Abstract
Free full text

Effects of BDNF, T3, and corticosterone on expression of the hypothalamic obesity gene network in vivo and in vitro
Abstract
Hypothalamic neuropeptides, neurotrophins, and systemic hormones modulate food intake and body composition. Although advances toward elucidating these interactions have been made, many aspects of the underlying mechanisms remain vague. Hypothalami from fat and lean chicken lines were assessed for differential expression of anabolic/orexigenic and catabolic/anorexigenic genes. Effects of triiodothyronine (T3), corticosterone (Cort), and brain-derived neurotrophic factor (BDNF) on expression of anabolic/orexigenic and catabolic/anorexigenic genes were tested in cultures of hypothalamic neurons. From this, we found that BDNF increased and T3 decreased gene expression for BDNF, leptin receptor (LEPR), pro-opiomelanocortin (POMC), thyrotropin releasing hormone (TRH), and agouti-related protein (AGRP). Thyroid hormone levels were manipulated during development to show that T3 inhibited BDNF, TRH, and BDNF receptor gene expression. Delivery of T3, Cort, T3 plus Cort, or vehicle in vivo continuously for 72 h indicated that Cort and T3 have overlapping roles in regulating TRH, LEPR, and POMC gene expression and that Cort and T3 regulate BDNF, neuropeptide Y, and AGRP in opposite directions. Collectively, these findings suggest that interactions between the neuropeptide BDNF and the hormones T3 and/or Cort may constitute a homeostatic mechanism that links hypothalamic energy regulation controlling body composition.
disrupted energy homeostasis leads to obesity affecting 100 million adults and 22 million children worldwide (50). This condition arises from multiple factors, including altered metabolism, genetic susceptibility, excess food intake, and insufficient energy expenditure (6, 32). Brain-derived neurotrophic factor (BDNF), thyroid hormone, and glucocorticoids are among many factors implicated in the etiology of obesity (12, 14, 15, 35, 41). Although it is known that hormones modulate and interact with neurotrophins and neuropeptides, the nature by which interactions alter hypothalamic gene expression remains vague. In rodents, humans, and chickens, the same hypothalamic neuropeptides regulate energy homeostasis and body fat mass (2, 4, 19, 26, 44, 47), demonstrating that the mechanisms regulating food intake and body weight are evolutionarily conserved. Under normal conditions, hypothalamic peptides function in a complimentary fashion to maintain energy homeostasis, with anabolic/orexigenic pathways increasing food intake and fat deposition and decreasing energy expenditure and catabolic/anorexigenic pathways eliciting the opposite responses (11, 21, 36, 44). We sought to test whether BDNF, l-triiodothyronine (T3), and glucocorticoids interact to modulate hypothalamic expression of genes involved in energy regulation.
BDNF haploinsufficient mice or mice with conditional depletion of BDNF in the brain after birth become obese (22, 41). In humans, decreased BDNF protein levels contribute to obesity, and increased BDNF is associated with anorexia (35, 38, 39). Furthermore, BDNF has been shown to increase thyrotropin-releasing hormone (TRH) in the hypothalamus (16), and T3 suppressed BDNF gene expression in neurons from the band of Broca (5), suggesting an interaction between the two pathways in development of obesity. In support of this interaction within the hypothalamus, BDNF is highly expressed in the hypothalamic ventromedial nucleus (48), an area previously demonstrated to be involved with satiety (2, 19), and T3 administration has been shown to also activate neurons in this nucleus (24). In addition, modestly elevated levels of T3 have been observed in morbidly obese euthyroid humans (34). Therefore, moderately increased T3 levels could suppress BDNF gene expression, contributing toward increased adiposity.
Another hormone, corticosterone (Cort), also interacts with thyroid hormones to regulate energy homeostasis, and these effects are often in opposition to one another. Elevated levels of thyroid hormone associated with hyperthyroidism decrease adipose tissue mass and increase food intake, whereas glucocorticoids have been shown to increase adipose tissue mass (15). These hormones have receptors in hypothalamic nuclei and modulate food intake and body weight by regulating transcriptional processes for anabolic/orexigenic and catabolic/anorexigenic neuropeptides (3, 7, 8, 15) and may interact to modulate BDNF expression.
To elucidate hormone/neuropeptide interactions, hypothalami from divergently selected fat and lean chicken lines, which accumulate differences in abdominal fat without simultaneous alterations in food intake (28, 46), were isolated, and levels of mRNA for genes known to modulate energy homeostasis were determined. These chicken lines were selected because their body composition diverges without differences in food intake. This allows for dissociation of food intake from development of adiposity, since both genetic and environmental factors contribute to energy homeostasis (18). Next, we demonstrated in vivo and in vitro that T3 and BDNF directly and reciprocally modulate hypothalamic expression of anabolic/orexigenic and catabolic/anorexigenic genes associated with genetic susceptibility for obesity. Finally, we show that many effects on mRNA levels were present when Cort or T3 levels were individually increased but absent when levels of both hormones were simultaneously elevated. A novel model for differential effects of T3, BDNF, and Cort on the hypothalamic obesity gene network is proposed.
METHODS
Animals and tissue collection.
Divergently selected fat and lean chicken lines, developed at the Institut Nationale Recherches Agronomique (INRA), were used. These animal lines were described previously (28). Selective breeding for 21 generations occurred by choosing animals with the greatest and lowest amount of abdominal fat with similar body weight at 9 wk of age. Animals were then bred to maintain genetic diversity within each line. In the current study, males from each line were reared in floor pens (4.4 × 3.9 meters), and fat and lean chickens were reared together in the same pen to eliminate environmental differences. They were provided ad libitum access to food and water using conventional starter (0–3 wk, 3,125 kcal/kg metabolizable energy and 20.9% protein) or grower pellet (3–11 wk, 3,025 kcal/kg metabolizable energy and 17.9% protein) diet. The light-dark cycle was as follows: 24 h of light for the first 2 days and then 14 h of light and 10 h of dark thereafter.
Hypothalami were dissected with the aid of an illuminated magnifying lens from 1-, 3-, 5-, and 7-wk-old chickens. Initial incisions were made just anterior to the occulomotor nerve (nervus occulomotorius) and posterior to the tuberculum olfactorium, based on published descriptions and diagrams (27). Next, lateral cuts were made ~2 mm from the midline to yield a rectangular piece of tissue. This was placed on its side, and a final cut was made at a depth immediately below the subseptal organ (organum subseptale) and parallel to the basal surface of the hypothalamus. Animals were killed (n = 4 for each age and group), and the hypothalamus was immediately dissected and frozen in liquid nitrogen and stored at −80°C until further processing. Commercial-coated tube radioimmunoassays were used to measure plasma levels of thyroid hormones [T3 and thyroxine (T4)] (MP Biomedicals, Solon, OH). All procedures were reviewed and approved by the Institutional Animal Care and Use Committees at the University of Maryland, University of Delaware, and INRA.
