Abstract
Free full text

7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development
Abstract
Eukaryotic gene expression is commonly controlled at the level of RNA polymerase II (RNAPII) pausing subsequent to transcription initiation. Transcription elongation is stimulated by the positive transcription elongation factor b (P-TEFb) kinase, which is suppressed within the 7SK small nuclear ribonucleoprotein (7SK snRNP). However, the biogenesis and functional significance of 7SK snRNP remain poorly understood. Here, we report that LARP7, BCDIN3, and the noncoding 7SK small nuclear RNA (7SK) are vital for the formation and stability of a cell stress-resistant core 7SK snRNP. Our functional studies demonstrate that 7SK snRNP is not only critical for controlling transcription elongation, but also for regulating alternative splicing of pre-mRNAs. Using a transient expression splicing assay, we find that 7SK snRNP disintegration promotes inclusion of an alternative exon via the increased occupancy of P-TEFb, Ser2-phosphorylated (Ser2-P) RNAPII, and the splicing factor SF2/ASF at the minigene. Importantly, knockdown of larp7 or bcdin3 orthologues in zebrafish embryos destabilizes 7SK and causes severe developmental defects and aberrant splicing of analyzed transcripts. These findings reveal a key role for P-TEFb in coupling transcription elongation with alternative splicing, and suggest that maintaining core 7SK snRNP is essential for vertebrate development.
Challenging the once predominantly held view that RNA polymerase II (RNAPII) recruitment to promoters is the rate-limiting event in regulating eukaryotic transcription, several recent studies indicate that RNAPII pauses subsequently to transcription initiation at a large number of genes in Drosophila and in several cell lineages in humans (1). A paradigm for this type of control is the regulation of HIV-1 transcription, which is paused because of concerted actions of the negative transcription elongation factors (N-TEFs) (1, 2). The block is reversed by the positive transcription elongation factor b (P-TEFb), consisting of a heterodimer between the cyclin-dependent kinase 9 (Cdk9) and one of the four C-type cyclins (CycT1/T2a(b)/K) (2), which phosphorylates Ser2 within the multiple heptapeptide repeats YSPTSPS of the carboxy-terminal domain (CTD) of the Rpb1 subunit of RNAPII, as well as components of N-TEFs. Of note, P-TEFb is critical for expressing early embryonic genes in C. elegans (3), and experiments in yeast and Drosophila demonstrate that it is also vital for proper 3′ end pre-mRNA processing (4, 5). Thus, regulating transcription elongation is a key checkpoint for modulating eukaryotic gene expression.
P-TEFb exists in 2 mutually exclusive complexes that differ in their kinase activity (2). A transcriptionally inactive complex [referred to herein as 7SK small nuclear ribonucleoprotein (7SK snRNP)] contains P-TEFb, hexamethylene bisacetamide-induced protein (HEXIM) 1 (HEXIM1) and/or HEXIM2, the noncoding 7SK small nuclear RNA (7SK) and the La-related protein 7 (LARP7) (6–12). Therein, 7SK is a molecular scaffold, which binds HEXIM1, HEXIM2, and LARP7 directly, enabling the sequestration and repression of P-TEFb. Another major fraction of P-TEFb constitutes the Brd4/P-TEFb complex, which represents the transcriptionally active P-TEFb and could be recruited to target genes because of the binding of Brd4 to acetylated histones or the mediator (13, 14). The dynamic partitioning of P-TEFb between the complexes constitutes a functional equilibrium that can be perturbed. For example, HIV-1 Tat increases the pool of active P-TEFb for optimal viral transcription by hijacking it from 7SK snRNP (15), and the disintegration of 7SK snRNP leads to cardiac hypertrophy or tumorigenesis (16, 11).
At present, little is known about the assembly and functional significance of maintaining 7SK snRNP in vivo. Here, we identify LARP7 and Bicoid-interacting 3, homolog (Drosophila) (BCDIN3) as vital components of 7SK snRNP, which in concert with 7SK govern the biogenesis and integrity of core 7SK snRNP. We further find that besides transcription elongation, 7SK snRNP also controls alternative splicing via suppressing P-TEFb. Finally, we demonstrate that core 7SK snRNP is essential for zebrafish embryonic development.
Results
LARP7 and BCDIN3 Are Present in 7SK snRNP and Interact with P-TEFb and HEXIM1 in RNase A- and Cell Stress-Dependent Manner.
