent, skn-1 independent manner. Experiment #2 is displayed in Figure 4C. The mean lifespan values were calculated by a log-rank (Mantel-Cox) statistical test from triplicate samples of 30 worms each. n, number of observed dead worms/number of total worms. Combined P-values were calculated using Fisher's combined probability test.
Table S9 sDR increases worm lifespan in an AMPK/aak-2 and clk-1 dependent, hsf-1 independent manner. Experiment #2 is displayed in Figure 4C. Experiment #1 is displayed in Figure 5. The mean lifespan values were calculated by a log-rank (Mantel- Cox) statistical test from triplicate samples of 30 worms each. n: number of observed dead worms/number of total worms. Combined P-values were calculated using Fisher's combined probability test. Note that experiment #2 of Table S9 was performed at the same time as experiment #2 of Table S8.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Anisimov VN, Berstein LM, Egormin PA, Piskunova TS, Popovich IG, Zabezhinski MA, Kovalenko IG, Poroshina TE, Semenchenko AV, Provinciali M, Re F, Franceschi C. Effect of metformin on life span and on the development of spontaneous mammary tumors in HER-2/neu transgenic mice. Exp. Gerontol. 2005;40:685–693. [Abstract] [Google Scholar]
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. [Europe PMC free article] [Abstract] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. [Europe PMC free article] [Abstract] [Google Scholar]
- Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. [Abstract] [Google Scholar]
- Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech. Ageing. Dev. 2007;128:546–552. [Abstract] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr. Biol. 2005;15:2063–2068. [Abstract] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. [Abstract] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. [Abstract] [Google Scholar]
- Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc. Natl. Acad. Sci. USA. 2006;103:7901–7905. [Europe PMC free article] [Abstract] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. [Abstract] [Google Scholar]
- Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr. Biol. 1999;9:493–496. [Abstract] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. [Abstract] [Google Scholar]
- Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. [Abstract] [Google Scholar]
- Curtis R, O’Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–126. [Abstract] [Google Scholar]
- Dagon Y, Avraham Y, Magen I, Gertler A, Ben-Hur T, Berry EM. Nutritional status, cognition, and survival: a new role for leptin and AMP kinase. J. Biol. Chem. 2005;280:42142–42148. [Abstract] [Google Scholar]
- Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl. Acad. Sci. USA. 2007;104:7217–7222. [Europe PMC free article] [Abstract] [Google Scholar]
- Dhahbi JM, Mote PL, Fahy GM, Spindler SR. Identification of potential caloric restriction mimetics by microarray profiling. Physiol. Genomics. 2005;23:343–350. [Abstract] [Google Scholar]
- Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cell. Mol. Life Sci. 2007;64:752–767. [Abstract] [Google Scholar]
- Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, Hekimi S. Structural and functional conservation of the Caenorhabditis elegans timing gene. Clk-1. Science. 1997;275:980–983. [Abstract] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. [Abstract] [Google Scholar]
- Gems D, Pletcher S, Partridge L. Interpreting interactions between treatments that slow aging. Aging Cell. 2002;1:1–9. [Abstract] [Google Scholar]
- Giannakou ME, Goss M, Partridge L. Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: not required, but its activity modulates the response. Aging Cell. 2008;7:187–198. [Abstract] [Google Scholar]
- Goodrick CL. Body weight increment and length of life: the effect of genetic constitution and dietary protein. J. Gerontol. 1978;33:184–190. [Abstract] [Google Scholar]
- Goodrick CL, Ingram DK, Reynolds MA, Freeman JR, Cider N. Effects of intermittent feeding upon body weight and lifespan in inbred mice: interaction of genotype and age. Mech. Ageing. Dev. 1990;55:69–87. [Abstract] [Google Scholar]
- Greer EL, Brunet A. Signaling networks in aging. J. Cell Sci. 2008;121:407–412. [Abstract] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK–FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007;17:1646–1656. [Europe PMC free article] [Abstract] [Google Scholar]
- Gruber J, Tang SY, Halliwell B. Evidence for a trade-off between survival and fitness caused by resveratrol treatment of Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2007;1100:530–542. [Abstract] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Sternberg PW. The 1-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168:1937–1949. [Europe PMC free article] [Abstract] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. [Europe PMC free article] [Abstract] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. [Abstract] [Google Scholar]