Neuronal cell culture.
Hypothalami were dissected in a saggital plane using the anterior commisure and the oculomotor nerve as neuroanatomical markers from embryonic day 20 chicken embryos (Ross broiler strain). Embryonic day 20 hypothalami are primarily independent of myelination, which begins around embryonic day 14 but occurs primarily after hatch since only 5% myelination is observed in 3-day-old chicks (43). Dissections were conducted in HBSS supplemented with 10 mM HEPES and sodium pyruvate (0.011 μl/ml). Neurons were dissociated in 0.05% trypsin in minimum essential medium for 60 min and then treated with 0.1% soybean trypsin inhibitor in Dulbecco/Vogt modified Eagle's minimal essential medium for 15 min. Neurons were cultured in neurobasal medium supplemented with 2% B-27, 0.25% l-glutamine, and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA) (2 × 106 cells/well) at 37°C for 4 days with 10−9 M T3 or 10 ng BDNF (Invitrogen) added 0, 1, 3, 6, or 24 h before harvesting. Culture plates were coated with poly-l-lysine before use. Cells were harvested with 0.05% trypsin, centrifuged, and snap-frozen in liquid nitrogen.
Thyroid hormone manipulations in vivo.
Fertile chicken eggs were injected two times, once on embryonic day 18 and once on embryonic day 19, with 0.1 ml of 10−7 M T3, 0.075 g/ml methimazole (MMI; a thyroid hormone synthesis inhibitor), or a similar volume of water in the egg albumen. A hole was drilled through the shell at the bottom of the egg for injection, and then covered with wax until the injection on the next day, with the wax being replaced each time. Hypothalami were then collected on embryonic day 20 as described above for the neuronal cell cultures.
In a second study, male chickens were given a subcutaneous implant of an osmotic minipump (Alzet, model 2001; ALZA, Mountain View, CA) at 29 days of age to continuously release hormone for up to 7 days at a rate of 1.0 μl/h. Cort and T3 hormones (Sigma Chemicals, St. Louis, MO) were dissolved in solutions of 50% dimethyl sulfoxide and 50% propylene glycol, so as to deliver 600 μg Cort·kg−1·day−1 and 192 μg T3·kg−1·day−1, respectively. Animals were given ad libitum access to food and water. The light-dark cycle was as follows: 24 h of light for the first 2 days and then 14 h of light and 10 h of dark thereafter. Animals were killed after 72 h of hormone or vehicle treatment, with the hypothalamus being immediately dissected, snap-frozen in liquid nitrogen (n = 4 for group), and stored at −80°C until further processing.
RNA extraction and real-time reverse transcription PCR.
Whole hypothalamic tissue RNA was extracted using RNeasy Midi kits, and RNA from cell cultures was extracted using RNeasy Mini kits (Qiagen, Valencia, CA). RNA was quantified using ultraviolet absorbance (260/280 nm) and with a bioanalyzer (Agilent Technologies, Palo Alto, CA). Superscript III reverse transcriptase (Invitrogen) and an oligo(dT) primer were used to create cDNA from 400 ng of RNA for cell culture and 1 μg of RNA for whole hypothalami experiments. mRNA levels were quantified with Sybr green real-time reverse transcription PCR (qPCR) master mix containing the following: 1 unit of Taq polymerase, 20× SYBR green I, 400 nM fluoroscein, 10 mM dNTPs, 25 mM MgCl2, 10× PCR buffer, and water.
The qPCR output provided a cycle threshold (Ct) value. The ΔΔCt value was generated using geNorm software and methods previously described (49). Briefly, the data were first transformed to a ΔCt value by subtracting the sample Ct value from the sample with the highest expression level to control for amplification efficiency. The ΔΔCt value was then calculated by normalizing gene expression to two housekeeping genes, β-actin and glyceraldehyde-3-phosphate dehydrogenase, with the provided software (URL: http://medgen.ugent.be/~jvdesomp/genorm/).
Primer design, validation, and identification of glucocorticoid and thyroid hormone response elements.
Primers were 18–30 nucleotides in length with a melting temperature between 58–64 or 69–72°C and were designed using Primer Express (version 2.0; Applied Biosystems, Foster City, CA). The amplicons were between 100 and 150 bp in length. Forward and reverse primer sequences are listed in Table 1. Glucocorticoid and thyroid hormone response elements were identified using TESS (Transcriptional Element Search System; University of Pennsylvania, URL: http://www.cbil.upenn.edu/tess). Default parameters were used. Sequences were first identified in NCBI having the following annotations: agouti-related protein (AGRP), NM_001031457.1; BDNF, NM_001031616.1; corticotropin-releasing hormone (CRH), XM_418279.2; leptin receptor (LEPR), NM_204323.1; neuropeptide Y (NPY), NM_205473.1; pro-opiomelanocortin (POMC), NM_001031098.1; BDNF tyrosine kinase receptor B (TrkB), NM_205231.1; and TRH, NM_001030383.1. The sequence was blasted to the chicken genome using Ensembl (URL: http://www.ensembl.org), and 2,000 bp of available sequence upstream of the 5′-region of exon 1 for each gene was identified and searched for a glucocorticoid (GRE) or thyroid hormone (TRE) response element sequence. Potential GRE- or TRE-binding sites were scored using the TESS standard parameters and identified to be either a good match, log-likelihood score within one of the best possible score, or a mismatch. All sites with a good match are listed in Table 2.
Table 1.