We immunopurified P-TEFb- or HEXIM1-associated factors from whole cell extracts (WCEs) of F1C2 or HH8 HeLa-based cell lines that stably expressed FLAG-tagged Cdk9 (Cdk9.F) or HEXIM1 (F.HEXIM1) proteins, respectively (6, 9), and performed mass spectrometry of prominent bands. In agreement with recent work (11, 12, 17), we identified LARP7 and BCDIN3 as P-TEFb and HEXIM1 interacting partners (Table S1 and Table S2). Of note, the depletion of LARP7 destabilizes 7SK for 7SK snRNP disintegration and malignant transformation of a mammary epithelial cell line (11), and BCDIN3 is a 7SK γ-methylphosphate capping enzyme (MePCE) essential for its stability (17). We next confirmed the proteomic results by Western blotting by using antibodies against endogenous LARP7, BCDIN3, P-TEFb and HEXIM1 proteins, and found that LARP7 and BCDIN3 were present in Cdk9.F and F.HEXIM1 immunoprecipitates (Fig. S1A). Furthermore, we determined that 7SK degradation in WCEs by RNase A and exposure of cells to stress-inducing agent actinomycin D (ActD) released LARP7 and BCDIN3 from the immunoprecipitates (Fig. S1 B and C, lanes 4–6). Thus, LARP7 and BCDIN3 are part of 7SK snRNP, which dissociate from P-TEFb and HEXIM1 upon 7SK degradation and cell stress.
LARP7, BCDIN3, and 7SK Form Core 7SK snRNP.
To test whether the interaction between LARP7 and BCDIN3 within 7SK snRNP is also 7SK- and cell stress-dependent, we established F.L7.293 and F.B3.293 293-based cell lines that expressed FLAG-tagged LARP7 (F.LARP7) and BCDIN3 (F.BCDIN3), respectively. Immunopurification of F.LARP7 or F.BCDIN3 from WCEs and analysis of the complexes by Western blotting and RT-PCR revealed that F.LARP7 and F.BCDIN3 were part of 7SK snRNP and that their interaction with P-TEFb and HEXIM1 was disrupted upon RNase A or ActD treatments (Fig. 1 A and B). In contrast, LARP7 and BCDIN3 remained associated under both conditions, and cell stress did not release 7SK from them (Fig. 1 A and B, lanes 6–8). Thus, unlike the 7SK-dependent bindings between P-TEFb and HEXIM1 in 7SK snRNP, LARP7 and BCDIN3 bind via protein–protein interactions stably and independently of other known components of 7SK snRNP. That 7SK associates with LARP7 and BCDIN3 under cell stress suggests extensive RNA–protein interactions within this 7SK snRNP subcomplex. We conclude that LARP7, BCDIN3, and 7SK constitute a cell stress-resistant core 7SK snRNP.
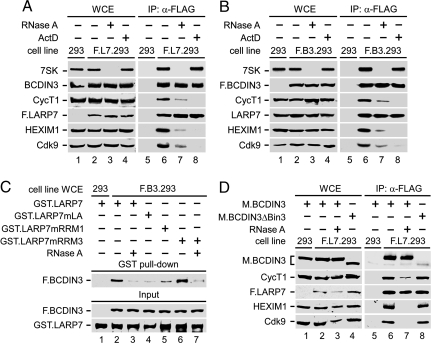
Identification of core 7SK snRNP and elucidation of its assembly. (A and B) WCEs of 293, F.L7.293, or F.B3.293 cells were subjected to IP with α-FLAG. Levels of F.LARP7, F.BCDIN3, and their associating factors in WCEs (Left) and immunoprecipitates (Right) were detected by RT-PCR and Western blotting, respectively. WCEs or cells before lysis were incubated (+) or not (−) with RNase A or ActD, respectively. (C) GST.LARP7 chimeras were incubated with WCEs of 293 or F.B3.293 cells (Top). WCEs were incubated (+) or not (−) with RNase A. Levels of bound F.BCDIN3 proteins (Middle) and those of F.BCDIN3 and GST chimeras present in the binding reactions (Bottom) were analyzed by Western blotting. (D) Levels of indicated proteins in WCEs or α-FLAG immunoprecipitations of 293 or F.L7.293 cells are shown, which were analyzed by Western blotting. WCEs were incubated (+) or not (−) with RNase A.
7SK-Binding La and RRM1 Motifs in LARP7 and the C-Terminal Bin3 Region in BCDIN3 Are Critical for the 7SK-Mediated Assembly of Core 7SK snRNP.
To understand the assembly of core 7SK snRNP, we noted that unlike BCDIN3, LARP7 contains 3 predicted RNA-binding motifs (Fig. S2A). Whereas its N-terminal region is composed of a La (18) and a canonical RNA recognition motif (RRM) 1 (RRM1) (19), its C-terminal region contains an atypical RRM3 (20). In agreement with recent studies (11, 12), LARP7 bound 7SK in vitro as determined by electrophoretic mobility shift assay (Fig. S2B, lane 2). Next, we individually mutated the predicted RNA-binding motifs to yield LARP7mLa, LARP7mRRM1, and LARP7mRRM3 proteins (for details, see Fig. S2A) and found that in contrast to RRM3, La and RRM1 motifs were critical for binding 7SK (Fig. S2B, lanes 3–5).