- Hardie DG. AMP-activated protein kinase as a drug target. Annu. Rev. Pharmacol. Toxicol. 2007;47:185–210. [Abstract] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 2001;11:1975–1980. [Abstract] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J. Gerontol. A. Biol. Sci. Med. Sci. 2006;61:444–460. [Abstract] [Google Scholar]
- Honda S, Ishii N, Suzuki K, Matsuo M. Oxygen-dependent perturbation of life span and aging rate in the nematode. J. Gerontol. 1993;48:B57–B61. [Abstract] [Google Scholar]
- Hosono R, Nishimoto S, Kuno S. Alterations of life span in the nematode Caenorhabditis elegans under monoxenic culture conditions. Exp. Gerontol. 1989;24:251–264. [Abstract] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp. Gerontol. 2002a;37:1371–1378. [Abstract] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. No reduction of metabolic rate in food restricted Caenorhabditis elegans. Exp. Gerontol. 2002b;37:1359–1369. [Abstract] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp. Gerontol. 2003;38:947–954. [Abstract] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. [Abstract] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. [Abstract] [Google Scholar]
- Hwang JT, Kwak DW, Lin SK, Kim HM, Kim YM, Park OJ. Resveratrol induces apoptosis in chemoresistant cancer cells via modulation of AMPK signaling pathway. Ann. NY. Acad. Sci. 2007;1095:441–448. [Abstract] [Google Scholar]
- Ingram DK, Zhu M, Mamczarz J, Zou S, Lane MA, Roth GS, deCabo R. Calorie restriction mimetics: an emerging research field. Aging Cell. 2006;5:97–108. [Abstract] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. [Abstract] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. [Abstract] [Google Scholar]
- Kaltenbach LS, Updike DL, Mango SE. Contribution of the amino and carboxyl termini for PHA-4/FoxA function in Caenorhabditis elegans. Dev. Dyn. 2005;234:346–354. [Abstract] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. [Europe PMC free article] [Abstract] [Google Scholar]
- Kawli T, Tan MW. Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat. Immunol. 2008;9:1415–1424. [Abstract] [Google Scholar]
- Kiefer JC, Smith PA, Mango SE. PHA-4/FoxA cooperates with TAM-1/TRIM to regulate cell fate restriction in the C. elegans foregut. Dev Biol. 2007;303:611–624. [Europe PMC free article] [Abstract] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech. Ageing. Dev. 1977;6:413–429. [Abstract] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. [Abstract] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1998;95:13091–13096. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee H, Cho JS, Lambacher N, Lee J, Lee SJ, Lee TH, Gartner A, Koo HS. The Caenorhabditis elegans AMP-activated protein kinase AAK-2 is phosphorylated by LKB1 and is required for resistance to oxidative stress and for normal motility and foraging behavior. J. Biol. Chem. 2008;283:14988–14993. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. [Abstract] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 2008;77:727–754. [Abstract] [Google Scholar]
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. [Europe PMC free article] [Abstract] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–3031. [Abstract] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech. Ageing. Dev. 2005;126:913–922. [Abstract] [Google Scholar]
- Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 2008;7:199–206. [Europe PMC free article] [Abstract] [Google Scholar]
- Miyadera H, Amino H, Hiraishi A, Taka H, Murayama K, Miyoshi H, Sakamoto K, Ishii N, Hekimi S, Kita K. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J. Biol. Chem. 2001;276:7713–7716. [Abstract] [Google Scholar]
- Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. [Europe PMC free article] [Abstract] [Google Scholar]
- Narbonne P, Roy R. Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development. 2006;133:611–619. [Abstract] [Google Scholar]
- Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J. Nutr. 1993;123:269–274. [Abstract] [Google Scholar]
- Pallottini V, Montanari L, Cavallini G, Bergamini E, Gori Z, Trentalance A. Mechanisms underlying the impaired regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in aged rat liver. Mech. Ageing. Dev. 2004;125:633–639. [Abstract] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. [Abstract] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. [Europe PMC free article] [Abstract] [Google Scholar]