Primer sequences used for quantitative real-time PCR
| Gene | Sense Primer | Antisense Primer |
|---|---|---|
| AGRP | 5′-AGAGCGGACCGTGAGGACACTT-3′ | 5′-GTTGGCATTTCCTCCCAAAGGA-3′ |
| AMPK | 5′-TCTCCGCGGTGGATTACTGT-3′ | 5′-AGCAGCACGTTCTCTGGTTTC-3′ |
| BDNF | 5′-TGGGTAACAGCAGCGGAGAA-3′ | 5′-TATTGCTTCAGTTGGCCTTTAG-3′ |
| CART | 5′-CACCTGCCCGAACTTCTTCTCGTA-3′ | 5′-CCCGAGAGAAGGAGCTGATCGA-3′ |
| CCKr | 5′-CGCACCGTCACCAACTCTTT-3′ | 5′-GAAGACGAAGGTGCCCATGA-3′ |
| CRH | 5′-CACAGCAACAGGAAACTGATGGAAA-3′ | 5′-AAAGAGGTGACATCAGAGCAGCACTATG-3′ |
| LEPR | 5′-AAAACCCAGAGCGTAGCGTCCAA-3′ | 5′-TTGCTTACGCGATCGTTCACAAG-3′ |
| MC4R | 5′-CGGGAGGCTGCTATGAACAA-3′ | 5′-AGCTGATGATGCCCAGAGTCA-3′ |
| MCH | 5′-GATTCCAGACATGACTTCTCAAATCATGGT-3′ | 5′-TCAGTGTCAGCTGGAAAAGCAATGG-3′ |
| NPY | 5′-GGGAAAGCACAGAAAACATTCC-3′ | 5′-AAATCCCATCACCACATCGAA-3′ |
| Orexin | 5′-CACGCTGAGAAGGACCTGACCAA-3′ | 5′-CCAGGGCCACAGGGAGGTATTTAA-3′ |
| POMC | 5′-AGGGACCTCAGGGATCATCAA-3′ | 5′-TGTTCAAGGGCAGGTTGGA-3′ |
| TRH | 5′-AGCATCTTTTGGAGACATTCAG-3′ | 5′-CAGCTCCAGGTAGTTGACAAGGT-3′ |
| TrkB | 5′-GTCCTGGGTGCTCACTAACCTT-3′ | 5′-TTATGGTTAACGAGGCAGGATTC-3′ |
| GAPDH | 5′-AAGGAGTGAGCCAAGCACACA-3′ | 5′-TCACTGCAGGATGCAGAACTG-3′ |
| β-Actin | 5′-CCCAAAGCCAACAGAGAGAAG-3′ | 5′-ACCATCACCAGAGTCCATCAC-3′ |
AGRP, agouti-related protein; AMPK, AMP-activated protein kinase; BDNF, brain-derived neurotrophic factor; CART, corticosterone- and amphetamine-regulated transcript; CCKr, cholecystokinin receptor; CRH, conrtcotropin-releasing hormone; LEPR, leptin receptor; MC4R, melanocortin 4 receptor; MCH, melanin-concentrating hormone; NPY, neuropeptide Y; POMC, pro-opiomelanocortin; TRH, thyrotropin-releasing hormone; TrkB, BDNF receptor; GAPDH, glycealdehyde-3-phosphate dehydrogenase.
Table 2.
Potential GRE and TRE located upstream of the 5′-start site for each gene
| Gene | TRE | # | bp from 5′ start | GRE | # | bp from 5′ start |
|---|---|---|---|---|---|---|
| AGRP | Yes | 3 | −345: TGACCT, −709: AGGTCA, −777: AGGACA | Yes | 36 | −15: CAGAG, −60: CAGAG, −71: GAAGA, −89: CTCTG, −252: TGTCCC, −264: AGAACA, −315: TGTGCC, −318: TGTTGT, −325: TGTTGT, −372: CTCTG, −452: TGTGGCACA, −479: TCTTCT, −538: GGGACA, −571: GGGACA, −593: CTCTG, −712: CAGAG, −741: CAGAG, −765: CAGAG, −777: AGGACA, −872: CAGAG, −1011: GGGACA, −1104: GGGACA, −1141: CAGAG, −1215: CAGAG, −1224: CTCTG, −1320: CTCTG, −1336: CAGAG, −1420: CTCTG, −1476: GGACA, −1615: CAGAG, −1730: TGTGAT, −1782: TGTGAT, −1808: TGTGAT, −1834: TGTGAT, −1912: TGTGAT, −1990: TGTGAT |
| BDNF | Yes | 2 | −727: TGTCCT, −594: TGACCT | Yes | 29 | −8: AGAAGA, −41: TCTTCT, −115: CTCTG, −168: CAGAG, −260: GGCACA, −276: ATCACA, −304: AGTTCA, −352: AGTTCT, −509: CTGTTCT, −578: CAGAG, −638: TGTTCT, −656: AGTTCT, −727: TGTCCT, −802: CTCTG, −820: AGAACAG, −874: TGTTGT, −916: CTGTTCT, −952: AGAACT, −1001: AGAACT, 1025: TGTCCC, −1069: CAGAG, −1195: CTGTTCT, −1220: TGTGAT, −1254: CTCTG, −1289: TGTTGT, −1346: AGAACT, −1422: TGAACT, −1579: TGTGAT, −1852: ATCACA |
| CRH | Yes | 1 | −315: TGACCT | Yes | 10 | −48: AGTTCT, −206: CAGAG, −1734: TGTTCT, −1738: TGTTGT, −1816: CAGAG, −1839: TGTGAT, −1867: GGCACA, −1878: TGTTCT, −1976: CTCTG, −1983: GACACA |
| LEPR | Yes | 3 | −257: TGTCCT, −680: TGTCCT, −1369: TGTCCT | Yes | 15 | −131: CAGAG, −257: TGTCCT, −286: TGTTCT, −341: CAGAG, 407: TGTGCC, −682: TGTGTC, −782: CAGAG, −805: CTCTG, −846: CAGAG, −1164: AGAACA, −1182: CAGAG, −1210: AGAACT, −1369: TGTCCT, −1610: TGTGAT, −1809: TGTGTC |
| NPY | Yes | 2 | −1803: TGTCCT, −1964: TGACCT | Yes | 16 | −607: AGGACA, −635: GGCACA, −644: TGTTGT, −687: CAGAG, −720: TGTGCC, −867: CAGAG, −1081: CTCTG, −1123: AGAACT, −1370: AGAAGA, −1462: ATGTTCT, −1529: GGGACA, −1589: GACACA, −1650: AGTTCA, −1803: TGTCCT, −1853: CAGAG, −1896: AGTGTTCT |
| POMC | No | 0 | Yes | 13 | −160: CAGAG, −378: CAGAG, −733: CTCTG, −812: TGTGCC, −924: CTCTG, −1033: CTCTG, −1119: CTCTG, −1190: CAGAG, −1264: CTCTG, −1297: GGGACA, −1586: CAGAG, −1901: TGTGAT, −1985: TGTGCCACA | |
| TRH | Yes | 3 | −184: AGGTCA, −1555: TGTCCT, −1836: AGGACA | Yes | 27 | −22: CAGAG, −44: CAGAG, −138: CTCTG, −169: CAGAG, −210: AGTTCT, −328: AGAAGA, −348: CTCTG, −595: ACAACA, −646: AGAAGA, −713: TGTGAT, −824: AGAAGA, −893: AGAACT, −931: CTCTG, −1017: AGAACA, −1115: TCTTCT, −1149: AGAACT, −1232: AGAAGA, −1318: ACAACA, −1386: AGTTCA, −1480: CTCTG, −1532: TGAACT, −1540: CTCTG, −1555: TGTCCT, −1699: AGAACA, −1720: CTCTG, −1836: AGGACA, −1896: TGTGTC |
| TrkB | Yes | 3 | −1067: AGGTCA, −1108: AGGTCA, −1820: AGGACA | Yes | 19 | −273: AGAAGA, −509: GGGACA, −786: GGGACA, −1080: GACACA, −1130: TGTGAT, −1137: ACAACA, −1232: AGTTCA, −1358: CTCTG, −1368: TGTTCT, −1447: AGAACAG, −1511: TGTGTC, −1531: AGAACAG, −1546: ATGTTCT, −1586: CAGAG, −1722: AGAAGA, −1820: AGGACA, −1905: TGTGTC, −1911: CAGAG, −1945:ACAACA |
GRE, glucocorticoid response element; TRE, thyroid hormone response element. #No. of potential GREs or TREs identified and the base pair number from the 5′-region of exon 1 for each gene. Items in bold indicate sites identified as both a GRE and TRE (AGGACA or TGTCCT).