That LARP7 bound 7SK suggested that the assembly of core 7SK snRNP could be mediated via 7SK. To address this notion, we performed in vitro GST pull-down experiments by using the recombinant wild-type or mutant GST.LARP7 chimeras as in Fig. S2, which were incubated with WCEs of 293 cells or F.B3.293 cells stably expressing F.BCDIN3 in the absence or presence of RNase A (Fig. 1C). Importantly, F.BCDIN3 bound GST.LARP7 in vitro, and this interaction was abrogated in the presence of RNase A (Fig. 1C, lanes 1–3). Moreover, consistent with the requirements of La and RRM1 motifs in LARP7 for the 7SK binding, GST.LARP7mLa and GST.LARP7mRRM1 failed to bind F.BCDIN3 efficiently. In contrast, GST.LARP7mRRM3 bound F.BCDIN3 in RNA-dependent manner (Fig. 1C, lanes 4–7). Thus, La and RRM1 motifs in LARP7 are required for binding 7SK, which mediates the assembly of core 7SK snRNP.
To determine which region in BCDIN3 is involved in this assembly, we expressed the Myc-tagged wild-type (M.BCDIN3) and mutant BCDIN3 protein without the evolutionary conserved C-terminal Bin3 region (M.ΒCDΙΝ3ΔΒin3) in F.L7.293 cells, and we analyzed their sequestration into the immunopurified F.LARP7-containing 7SK snRNP by Western blotting (Fig. 1D). Predictably, M.BCDIN3 was assembled into the 7SK snRNP and was not released from it in the presence of RNase A, confirming the stable nature of an already assembled LARP7–BCDIN3 complex (Fig. 1D, lanes 6, 7). In contrast, M.BCDIN3ΔBin3 failed to be part of 7SK snRNP, indicating that the Bin3 region in BCDIN3 is critical for its interaction with LARP7 (Fig. 1D, lane 8). These results demonstrate that the C-terminal Bin3 region in BCDIN3 is also required for the assembly of core 7SK snRNP.
La, RRM1, and RRM3 Motifs in LARP7 and the C-terminal Bin3 Region in BCDIN3 Are Required for Their Sequestration into 7SK snRNP.
Using immunoprecipitation experiments, we found that the critical motifs and regions in LARP7 and BCDIN3 identified above were also important for their sequestration into the endogenous P-TEFb-containing 7SK snRNP (Fig. S2 C and D). Notably, when compared to mutations of RNP1 within RRM1 of LARP7, mutation of the conserved tyrosine residue at position 128 in RNP2 only modestly affected this interaction (Fig. S3). In addition, disruption of RRM3 in LARP7, which did not affect its 7SK or BCDIN3 binding in vitro, prevented assembly of this mutant LARP7 protein into 7SK snRNP (Fig. S2C, lane 6). Given the recent work, which demonstrated that the LARP7 C-terminal region containing RRM3 was sufficient to bind Cdk9 in vitro (12), it is likely that cooperative LARP7–7SK and LARP7–Cdk9 interactions ensure its presence in 7SK snRNP. Taken together, LARP7 requires La, RRM1, and RRM3 motifs for its sequestration into P-TEFb-containing 7SK snRNP, whereas the C-terminal Bin3 region in BCDIN3 is critical for this binding.
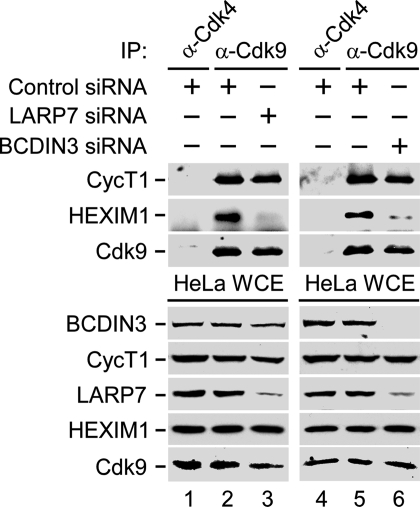
Depletion of LARP7 or BCDIN3 Disintegrates 7SK snRNP and Derepresses P-TEFb for Stimulating Transcription.
