Abstract
Free full text

Myeloid Neoplasia
Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies
Abstract
Disease alleles that activate signal transduction are common in myeloid malignancies; however, there are additional unidentified mutations that contribute to myeloid transformation. Based on the recent identification of TET2 mutations, we evaluated the mutational status of TET1, TET2, and TET3 in myeloproliferative neoplasms (MPNs), chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML). Sequencing of TET2 in 408 paired tumor/normal samples distinguished between 68 somatic mutations and 6 novel single nucleotide polymorphisms and identified TET2 mutations in MPN (27 of 354, 7.6%), CMML (29 of 69, 42%), AML (11 of 91, 12%), and M7 AML (1 of 28, 3.6%) samples. We did not identify somatic TET1 or TET3 mutations or TET2 promoter hypermethylation in MPNs. TET2 mutations did not cluster in genetically defined MPN, CMML, or AML subsets but were associated with decreased overall survival in AML (P = .029). These data indicate that TET2 mutations are observed in different myeloid malignancies and may be important in AML prognosis.
Introduction
Our understanding of the molecular pathogenesis of myeloid malignancies, most notably acute myeloid leukemia (AML) and chronic myeloid leukemia (CML), has largely resulted from the identification and characterization of recurrent chromosomal translocations.1 However, in many patients with myeloproliferative neoplasms (MPNs) and chronic myelomonocytic leukemia (CMML), recurrent clonal cytogenetic abnormalities are not observed. More recently, DNA resequencing studies of candidate genes,2 gene families,3,4 and the cancer genome5 in MPN, CMML, and AML have identified somatic mutations in FLT3,6 JAK2,7–13 MPL,14,15 and the RAS family of oncogenes.16 These discoveries demonstrate activation of signal transduction pathways is a common pathogenic event in myeloid malignancies and have led to the development of molecularly targeted therapies. However, with the exception of CML, these therapies have yet to substantively improve outcomes for patients with myeloid malignancies.17,18 This may reflect insufficient target inhibition, or, alternatively, this may indicate incomplete dependence on these activated pathways resulting from the presence of additional somatic mutations with prognostic, therapeutic, and biologic relevance.
The role of TET (Ten-Eleven Translocation) family gene members in hematopoietic transformation was thought to be restricted to the involvement of TET1 as a translocation partner MLL-translocated AML, until the recent identification of inactivating mutations in TET2 in MPN and MDS patients.19 We therefore sought to evaluate a large set of MPN, CMML, and AML samples for somatic TET2 alterations. We sequenced all coding exons of TET2 in 408 paired tumor/normal samples and then assessed the frequency of somatic TET2 mutations in 606 patients with MPN, CMML, and AML. We also investigated whether deletion or epigenetic inactivation of TET2 are observed in MPN and evaluated MPN patients for somatic mutations in TET1 and TET3.
Methods
Patients
DNA was isolated from peripheral blood and/or bone marrow from 606 MPN, CMML, and AML samples. Matched normal DNA was available for 408 samples, including 354 sporadic MPN samples, 26 CMML samples, and 28 affected members of MPN pedigrees. Blood/bone marrow DNA but not matched normal DNA was available for 198 samples, including 3 sporadic MPN samples, 20 affected members of MPN kindreds, 96 AML samples, 45 CMML samples, and 34 M7 AML samples (16 from the Eastern Cooperative Oncology Group). Approval was obtained from the institutional review boards at the Dana-Farber Cancer Institute and at Memorial Sloan-Kettering Cancer Center for these studies, and informed consent was provided according to the Declaration of Helsinki.
Sequence analysis of TET1, TET2, and TET3
DNA resequencing of all coding exons of TET1-3 was performed (primers/conditions are listed in supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Nonsynonymous alterations not present in single nucleotide polymorphism (database [db]SNP) were annotated as somatic mutations or SNPs based on sequence analysis of matched germ line DNA. Nonsynonymous alterations not in dbSNP nor determined to be somatic in paired samples or in recently reported data19 were censored. All somatic mutations were validated by resequencing nonamplified DNA.
Copy number analysis of TET1, TET2, and TET3
A total of 207 MPN tumor samples were analyzed using Affymetrix 250K StyI Arrays.20 The JAK2V617F-mutant AML cell lines HEL and SET2 were analyzed using Affymetrix 6.0 SNP Arrays.