Statistical analysis of data.
Differences in gene expression for all experiments were tested using a one-way (cell culture treatment) or two-way (line by age) ANOVA, with a PDIFF post hoc analysis used to identify differences between the groups (version 8.02, Statistical Analysis System; SAS Institute, Cary, NC). Values reported are means ± SE, and P <0.05 was required for statistical significance.
RESULTS
Hypothalamic anabolic/orexigenic and catabolic/anorexigenic gene expression differs in fat and lean animals.
Hypothalami were collected from fat and lean chickens at weeks 1, 3, 5, and 7 of age to measure differences in gene expression. Increased abdominal fat developed by week 3 in the fat relative to lean line (2.3 ± 0.1 and 1.0 ± 0.1% of body weight, respectively; P < 0.05) with no differences in abdominal fat at week 1 of age (0.4 ± 0.1 and 0.3 ± 0.1%, respectively), providing a baseline to compare physiological alterations before and after the onset of the phenotypes. Percent abdominal fat continued to diverge between the fat and lean lines until week 7 (4.4 ± 0.2 and 1.6 ± 0.1%, respectively; 88 ± 3.0 and 31 ± 2.0 g, respectively; P < 0.05). We measured hypothalamic mRNA levels for selected genes and plasma levels of T3 and T4. The genes selected were chosen because they are known to modulate energy homeostasis in mammals and are likely to play a similar role in birds (26, 44). From this initial screen, we identified seven genes with significant differences in expression between the divergently selected chicken lines: TrkB, TRH, LEPR, POMC, AGRP, NPY, and CRH (Fig. 1). Cocaine and amphetamine-regulated transcript (CART) approached significance (P = 0.055). T3 levels were modestly increased at week 1 in the fat line, which resembled data from morbidly obese euthyroid humans (34), as well as previous observations in the fat and lean lines (29). As differences in adiposity began to emerge, T3 differences disappeared between the lines. No differences were observed for T4 between the lines at any age (data not shown).
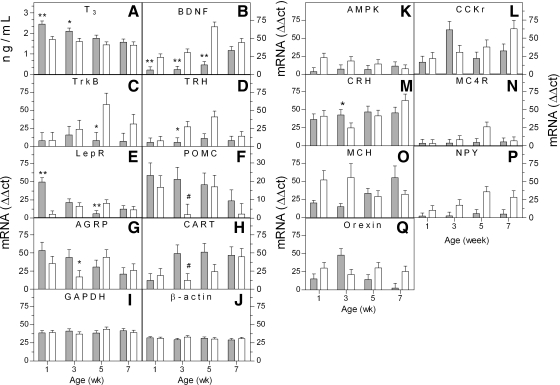
Hormone and neuropeptide levels in fat and lean Lines during development of adiposity. Plasma and hypothalami were collected from the fat (filled bar) and lean (open bar) chicken lines at 1, 3, 5, and 7 wk of age. A: plasma thyroid hormone levels in fat and lean lines. Endogenous triiodothyronine (T3) levels were increased in the fat relative to lean line at weeks 1 and 3 (n = 4). B–J: quantification of hypothalamic brain-derived neurotrophic factor (BDNFl; B), BDNF tyrosine kinase receptor B (TrkBl; C), thyrotropin-releasing hormone (TRHl; D), leptin receptor (LEPR; E) pro-opiomelanocortin (POMCl; F), agouti-related protein (AGRP; G), cocaine- and amphetamine-regulated transcript (CART; H), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; I), β-actin (J), α2-subunit of AMP-activated protein kinase (AMPK; K), cholecystokinin-B receptor (CCKr; L), corticotropin-releasing hormone (CRH; M), melanocortin-4 receptor (MC4R; N), melanin-concentrating hormone (MCH; O), neuropeptide Y (NPYl; P), and orexin (Q) by quantitative real-time reverse transcription (qPCR) from weeks 1, 3, 5, and 7 of age (n = 4). *P < 0.05, **P < 0.01, and #P < 0.055 fat relative to lean line for the same age. lOverall significant difference (P < 0.05) between lines for BDNF, TrkB, TRH, POMC, and NPY across all ages.
T3 and BDNF modulate anabolic/orexigenic and catabolic/anorexigenic gene expression in a complementary manner.
We next assessed whether the different levels of circulating T3 and hypothalamic BDNF mRNA noted between the fat and lean lines could account for any of the other differences in hypothalamic gene expression. Hypothalamic neuronal cultures were treated with 10−9 M T3 or 10 ng BDNF for 0, 1, 3, 6, and 24 h and assayed with qPCR for mRNA levels. Primary hypothalamic neurons were cultured for 4 days, since BDNF mRNA levels initially declined and then stabilized by day 3 in vitro (data not shown), and neurons grown 4 days in vitro have not yet formed synaptic connections with other neurons, implying that results reflect treatment effects on mRNA levels independent of stimulation received from synaptic interactions with other neurons.
Treating hypothalamic neurons with T3 suppressed expression of BDNF, TrkB, and TRH mRNA, relative to untreated controls (0 h) (Fig. 2, A, C, and E, respectively). Treatment of neurons with 10 ng of BDNF modulated expression of the same genes in the opposite direction, relative to T3 treatment (Fig. 2, B, D, and F, respectively). β-Actin was not different after either treatment (Fig. 2, M and N). This indicated that T3 and BDNF regulate expression of some common target genes in an opposite way. Because LEPR is one gene known to modulate many other genes influencing body composition and food intake (21), we determined whether similar effects on LEPR would be seen. T3 suppressed LEPR gene expression after 1, 3, 6, and 24 h (Fig. 2G), and BDNF increased expression of the LEPR gene after 6 h (Fig. 2H). This suggests that BDNF and T3 reciprocally modulate expression of a key gene in the hypothalamic obesity gene circuit.