Because LARP7 and BCDIN3 alongside 7SK represent core 7SK snRNP, we next postulated that their depletion would disintegrate 7SK snRNP. Indeed, besides diminishing the LARP7 or BCDIN3 protein levels (Fig. 2, lanes 3 and 6), expression of LARP7- or BCDIN3-siRNA in HeLa cells significantly reduced levels of 7SK by 75% and 77% as determined by RT-qPCR, respectively, whereas control or HEXIM1 siRNA had no effect (Fig. S4A, bars 1–4). BCDIN3-siRNA depleted LARP7 protein levels as well (Fig. 2, lane 6), and this depletion occurred posttranscriptionally (Fig. S5), suggesting that it caused LARP7 proteolysis. Importantly, destabilization of core 7SK snRNP via LARP7 or BCDIN3 diminution released HEXIM1 from P-TEFb (Fig. 2, lanes 3 and 6) and stimulated transcription from the integrated HIV-1 LTR by ≈3.5-fold or 7.2-fold, respectively (Fig. S4B, bars 1–3). Thus, depletion of the components of core 7SK snRNP derepresses P-TEFb and stimulates HIV-1 transcription.
Disintegration of 7SK snRNP Promotes Alternative Splicing via the Transcriptionally Active P-TEFb.
A large body of evidence indicates functional and physical coupling of transcription and processing of nascent pre-mRNA (21). To understand how transcription elongation modulates alternative splicing, we next tested whether derepression of P-TEFb impacts this coupling. Here, we used a well-established alternative splicing assay consisting of the pSVED-A Tot minigene (22) (Fig. 3A). Transcription from the cassette yields 2 alternatively spliced mRNAs that include or lack the EDA exon (EDA), which contains a suboptimal 3′ splice site. Strikingly, coexpression of LARP7- or BCDIN3-siRNA with pSVED-A Tot in HeLa cells enhanced the inclusion of EDA greatly as is evident from the reciprocal increase of EDA-including at the expense of EDA-less mRNAs (Fig. 3B, lanes 1–4). RT-qPCR revealed that the ratio between EDA-including and EDA-less mRNAs (EDA/ΔEDA) increased ≈5-fold or 6.2-fold under these conditions (Fig. S6A, bars 1–3). Importantly, these effects were counteracted by coexpressing siRNA against Brd4, an integral component of transcriptionally active P-TEFb (13, 14), suggesting that the enhanced EDA inclusion levels were P-TEFb-dependent (Fig. 3C and Fig. S6B, bars 1–6, and Fig. S6 C–E). These EDA levels were not stimulated by hnRNP A1, an inhibitory splicing factor (23) present in a distinct 7SK snRNP complex devoid of P-TEFb (24, 25), which is most likely also released from this 7SK snRNP upon depletion of LARP7 or BCDIN3. As expected, depletion of hnRNP A1 mRNA enhanced the inclusion of EDA (Fig. S7). Thus, the release of P-TEFb from 7SK snRNP promotes alternative splicing.
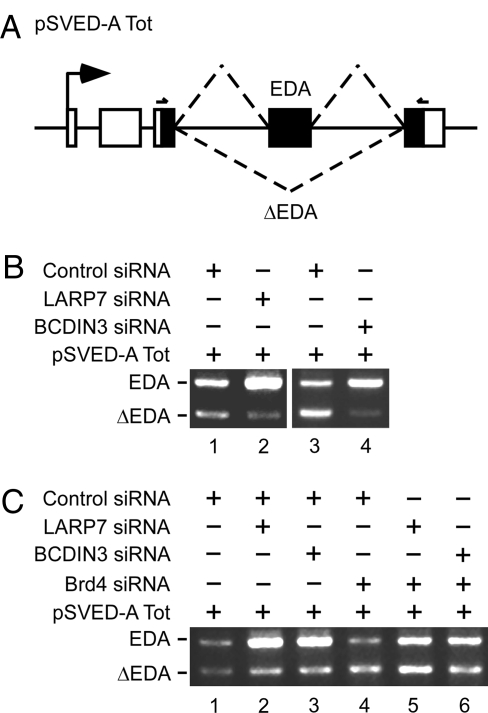
Depletion of LARP7 or BCDIN3 enhances the inclusion of EDA in P-TEFb-dependent manner. (A) Schematic of the pSVED-A Tot minigene. Arrow represents the transcription start site from the α-globin promoter. The third exon of the 3 α-globin exons (white rectangles) contains 3 fibronectin exons (black rectangles, exon number 32–34) with the introns (solid lines). Dashed lines represent the 2 possible mature EDA and ΔEDA mRNAs that include or lack the second fibronectin exon named EDA, respectively. The psv5′j/psv3′j primer pair used for RT-PCR analysis is indicated. (B and C) Levels of EDA and ΔEDA mRNAs were detected by RT-PCR by using total RNA samples isolated from HeLa cells that expressed siRNAs and pSVED-A Tot as indicated above the agarose gels.