- Piper MD, Bartke A. Diet and aging. Cell Metab. 2008;8:99–104. [Abstract] [Google Scholar]
- Piper MD, Partridge L. Dietary restriction in Drosophila: delayed aging or experimental artefact? PLoS Genet. 2007;3:e57. [Europe PMC free article] [Abstract] [Google Scholar]
- Retzlaff E, Fontaine J, Furuta W. Effect of daily exercise on life-span of albino rats. Geriatrics. 1966;21:171–177. [Abstract] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. [Europe PMC free article] [Abstract] [Google Scholar]
- Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. [Abstract] [Google Scholar]
- Roth GS, Ingram DK, Lane MA. Caloric restriction in primates and relevance to humans. Ann. N. Y. Acad. Sci. 2001;928:305–315. [Abstract] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. [Abstract] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr. Biol. 2008;18:1355–1364. [Europe PMC free article] [Abstract] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing. Dev. 2005;126:987–1002. [Abstract] [Google Scholar]
- Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity and lifespan in Drosophila melanogaster. Aging Cell. 2008;7:478–490. [Europe PMC free article] [Abstract] [Google Scholar]
- Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, Sutphin GL, Kennedy BK, Kaeberlein M. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008;7:394–404. [Europe PMC free article] [Abstract] [Google Scholar]
- Szewczyk NJ, Udranszky IA, Kozak E, Sunga J, Kim SK, Jacobson LA, Conley CA. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J. Exp. Biol. 2006;209:4129–4139. [Abstract] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. [Abstract] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. [Europe PMC free article] [Abstract] [Google Scholar]
- Valenzano DR, Terzibasi E, Genade T, Cattaneo A, Domenici L, Cellerino A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr. Biol. 2006;16:296–300. [Abstract] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell. 2005;9:605–615. [Abstract] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing. Dev. 2006;127:48–56. [Abstract] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J. Nutr. 1986;116:641–654. [Abstract] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. [Abstract] [Google Scholar]
- Zang M, Xu S, Maitland-Toolan KA, Zuccollo A, Hou X, Jiang B, Wierzbicki M, Verbeuren TJ, Cohen RA. Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes. 2006;55:2180–2191. [Abstract] [Google Scholar]
Articles from Aging Cell are provided here courtesy of Wiley
Full text links
Read article at publisher's site: https://doi.org/10.1111/j.1474-9726.2009.00459.x
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/j.1474-9726.2009.00459.x
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1111/j.1474-9726.2009.00459.x
Article citations
Hyperactive mTORC1/4EBP1 signaling dysregulates proteostasis and accelerates cardiac aging.
Geroscience, 09 Oct 2024
Cited by: 0 articles | PMID: 39379739
Non-canonical metabolic and molecular effects of calorie restriction are revealed by varying temporal conditions.
Cell Rep, 43(9):114663, 20 Aug 2024
Cited by: 0 articles | PMID: 39167490 | PMCID: PMC11427179
The C. elegans Myc-family of transcription factors coordinate a dynamic adaptive response to dietary restriction.
Geroscience, 46(5):4827-4854, 15 Jun 2024
Cited by: 0 articles | PMID: 38878153 | PMCID: PMC11336136
Understanding coenzyme Q.
Physiol Rev, 104(4):1533-1610, 09 May 2024
Cited by: 1 article | PMID: 38722242 | PMCID: PMC11495197
Review Free full text in Europe PMC
The integrated stress response protects against ER stress but is not required for altered translation and lifespan from dietary restriction in Caenorhabditis elegans.
Front Cell Dev Biol, 11:1263344, 14 Dec 2023
Cited by: 1 article | PMID: 38161330 | PMCID: PMC10755965
Go to all (349) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans.
Curr Biol, 17(19):1646-1656, 27 Sep 2007
Cited by: 497 articles | PMID: 17900900 | PMCID: PMC2185793
Dietary restriction involves NAD⁺ -dependent mechanisms and a shift toward oxidative metabolism.
Aging Cell, 13(6):1075-1085, 25 Sep 2014
Cited by: 58 articles | PMID: 25257342 | PMCID: PMC4244309
d-Allulose, a stereoisomer of d-fructose, extends Caenorhabditis elegans lifespan through a dietary restriction mechanism: A new candidate dietary restriction mimetic.
Biochem Biophys Res Commun, 493(4):1528-1533, 28 Sep 2017
Cited by: 18 articles | PMID: 28965946
5'-AMP-Activated Protein Kinase Signaling in Caenorhabditis elegans.
Exp Suppl, 107:375-388, 01 Jan 2016
Cited by: 2 articles | PMID: 27812988
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: CA09302
Grant ID: T32 CA009302
NIA NIH HHS (3)
Grant ID: R01 AG031198
Grant ID: R01 AG026648
Grant ID: R01 AG026648-01