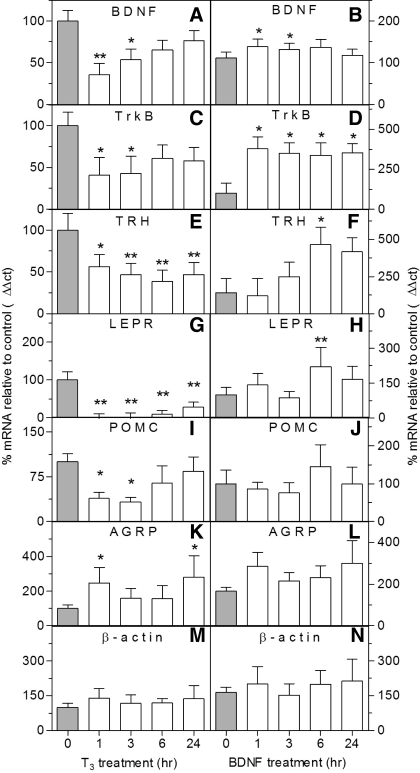
T3 and BDNF have opposite effects in vitro on hypothalamic genes known to regulate energy homeostasis. A, C, E, G, I, K, and M: T3 treatment decreased BDNF (A), TrkB (C), TRH (E), LEPR (G), and POMC (I) gene expression and increased expression of AGRP (K), but not β-actin (M), mRNA levels (n = 4–10). B, D, F, H, J, L, and N: BDNF treatment increased gene expression of BDNF (B), TrkB (D), TRH (F), and LEPR (H) relative to control, but not POMC (J), AGRP (L), and β-actin mRNA levels (N) (n = 4–6). Expression levels are normalized to a control value of 100%. *P < 0.05 and **P < 0.01, treatment vs. 0-h control.
Given that T3 and BDNF reciprocally modulated expression for the catabolic/anorexigenic genes BDNF, TrkB, TRH, and LEPR, we proposed that T3 would suppress the other catabolic/anorexigenic genes in our gene list, POMC and CRH, and increase the anabolic/orexigenic genes, AGRP and NPY, whereas BDNF would have the opposite effects. Indeed, T3 suppressed POMC mRNA at 1 and 3 h (Fig. 2I) and increased AGRP expression at 1 and 24 h (Fig. 2K). However, BDNF had no effect on these genes (Fig. 2, J and L, respectively). BDNF and T3 had no effect on NPY or CRH (data not shown). This suggests that T3 directly suppresses expression of the catabolic/anorexigenic gene, POMC, and increases expression of the anabolic/orexigenic gene, AGRP. BDNF was able to increase expression of most catabolic/anorexigenic genes directly (BDNF, TrkB, TRH, and LEPR).
We tested whether T3 was able to modulate these same genes during development, in vivo. The dose of MMI, a thyroid hormone synthesis inhibitor, used suppressed serum T4 levels to an undetectable level on embryonic day 17, embryonic day 19, and day 1 relative to control in a previous study using a similar injection paradigm (37). We injected T3 or MMI in developing chicken embryos on embryonic days 18 and 19 and collected hypothalami on embryonic day 20. We observed effects on BDNF, TrkB, and TRH mRNA levels (Fig. 3, A–C, respectively) consistent with those observed in vitro, but, surprisingly, both T3 and MMI decreased LEPR, increased POMC mRNA levels, and had no effect on AGRP (Fig. 3, D–F, respectively).
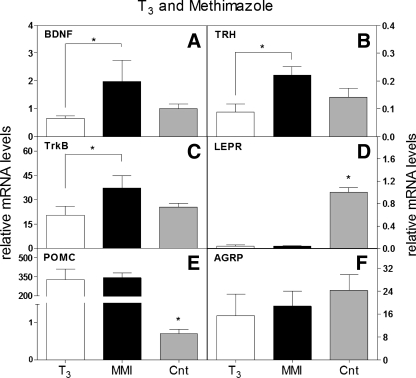
T3 reciprocally modulates catabolic/anorexigenic and anabolic/orexigenic genes. A-F: T3 was injected on embryonic days 18 and 19, and effects on hypothalamic gene expression were assessed on embryonic day 20. T3 suppressed BDNF (A), TrkB (B), TRH (C), and LEPR (D) gene expression, whereas methimazole (MMI) increased BDNF (A), TrkB (B), and TRH (C), but not LEPR (D), mRNA levels. T3 and MMI both increased POMC (E) and had no affect on AGRP (F) mRNA (n = 6). *P < 0.05 and **P < 0.01. CNT, control (water).
T3 and Cort play both overlapping and independent roles in modulating anabolic/orexigenic and catabolic/anorexigenic gene expression.
Although it is known that thyroid hormone and Cort act in opposition to one another, with Cort being considered an anabolic hormone and T3 a catabolic hormone, they have been shown to have synergistic effects when used in combination. For example, T3 treatment alone slightly enhanced protein synthesis in muscle, whereas Cort treatment increased protein breakdown, and Cort in conjunction with thyroid hormone enhanced protein breakdown in muscle sixfold (17). Under states of chronic stress, glucocorticoids have been shown to increase food intake, which may be because of a dysregulation of normal mechanisms that modulate food intake (1). When glucocorticoid signaling is no longer effective, increased food intake mimics the food intake phenotype observed when thyroid hormone levels are elevated; however, adiposity is still regulated in opposite directions. Therefore, we wanted to determine which hypothalamic genes known to regulate both food intake and body weight are modulated in the same or different directions after chronic 72-h treatment with Cort, T3, or Cort plus T3.
Hormones were delivered via osmotic minipump for 72 h. T3 decreased expression of the catabolic/anorexigenic neuropeptide genes TRH, BDNF, TrkB, LEPR, and POMC (Fig. 4, A–E). T3 also increased expression of AGRP, an anabolic/orexigenic neuropeptide (Fig. 4F). Delivering T3 via osmotic minipump had no effect on NPY or CRH (Fig. 4, G and H), as previously demonstrated in cell culture. This suggests that in vivo T3 treatment is able to suppress expression of catabolic/anorexigenic genes and increase expression of an anabolic/orexigenic gene. There was no change in β-actin gene expression after any of the treatments (Fig. 4I).