P-TEFb Promotes Alternative Splicing via Ser2-P RNAPII and SF2/ASF.
Next, we addressed the mechanism by which P-TEFb promotes alternative splicing in greater detail. First, we found that flavopiridol, a potent and highly specific P-TEFb inhibitor (2), effectively antagonized the enhancement of EDA inclusion levels achieved by depleting BCDIN3 (Fig. 4A, lanes 3 and 4), indicating that upon 7SK snRNP disintegration, the kinase activity of P-TEFb stimulates alternative splicing. Therefore, we speculated that P-TEFb may promote alternative splicing via phosphorylating the CTD of RNAPII at Ser2, which could increase the recruitment of splicing factors for facilitating EDA inclusion levels. Notably, Ser/Arg-rich (SR) proteins function in coupling transcription to pre-mRNA splicing (23), and the SR protein SF2/ASF stimulates inclusion of EDA (26). Thus, we next asked whether expression of SF2/ASF-siRNA would antagonize the stimulatory effect of P-TEFb on the EDA inclusion. SF2/ASF-siRNA, which reduced SF2/ASF mRNA levels (Fig. S8), inhibited EDA inclusion (Fig. 4B, lanes 1 and 2). Importantly, enhanced inclusion of EDA, upon expressing BCDIN3-siRNA, was effectively counteracted by SF2/ASF-siRNA (Fig. 4B, lanes 3 and 4). Collectively, these results suggest that P-TEFb promotes alternative splicing at the pSVED-A Tot minigene via Ser2-P RNAPII and SF2/ASF.
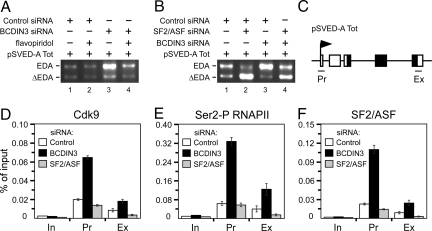
P-TEFb controls alternative splicing via Ser2-P RNAPII and SF2/ASF. (A and B) Levels of EDA and ΔEDA mRNAs were detected by RT-PCR by using total RNA samples isolated from HeLa cells that expressed siRNAs and pSVED-A Tot as indicated above the agarose gels. Cells were treated (+) or not (−) with 60 nM of flavopiridol for 24 h before the isolation of total RNA. (C) Schematic of the pSVED-A Tot minigene used in the ChIP-qPCR. Positions of the promoter (Pr) and exonic (Ex) regions of the minigene that were amplified with qPCR are indicated. (D–F) ChIP-qPCR analysis of the pSVED-A Tot minigene in HeLa cells treated with control (white bars), BCDIN3 (black bars), or SF2/ASF (gray bars) siRNAs. Chromatin was immunoprecipitated with the depicted antibodies. Results for 3 ChIP-qPCR assays are shown ± SEM and are shown as percent (%) of input binding.
To address this notion directly, we performed quantitative chromatin immunoprecipitation (ChIP-qPCR) assays (Fig. 4, panels C–F). Critically, in cells that expressed BCDIN3-siRNA, the occupancy of the Cdk9 subunit of P-TEFb at the promoter and exonic region of the minigene increased ≈3.2-fold and 2.1-fold, respectively, when compared to the occupancy in cells expressing control-siRNA (Fig. 4D). This enrichment of P-TEFb resulted in ≈5.2-fold and 3.1-fold increases in the presence of Ser2-P RNAPII at the same minigene regions, respectively (Fig. 4E). Importantly, the promoter and exonic occupancy of SF2/ASF was augmented by ≈4.9-fold and 2.4-fold, respectively, in cells that expressed BCDIN3-siRNA (Fig. 4F). In contrast, exonic occupancy of Cdk9 and Ser2-P RNAPII was reduced by 2.4-fold and 2.6-fold, respectively, in SF2/ASF-siRNA expressing cells (Fig. 4 D and E), suggesting the bidirectional coupling between transcription elongation and cotranscriptional splicing. These results are consistent with a recent report, which demonstrated that another SR protein SC35 was required for efficient transcription elongation, P-TEFb recruitment, and CTD Ser2 phosphorylation in a gene-specific manner (27). We conclude that disintegration of 7SK snRNP leads to the P-TEFb-mediated phosphorylation of the CTD of RNAPII at Ser2, thereby facilitating the recruitment of the stimulatory splicing factor SF2/ASF for promoting EDA inclusion levels.
Knockdown of larp7 or bcdin3 in Zebrafish Decreases 7SK Levels and Leads to Significant Defects in Development and Splicing.