Methylation-specific polymerase chain reaction
Methylation of 2 CpG islands in the promoter region of TET2 was assessed in 37 MPN patients and 4 JAK2V617F-positive leukemia cell lines (SET2, MBO2, HEL, UKE1). Methylation-specific polymerase chain reaction was performed as previously described (primers are listed in supplemental Table 1).21
Statistics
Statistical analyses were performed using MedCalc (MedCalc).
Results and discussion
Sequence analysis of all coding exons of TET2 in 408 paired tumor/normal samples identified 8 frameshift, 12 nonsense, and 37 nonsynonymous alterations not present in dbSNP. Analysis of germ line DNA distinguished between 31 somatic missense mutations and 6 unannotated SNPs (Table 1; supplemental Figure 1); all unannotated SNPs were observed in matched normal tissue in at least 2 samples. All frameshift and nonsense mutations were not present in matched normal tissue. The strategy of paired sequencing of normal and tumor tissue is critical for accurate annotation of candidate mutations as 2 novel SNPs, which were recently reported as TET2 mutations (Q1084P and Y867H)22 were present in the germ line in multiple patient samples consistent with their being unannotated SNPs. After defining the spectrum of somatic TET2 mutations in paired tumor/normal samples, we determined the frequency of TET2 mutations in MPN (7.6%, including 9.8% polycythemia vera, 4.4% essential thrombocythemia, and 7.7% primary myelofibrosis), CMML (42.1%), AML (12.1%), and acute megakaryoblastic leukemia (3.6%). We identified biallelic/homozygous TET2 mutations in 1 of 354 MPN patients and in 7 of 69 CMML patients (P < .001, Fisher exact test). Sequencing of TET2 in 48 affected members from 28 MPN kindreds identified somatic TET2 mutations in 7 affected persons. We also identified 4 germ line nonsynonymous variants in affected members of MPD kindreds present in dbSNP that could represent rare familial MPN alleles. However, 3 of these 4 SNPs were observed in only some affected members of kindred but not others, and the fourth variant (M1701I) is observed in many sporadic MPN, CMML, and AML cases. Somatic TET2 mutations were not noted in the 4 JAK2V617F-positive leukemic cell lines.
Table 1
Novel TET2 somatic missense mutations and unannotated SNPs in 4q24
| Alteration | Nucleotide change | Genomic coordinate | Amino acid change |
|---|---|---|---|
| Somatic mutation | 434 G→A | 106374983 | S145N |
| Somatic mutation | 935 A→G | 106375484 | N312S |
| Somatic mutation | 1379 C→T | 106375928 | S460F |
| Somatic mutation | 1997 A→G | 106376546 | D666G |
| Somatic mutation | 2821 C→T | 106377370 | P941S |
| Somatic mutation | 3403 G→A | 106377953 | C1135Y |
| Somatic mutation | 3575 T→G | 106383519 | C1194W |
| Somatic mutation | 3609 A→T | 106384192 | S1204C |
| Somatic mutation | 3639 C→T | 106384222 | R1214W |
| Somatic mutation | 3724 A→T | 106384307 | D1242V* |
| Somatic mutation | 3733 A→C | 106384316 | Y1245S |
| Somatic mutation | 3780 C→T | 106384363 | R1261C |
| Somatic mutation | 3781 G→A | 106384364 | R1261H* |
| Somatic mutation | 3862 G→T | 106400285 | G1289V |
| Somatic mutation | 4074 C→T | 106410247 | R1358C* |
| Somatic mutation | 4080 G→C | 106410253 | G1360R |
| Somatic mutation | 4248 G→T | 106413237 | V1417F |
| Somatic mutation | 5151 G→T | 106416269 | V1718L* |
| Somatic mutation | 5268 C→G | 106416386 | H1757D |
| Somatic mutation | 5283 A→T | 106416401 | Q1828L |
| Somatic mutation | 5430 T→C | 106416548 | C1811R |
| Somatic mutation | 5617 T→C | 106416735 | I1873T* |
| Somatic mutation | 5641 A→G | 106416759 | H1881R* |
| Somatic mutation | 5698 T→C | 106416816 | V1900A |
| Somatic mutation | 5754 C→T | 106416873 | A1919V |
| Somatic mutation | 5776 G→A | 106416894 | R1926H |
| Somatic mutation | 5780 G→A | 106416898 | R1927K |
| Somatic mutation | 5820 C→T | 106416938 | P1941S |
| Somatic mutation | 5896 G→A | 106417014 | R1966H |
| Somatic mutation | 5920 C→T | 106417038 | R1974M |
| Somatic mutation | 5998 G→A | 106417116 | R2000K |
| SNP | 100 C→T | 106374649 | L34F |
| SNP | 520 C→A | 106375069 | P174H |
| SNP | 2599 T→C | 106377148 | Y867H |
| SNP | 3418 A→T | 106377767 | E1073V |
| SNP | 3451 A→C | 106377800 | Q1084P |
| SNP | 5166 C→T | 106416284 | P1723S |
Novel unannotated SNPs in 4q24. SNPs were defined as missense mutations that were seen in more than one tumor and paired buccal sample.