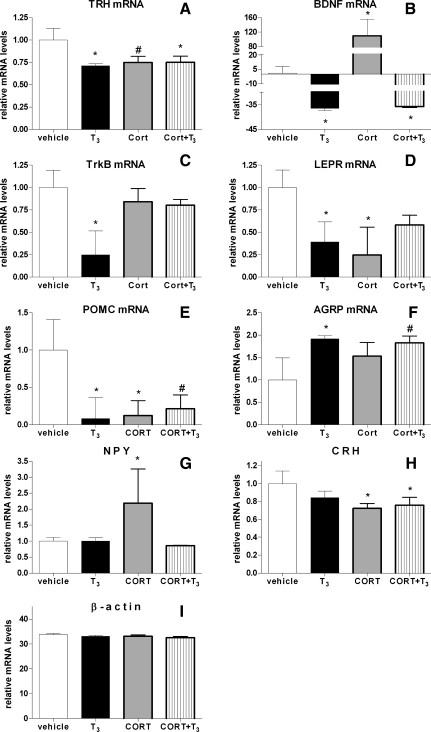
Anabolic/orexigenic and catabolic/anorexigenic gene expression in the hypothalamus after 72 h of hormone treatment. Corticosterone (Cort) and T3 were administered for 72 h using osmotic minipumps, and then hypothalami were collected. A-H: quantification of hypothalamic BDNF (A), TrkB (B), TRH (C), LEPR (D), POMC (E), AGRP (F), NPY (G), CRH (H), and β-actin (I) mRNA levels (n = 4) by qPCR analysis. *P < 0.05, hormone treatment relative to vehicle control.
We expected to observe the opposite effects as those seen with T3 with Cort treatment. Surprisingly, for many of the genes, we observed a similar expression pattern as seen after T3 treatment. For example, we observed that Cort decreased mRNA levels for the catabolic/anorexigenic neuropeptides TRH, LEPR, and POMC (Fig. 4, A, D, and E). On the other hand, opposite to that observed with T3, Cort increased levels of BDNF mRNA (Fig. 4B). Cort had no effect on TrkB or AGRP (Fig. 4, C and F, respectively), and, unlike T3, Cort increased mRNA levels for the anabolic/orexigenic neuropeptide NPY and decreased expression for the catabolic/anorexigenic neuropeptide CRH (Fig. 4, G and H).
Delivering Cort in conjunction with T3 had surprising effects on anabolic/orexigenic and catabolic/anorexigenic gene expression in the hypothalamus. Similar to T3 treatment alone, T3 plus Cort decreased expression of TRH and BDNF mRNA (Fig. 4, A and B). Decreased POMC and increased AGRP expression had only a trend toward significance, which is surprising since T3 or Cort alone was able to decrease POMC mRNA levels, and T3 alone was able to increase AGRP mRNA levels (Fig. 4, E and F). Surprisingly, simultaneous treatment with Cort and T3 had no effect on TrkB, LEPR, and NPY (Fig. 4, C, D, and G, respectively). As predicted, Cort treatment, even in conjunction with T3 delivery, decreased expression of CRH (Fig. 4H).
GRE and TRE hybrid response elements identified in anabolic/orexigenic and catabolic/anorexigenic genes.
Glucocorticoid and thyroid hormone hybrid response elements were previously shown to selectively bind thyroid hormone receptor complexes (51). Therefore, we sought to identify whether endogenous hybrid response elements containing either or both glucocorticoid and thyroid hormone response elements (GRE and TRE, respectively) existed in the 5′-upstream region of the anabolic/orexigenic and catabolic/anorexigenic genes differentially regulated by Cort and/or T3 treatment. Putative response elements containing both a GRE and TRE were identified in the 2,000-bp region upstream of exon 1 for the following genes: AGRP, BDNF, CRH, LEPR, NPY, POMC, TRH, and TrkB (Table 2). POMC was the only gene that did not have a TRE site in this region. AGRP, LEPR, TRH, and TrkB contained the most TRE sites, and AGRP contained the most GRE sites, regardless Cort treatment over 72 h did not alter AGRP expression, suggesting that regulation of gene expression may occur before 72 h or that the GRE sites in the AGRP gene are not functional. POMC was the only gene that did not contain a TRE, yet T3 treatment was able to suppress gene expression, suggesting that inhibition of POMC expression does not occur directly or through this region of its gene. Six out of the eight genes (AGRP, BDNF, TrkB, NPY, LEPR, and TRH) had potential hybrid response elements for both the glucocorticoid and thyroid hormone receptors (Table 2), perhaps providing an explanation for the gene expression pattern observed when the levels of both hormones were elevated simultaneously. However, because AGRP did not respond to Cort treatment, the hybrid response element identified in AGRP is likely to function primarily as a TRE and not a GRE.
DISCUSSION
Many neuropeptides in the hypothalamus have been implicated in energy homeostasis, some increasing fat deposition (anabolic/orexigenic) and some decreasing fat deposition (catabolic/anorexigenic). Here we dissociated genetic from environmental influences on obesity by utilizing divergently selected fat and lean lines of chickens in which food intake does not differ (28, 46). After screening 14 hypothalamic genes known to modulate body composition and food intake, we identified key candidate genes likely to influence the genetic predisposition for increased adiposity. These included BDNF, TrkB, TRH, POMC, NPY, and LEPR. BDNF and T3 reciprocally modulated anabolic/orexigenic and catabolic/anorexigenic gene expression in the hypothalamus. To our knowledge, this is the first example of a neuropeptide and a hormone working in a complementary fashion to regulate genes in the hypothalamic obesity gene network.
BDNF and T3 affected hypothalamic gene expression in a complementary fashion.
Our results show that T3 was able to increase anabolic/orexigenic gene expression (AGRP) and decrease catabolic/anorexigenic gene expression (BDNF, TrkB, TRH, LEPR, and POMC) both in vivo and in vitro. BDNF was able to directly modulate catabolic/anorexigenic genes in vitro in the opposite direction as T3, with the exception of POMC and AGRP. However, it has been previously shown that leptin can suppress AGRP and increase POMC gene expression (25), and, since T3 and BDNF reciprocally modulated LEPR gene expression, there may be indirect modulation of AGRP and POMC gene expression via LEPR. We propose a model based on our in vivo and in vitro data showing reciprocal regulation of the hypothalamic obesity gene circuit by T3, BDNF, and Cort (Fig. 5). Collectively, our results suggest that levels of BDNF, T3, and Cort may explain how some individuals are more susceptible to develop an obese phenotype without altering food intake, as observed in the euthyroid morbidly obese patients with moderately elevated T3 levels (34).