Finally, by using zebrafish as a model system, we asked whether larp7 or bcdin3 are essential genes in vivo. We first analyzed the expression pattern of larp7 and bcdin3 by RNA in situ hybridization and found that they were expressed ubiquitously at early developmental stages (Fig. 5 A and B, panel 1). Expression of both genes became enriched in anterior regions, including developing brain, at the postsomitogenesis stage (Fig. 5 A and B, panel 2). The in situ signals were specific to larp7 or bcdin3, because the sense probes did not yield any signals (Fig. 5 A and B, panels 3 and 4). To determine whether larp7 or bcdin3 influence zebrafish development, we knocked down their activity by using morpholinos (MO) that selectively blocked the splicing of their pre-mRNAs (Fig. S9). Strikingly, both larp7 MO (>90%, n > 100) and bcdin3 MO (>90%, n > 50), but not a control MO, resulted in severe developmental defects, which was obvious at 24 h postfertilization (hpf), and included altered body axis formation and selective darkening of the brain region indicative of neurodegeneration (Fig. 5C). The major axial polarity including the anteroposterior and dorsoventral axes, however, appeared largely normal in the morphants, which also possessed recognizable eye primordia. Although some embryos survived to 48 hpf, they exhibited the morphological defects consistent with the stage at 24 hpf, which were degeneration at brain region and no obvious signs of organogenesis.
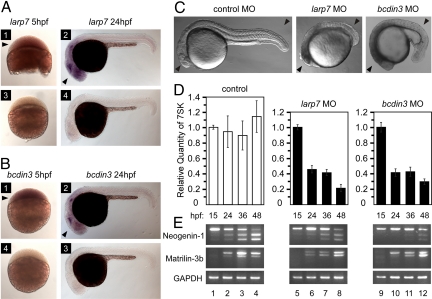
Knockdown of larp7 and bcdin3 in zebrafish caused developmental defects, decreased 7SK levels, and aberrant splicing. (A and B) RNA in situ hybridizations of whole-mount zebrafish embryos by using larp7 and bcdin3 antisense (1, 2) and sense probes (3, 4) at the indicated time points. Black arrowheads in panels 1 and 2 indicate larp7 and bcdin3 expressions in blastomeres and their enriched expressions in developing brain/head, respectively. (C) Zebrafish embryos injected with either larp7 MO or bcdin3 MO show developmental defects at 24 hpf compared with embryo injected with a control MO. Black arrowheads indicate the developing brain/head and trunk/tail. (D) Relative quantities of 7SK levels were determined by RT-qPCR at the indicated time points below the graph. The error bars represent the mean ± SD. (E) Splicing pattern analysis for the 2 genes indicated on the left at different time points as shown above the agarose gels. Levels of splicing variants were determined by RT-PCR by using the samples as in D and gene-specific primers.
Next, we asked whether knockdown of larp7 and bcdin3 would destabilize 7SK in vivo. Indeed, both knockdowns led to reduced 7SK levels as determined by RT-qPCR (Fig. 5D). 7SK destabilization was evident at 24hpf and thus correlated with the appearance of developmental defects in the morphants, suggesting that zebrafish LARP7 and BCDIN3 may ensure proper development via maintaining 7SK snRNP. Finally, we tested whether the knockdown of larp7 or bcdin3 would also affect splicing in vivo. We compared patterns of splicing variants for neogenin-1 and matrilin-3b, 2 genes that are expressed predominantly in the developing brain and cartilaginous tissues, respectively, and whose splicing patterns are developmentally regulated (28, 29). When compared to control embryos, knockdown of larp7 and bcdin3 caused significant and similar changes in the splicing patterns of both genes as determined by RT-PCR (Fig. 5E). For example, an obvious decrease in amounts of the longest neogenin-1 splicing variant relative to the shorter variants in control embryos during development was equally delayed in both knockdown embryos. The changes coincided with the decrease in the levels of 7SK, indicating that 7SK snRNP disintegration could mediate these effects. In sum, larp7 and bcdin3 are essential genes for zebrafish development and are required for 7SK stability and proper alternative splicing in vivo.
Discussion
7SK snRNP provides the pivotal inhibitory control of P-TEFb (2), the central transcriptional kinase that stimulates the transition from abortive to productive transcription elongation. Here, we provide important insights into the biogenesis and stability of 7SK snRNP. We identified core 7SK snRNP, consisting of LARP7, BCDIN3, and 7SK, of which assembly was mediated by 7SK and depended on LARP7–7SK and LARP7–BCDIN3 interactions. Of note, depletion of each protein component of core 7SK snRNP destabilized 7SK or even one another, further underscoring physical interconnectedness between them. Critically, inactivation of larp7 or bcdin3 also destabilized 7SK in our in vivo zebrafish model system. Overall, these findings suggest that in addition to the capping of 7SK by BCDIN3 and binding its 3′ poly(U) tail by LARP7 (17, 11), the extensive RNA–protein and protein–protein interactions within core 7SK snRNP guarantee its maintenance. Hence, core 7SK snRNP cannot be disintegrated when exposed to ActD, which explains the stability of 7SK under this cell stress-inducing condition and allows core 7SK snRNP to be a platform for the dynamic and reversible remodeling of 7SK snRNP as observed before (6, 24, 25).
Our findings on alternative splicing suggest the important role for P-TEFb in cotranscriptional pre-mRNA processing. Two major but not mutually exclusive mechanisms, by which transcription and alternative splicing could be coupled, have been suggested (21). These include the recruitment of splicing factors to the transcribing RNAPII and the kinetic coupling, which involves the rate of transcription elongation and thus the time in which splice sites are offered to the splicing apparatus. Although we do not know whether P-TEFb affected the speed of elongating RNAPII on the minigene, our results suggest that its kinase activity stimulates alternative splicing primarily via the recruitment mechanism. We propose that P-TEFb couples transcription elongation and alternative splicing via Ser2-P RNAPII. In turn, this event facilitates the recruitment of SF2/ASF and possibly other SR proteins together with U1 snRNP to the elongating RNAPII, which may enable them to employ their RRMs to scan the nascent pre-mRNA for splicing signals such as exonic splicing enhancer (ESE) element and 5′ splice site, respectively, as soon as they exit from Ser2-P RNAPII. Thus, they could stimulate the early steps in spliceosome assembly, leading to the promotion of alternative splicing. In support of this model, all components of the U1 snRNP complex and SR proteins are the major splicing factors that bind RNAPII, and SF2/ASF interacts with Ser2-P RNAPII (23). In addition, EDA contains a suboptimal 3′ splice site (22), and its splicing is stimulated by SF2/ASF and depends on the presence of ESE element located within EDA (26). Importantly, we further demonstrated that the splicing patterns of the 2 developmentally-regulated genes in the zebrafish larp7 or bcdin3 knockdown embryos changed greatly and in a similar manner. That these knockdowns led to predominantly longer neogenin-1 or shorter matrilin-3b splicing variants could be attributed to the favored usage of suboptimal splice sites by the splicing machinery upon the recruitment of P-TEFb and the appropriate splicing factors to these genes.
Another exciting aspect of our work is the fact that the protein components of core 7SK snRNP are essential for proper vertebrate development. Because 7SK levels were reduced in larp7 and bcdin3 knockdowns, and both genes were ubiquitously expressed in developing embryos, numerous morphological abnormalities likely resulted from a widespread disintegration of 7SK snRNP. Notably, LARP7 and BCDIN3 mRNA levels were enriched in the developing brain, whose gene expression is controlled extensively at the level of alternative splicing. Thus, it is conceivable that the observed neurodegeneration could in part be a manifestation of extensive pre-mRNA splicing defects. In addition, extensive developmental defects may stem from the derepression of P-TEFb, which could in turn promote expression of otherwise silent developmental control genes that may contain paused RNAPII molecules. Importantly, transient and global inhibition of P-TEFb has been proposed to be essential for maintaining germ cell identity in C. elegans (30, 31) and Drosophila (32), and transcription elongation controls cell fate specification in the Drosophila embryo (33). We therefore speculate that 7SK snRNP disintegration would lead to inappropriate differentiations and loss of identities of germ cells and multiple somatic cell lineages, causing developmental defects observed here. Hence, 7SK snRNP may respond to developmental signals during embryogenesis and release active P-TEFb for the rapid temporal and spatial control of target transcript elongation and cotranscriptional processing.
Methods
Immunoprecipitation, GST Pull-Down, and Electrophoretic Mobility Shift Assay.
Immunoprecipitations were performed as described (9). GST-LARP7 proteins bound to the Glutathione–Sepharose beads were incubated for 2 h at 4 °C with WCE of F.B3.293 in 500 μl reaction volume that contained buffer D with 0.1M KCl. After washing with the buffer D containing 0.3M KCl, the bound proteins were separated by SDS/PAGE and detected by Western blotting. EMSAs were performed as described (10).
Splicing Variant Analysis.