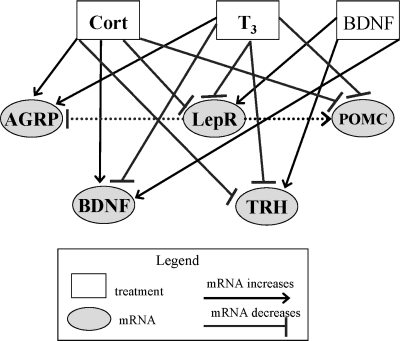
Model for BDNF, T3, and Cort interactions modulating the hypothalamic obesity gene network. We propose that elevated systemic T3 levels decrease BDNF, TRH, LEPR, and POMC mRNA levels but increase AGRP gene expression in the hypothalamus. In contrast, we propose that BDNF reciprocally modulates expression of the same genes by increasing BDNF, TRH, and LEPR mRNA levels. BDNF may also indirectly modulate POMC and AGRP mRNA levels by first increasing LEPR or TRH expression. In addition, we propose that systemic glucocorticoids (Cort) can increase hypothalamic expression of AGRP and BDNF and decrease expression of LEPR, POMC, and TRH. These effects culminate in a neuropeptide/endocrine homeostatic mechanism that contributes to the regulation of metabolism and energy expenditure.
Our results from the divergently selected fat and lean chicken lines parallel the cell culture and osmotic minipump data, suggesting that BDNF or T3 levels may affect hypothalamic gene expression during the development of adiposity. Corresponding with predictions presented in our model, plasma T3 was increased and BDNF expression decreased in the fat line relative to the lean line before and at the onset of adiposity divergence (weeks 1 and 3). An interaction between BDNF and T3 was also demonstrated in vitro. Hypothalamic TRH mRNA levels were lower in the fat line at week 3, suggesting that increased BDNF expression in the lean line could increase expression of TRH, or increased T3 in the fat line could inhibit expression of TRH, similar to the in vitro data. Finally, hypothalamic BDNF mRNA levels continued to increase at week 5 in the lean line relative to the fat line, which corresponds to an increase in TrkB and LEPR gene expression. In vitro, BDNF increased TrkB and LEPR mRNA levels. Our findings support a novel pathway that contributes to hypothalamic control of constitutive differences in the capacity for body fat deposition.
Complex effects of Cort and thyroid hormone on hypothalamic gene expression.
Cort and T3 had overlapping effects on hypothalamic mRNA levels for genes known to modulate food intake and body weight, specifically TRH, LEPR, and POMC. We also found that Cort increased and T3 decreased BDNF gene expression. Some genes were only regulated by one hormone; Cort increased NPY mRNA levels, whereas T3 had no effect; T3 decreased TrkB mRNA, whereas Cort had no effect on TrkB expression; and T3 increased AGRP mRNA, whereas Cort had no effect. These effects agree with increased Cort levels resulting in anabolic/orexigenic activity by increasing expression of the anabolic/orexigenic gene NPY. On the other hand, increased T3 levels generally increase catabolic activity, which would explain the downregulation of the anabolic/orexigenic gene AGRP.
Glucocorticoids and thyroid hormones are known to modulate adipose mass and food intake. Increased glucocorticoid levels decrease insulin levels, leptin levels, and POMC mRNA levels but increase AGRP and NPY mRNA in the hypothalamus (33). Thyroid hormone increases leptin hormone and NPY mRNA levels and decreases CART and POMC mRNA (20). As predicted, we found that Cort decreased POMC levels and increased NPY levels, whereas T3 treatment decreased POMC. Surprisingly, Cort had no effect on AGRP mRNA levels, and T3 had no effect on NPY mRNA levels. These unexpected differences may be a species-specific phenomenon or attributable to the duration of the treatment.
Thyroid hormone and glucocorticoids modulate adiposity in the opposite direction, but both have been shown to increase food intake. Thus we wanted to determine how increased levels of both Cort plus T3 would modulate gene expression patterns. This allows elucidation for whether T3 or Cort predominantly regulates gene expression for anabolic/orexigenic and catabolic/anorexigenic genes, perhaps providing an explanation for the opposite effects observed on body weight or similar effects on food intake. BDNF mRNA levels were regulated by both T3 and Cort, with T3 inhibiting and Cort increasing expression levels. When treating with T3 plus Cort, BDNF mRNA levels resembled the inhibitory pattern observed after treating with T3 alone. NPY mRNA levels were increased after Cort treatment, not changed after T3 treatment alone, and Cort plus T3 ablated the increased gene expression. This suggests that T3 is the primary regulator for BDNF gene expression and that T3 can also block the increased expression of NPY mRNA observed when Cort was added alone. On the other hand, CRH gene expression was decreased when either Cort was added alone and when Cort was added with T3, as predicted.
Last, identification of putative transcription factor binding sites revealed that BDNF, TrkB, NPY, AGRP, TRH, and LEPR have sites that may function as a dual TRE/GRE. However, the functionality of these sites is currently unknown. If the sites are active, it is possible that, if the GRE site induces transcription, then T3 may inhibit transcription by competitively binding to the site, or perhaps binding with a higher affinity. TRH and LEPR are least likely to have a functional GRE/TRE since Cort treatment alone had no effect on TRH mRNA and since Cort plus T3 only partially decreased LEPR gene expression. It must be noted that the functionality of these elements remains to be confirmed.
Paradoxically, T3 decreased POMC gene expression despite the lack of TRE sites within 2,000 bp upstream of exon 1. It is also possible that T3 could increase expression of POMC mRNA indirectly by increasing expression of another, unknown, protein. However, we observed that POMC mRNA levels increased after treating primary neuronal cultures for 1 and 3 h, suggesting direct effects on POMC gene expression by T3. The most plausible explanation is that regulatory TREs are located elsewhere in the POMC gene. It is uncertain why MMI-treated embryos would increase POMC expression relative to the observed decrease in expression observed later in life, as demonstrated by osmotic minipump treatment. The different experimental paradigms used to evaluate thyroid hormone effects on gene expression in the hypothalamus are suggestive of direct effects; however, the link between direct effects suggested by cell culture relative to possible indirect effects in the osmotic minipump experiment should be interpreted conservatively. Moreover, technical considerations limited our ability to test multiple doses and durations of thyroid hormone treatment in the minipump experiment. Nonetheless, effects of thyroid hormone on hypothalamic gene expression were demonstrated in vivo.
Potential role for regulating energy balance.