Total RNA was isolated by using TRIzol Reagent (Invitrogen). Reverse transcription was performed with the M-MLV reverse transcriptase (Invitrogen) by using random hexamers or gene-specific primers. RT-PCR analysis of the pSVED-A Tot splicing variants was performed by using the psv5′j/psv3′j primer pair described previously (22). RT-qPCR of the variants used primer pairs that discriminated between the 2 mRNAs. The EDA-including variant was amplified by using psv5′j/EDAR primer pair, whereas the EDA-less variant was amplified by using f32–34F/f34R primer pair. f32–34F, EDAR, and f34R primers annealed to the sequences of fibronectin mRNA encoded by the exons 32 and 34, 33 (EDA), and 34, respectively. For primer sequences, see Table S3. RT-PCR analysis of neogenin-1 and matrilin-3b splicing variants was done by using the znsp3/znsp3′ and m3bz4/m3bz2 primer pairs, respectively, which were described previously (28, 29).
Morpholino (MO) Synthesis and Injections.
Control, larp7, and bcdin3 MO antisense oligonucleotides were purchased (Gene Tools, LCC) and designed to complement the exon 2/intron 2 junction. For MO sequences, see Table S4. For knockdown experiments, 2–3 nl of the 1.25 mM control MO, 1 mM larp7 MO, and 1.25 mM bcdin3 MO were injected into the yolk of one- to eight-cell-stage embryos as previously described (34). MO efficacies were assessed by RT-PCR as indicated (Fig. S9). For primer sequences, see Table S5.
For additional details, see SI Methods, Table S6, and Table S7.
Acknowledgments.
We thank Drs. Benoit Coulombe and Qiang Zhou for the reagents, Drs. Steven C. Hall and H. Ewa Witkowska of the University of California at San Francisco (UCSF) Mass Spectrometry Core Facility for their assistance in mass spectrometry acquisition and data analysis, and members of the Peterlin laboratory for discussions. The work was supported by National Institutes of Health Grants AI058708 and GM082250 (to B.M.P.) and NS042626 (to S.G.). The UCSF Mass Spectrometry Core Facility acknowledges support from a Cancer Center Support Grant (P30 CA82103) funded by the National Cancer Institute and from the Sandler Family Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903188106/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0903188106
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2683122?pdf=render
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/106/19/7798
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/106/19/7798
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/106/19/7798.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Catalytic activity of the Bin3/MePCE methyltransferase domain is dispensable for 7SK snRNP function in Drosophila melanogaster.
Genetics, 226(1):iyad203, 01 Jan 2024
Cited by: 1 article | PMID: 37982586
Role of long noncoding RNA in regulating HIV infection-a comprehensive review.
mBio, 15(2):e0192523, 05 Jan 2024
Cited by: 1 article | PMID: 38179937 | PMCID: PMC10865847
Review Free full text in Europe PMC
Inhibition of CDK12 elevates cancer cell dependence on P-TEFb by stimulation of RNA polymerase II pause release.
Nucleic Acids Res, 51(20):10970-10991, 01 Nov 2023
Cited by: 1 article | PMID: 37811895 | PMCID: PMC10639066
Drosophila Amus and Bin3 methylases functionally replace mammalian MePCE for capping and the stabilization of U6 and 7SK snRNAs.
Sci Adv, 9(50):eadj9359, 15 Dec 2023
Cited by: 0 articles | PMID: 38100593 | PMCID: PMC10848712
Targeting CDK9 with selective inhibitors or degraders in tumor therapy: an overview of recent developments.
Cancer Biol Ther, 24(1):2219470, 01 Dec 2023
Cited by: 5 articles | PMID: 37272701 | PMCID: PMC10243401
Review Free full text in Europe PMC
Go to all (114) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The 7SK/P-TEFb snRNP controls ultraviolet radiation-induced transcriptional reprogramming.
Cell Rep, 35(2):108965, 01 Apr 2021
Cited by: 19 articles | PMID: 33852864
PPM1G Binds 7SK RNA and Hexim1 To Block P-TEFb Assembly into the 7SK snRNP and Sustain Transcription Elongation.
Mol Cell Biol, 35(22):3810-3828, 31 Aug 2015
Cited by: 35 articles | PMID: 26324325 | PMCID: PMC4609742
Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.
Nucleic Acids Res, 44(16):7527-7539, 01 Jul 2016
Cited by: 77 articles | PMID: 27369380 | PMCID: PMC5027500
Review Free full text in Europe PMC
Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein.
J Biol Chem, 287(43):36609-36616, 05 Sep 2012
Cited by: 141 articles | PMID: 22952229 | PMCID: PMC3476326
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P30 CA082103
Grant ID: P30 CA82103
NIAID NIH HHS (2)
Grant ID: AI058708
Grant ID: P01 AI058708
NIGMS NIH HHS (3)
Grant ID: GM082250
Grant ID: P50 GM082250
Grant ID: P50 GM082250-02
NINDS NIH HHS (2)
Grant ID: NS042626
Grant ID: R01 NS042626