During the development of adiposity, it is plausible that interactions between T3 and Cort program an initial set point within the hypothalamus to establish long-term levels of body fat stores, possibly by regulating the maturation of homeostatic signaling circuitry (e.g., neural, hormonal, or metabolic). For example, in the fat and lean animals, one mechanism altered during the development of adiposity could be initially signaled by T3 or BDNF and secondarily by metabolic parameters, such as glucose utilization (10, 30, 45), lipogenesis (42), or fuel partitioning (31). Currently, the rate of neuropeptide release in response to T3 or BDNF is unknown and may modulate neural circuitry more than protein or mRNA production. Furthermore, interactions between BDNF and LEPR signaling may modulate fat deposition and metabolism. In the mouse, leptin increased hypothalamic BDNF mRNA levels (23), and here we show that BDNF increased hypothalamic LEPR mRNA. In the fat line relative to the lean line, liver leptin levels are reportedly increased 1.7-fold at 15 wk of age (9). However, a controversy about the validity of this identified chicken leptin sequence exists (13), suggesting that the results from this previous report should be interpreted cautiously. Regardless, the avian LEPR has been sequenced accurately (40). Thus BDNF and LEPR levels may modulate energy expenditure and energy balance to influence adipose deposition by affecting hypothalamic signaling networks (Fig. 5). Therefore, T3 acts in a reciprocal manner to inhibit BDNF, TrkB, LEPR, and even POMC to compensate for these changes in energy expenditure, thereby establishing a feedback loop that is enforced further by increasing AGRP expression. We propose that BDNF and T3 levels may serve as a fine tipping point for determining how the hypothalamic obesity circuitry develops, along with interactions between glucocorticoids and thyroid hormone. Elevated levels of one hormone may not necessarily modulate gene expression in the same manner as it would if the other hormone is concurrently elevated. Here we begin to elucidate the underpinnings for how hormone interactions may modulate anabolic/orexigenic and catabolic/anorexigenic gene expression in the hypothalamus.
Perspectives and Significance
There are hundreds of genes expressed in the brain that contribute to obesity. Among these, genes that are evolutionarily conserved to regulate physiological processes are strong candidates for playing principal roles in maintaining homeostasis or contributing to the onset of obesity. Given this, research in lower vertebrates is important because it provides a way to identify the primary control genes in more complex systems by first identifying which processes are conserved in divergent species. To begin to tease apart the evolutionarily conserved hypothalamic signaling pathways associated with obesity, we investigated systems present in birds and mammals, neurotrophins, and genes associated with thyroid hormone or glucocorticoid signaling. We have shown that these hormones alter more than one hypothalamic gene simultaneously, thus demonstrating the complexity of the hypothalamic obesity gene network. We have demonstrated that BDNF and T3 reciprocally modulate expression of many genes in this network, suggesting that BDNF and T3 levels may act as a fine tipping point for determining whether hypothalamic gene expression and adiposity increase or decrease over development. The implications of this are large, since thyroid hormone supplements are commonly given to treat thyroid hormone disorders, which may disrupt the balance between BDNF and T3 signaling and ultimately disrupt expression of many genes in the hypothalamus that serve to maintain energy regulation within a healthy range. These interactions between T3 and BDNF also suggest that obesity therapeutics could be developed to simultaneously target BDNF and T3 to inhibit or reverse further fat deposition.
GRANTS
This work was supported by a grant from the United States Department of Agriculture Initiative for Future Agricultural and Food Systems (Award No. 00-52100-9614; to L. A. Cogburn, T. E. Porter, and J. Simon), a grant from the Cosmos Club Foundation (to M. S. Byerly), and National Institutes of Health (NIH) National Research Service Award (F31 DK-743802; to M. S. Byerly) and a NIH graduate student training fellowship (MH-20048; to M. S. Byerly).
Notes
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
Articles from American Journal of Physiology - Regulatory, Integrative and Comparative Physiology are provided here courtesy of American Physiological Society
Full text links
Read article at publisher's site: https://doi.org/10.1152/ajpregu.90813.2008
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2698606
Free to read at intl-ajpregu.physiology.org
http://intl-ajpregu.physiology.org/cgi/content/abstract/296/4/R1180
Free after 12 months at intl-ajpregu.physiology.org
http://intl-ajpregu.physiology.org/cgi/content/full/296/4/R1180
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1152/ajpregu.90813.2008
Article citations
Effect of Dietary Benzoic Acid and Oregano Essential Oil as a Substitute for an Anti-Coccidial Agent on Growth Performance and Physiological and Immunological Responses in Broiler Chickens Challenged with <i>Eimeria</i> Species.
Animals (Basel), 14(20):3008, 17 Oct 2024
Cited by: 0 articles | PMID: 39457937 | PMCID: PMC11504159
Selective footprints and genes relevant to cold adaptation and other phenotypic traits are unscrambled in the genomes of divergently selected chicken breeds.
J Anim Sci Biotechnol, 14(1):35, 24 Feb 2023
Cited by: 7 articles | PMID: 36829208 | PMCID: PMC9951459
Effect of thermal conditioning on serum electrolytes, metabolites, corticosterone and expression of CRH gene in selected chicken strains.
J Appl Genet, 63(4):729-741, 01 Aug 2022
Cited by: 0 articles | PMID: 35913614
Insulin immuno-neutralization decreases food intake in chickens without altering hypothalamic transcripts involved in food intake and metabolism.
Poult Sci, 96(12):4409-4418, 01 Dec 2017
Cited by: 2 articles | PMID: 29053815 | PMCID: PMC5850116
Regulation of Agouti-Related Protein and Pro-Opiomelanocortin Gene Expression in the Avian Arcuate Nucleus.
Front Endocrinol (Lausanne), 8:75, 13 Apr 2017
Cited by: 21 articles | PMID: 28450851 | PMCID: PMC5389969
Review Free full text in Europe PMC
Go to all (29) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database (Showing 7 of 7)
- (1 citation) RefSeq - NM_205231.1
- (1 citation) RefSeq - NM_001030383.1
- (1 citation) RefSeq - XM_418279.2
- (1 citation) RefSeq - NM_205473.1
- (1 citation) RefSeq - NM_001031616.1
- (1 citation) RefSeq - NM_001031457.1
- (1 citation) RefSeq - NM_001031098.1
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system.
Endocrinology, 154(10):3643-3651, 26 Jul 2013
Cited by: 99 articles | PMID: 23892476
Layer and broiler chicks exhibit similar hypothalamic expression of orexigenic neuropeptides but distinct expression of genes related to energy homeostasis and obesity.
Brain Res, 1273:18-28, 01 Apr 2009
Cited by: 40 articles | PMID: 19345199
Effects of adrenalectomy on AGRP, POMC, NPY and CART gene expression in the basal hypothalamus of fed and fasted rats.
Brain Res, 958(1):130-138, 01 Dec 2002
Cited by: 105 articles | PMID: 12468037
The TRH neuron: a hypothalamic integrator of energy metabolism.
Prog Brain Res, 153:209-235, 01 Jan 2006
Cited by: 182 articles | PMID: 16876577
Review
Funding
Funders who supported this work.
NIDDK NIH HHS (4)
Grant ID: F31 DK-743802
Grant ID: F31 DK074380-02
Grant ID: F31 DK074380-01A1
Grant ID: F31 DK074380
NIMH NIH HHS (1)
Grant ID: MH-20048