We did not identify methylation at either of 2 CpG islands of the TET2 promoter in 37 MPN samples or in 4 JAK2V617F-positive leukemic cell lines (supplemental Figure 2). Copy number analysis of 207 MPN patients identified 3 patients with heterozygous deletions of one copy of the region containing TET2 (4q24), suggesting that TET2 mutations are more common than large deletions in MPN patients. Sequencing data from these 3 patients revealed that one patient had a homozygous somatic missense mutation, consistent with heterozygous mutation followed by deletion of the remaining copy of TET2 (supplemental Figure 3). The HEL cell line had a heterozygous deletion of the TET2 locus. One MPN patient had a large deletion on chromosome 10, which included the TET1 locus (10q21.3). Furthermore, although we identified several novel SNPs in TET1 and TET3 (supplemental Table 3), we did not identify somatic TET1 or TET3 mutations in 96 MPN patients. No MPN samples or cell lines had loss of the TET3 locus (2p13.1) or amplifications of TET1, TET2, and TET3.
The frequency of TET2 mutations did not differ between JAK2V617F-positive (16.4%) and JAK2V617F-negative (2.5%) MPN (P = .08, Fisher exact test). Likewise, TET2 mutations were equally frequent in MPN patients with and without the recently described JAK2V617F-positive MPN predisposition haplotype (P = .9).20 We did not note a correlation between TET2 alterations and mutations in FLT3, JAK2, and RAS in CMML, nor did we observe a correlation between TET2 mutations and specific cytogenetic subgroups MLL, FLT3, CEBPA, or NPM1 mutations, or a history of antecedent MPN/MDS in AML (P > .5, Fisher exact test). However, we did note that TET2 mutations are associated with decreased overall survival in AML compared with TET2-wild-type AML patients (P = .03, Figure 1D; supplemental Table 2).
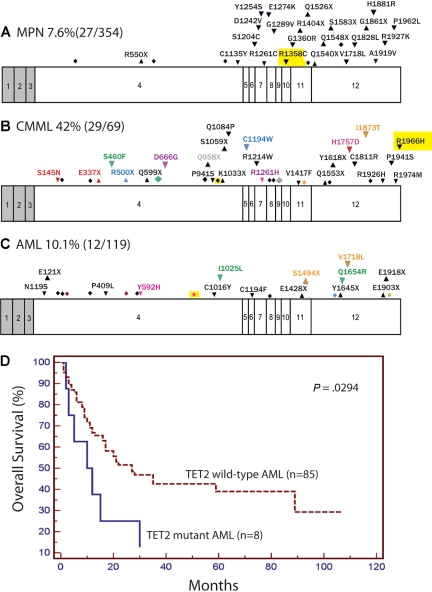
Exons of TET2 with locations of mutations and effect of mutations on overall survival in AML. Locations of mutations in MPN (A), CMML (B), and AML/acute megakaryoblastic leukemia samples (C) as well Kaplan-Meier estimates of overall survival in AML (D) are shown. Shaded regions represent non–protein-coding exons and introns are not shown. Exons are drawn to relative scale. Missense mutations (down arrowheads), nonsense mutations (up arrowheads), and frameshifts (diamonds) are shown at their approximate location along the exon. Mutations occurring within the same patient sample are represented in the same color. Mutations that were homozygous are highlighted in yellow.
In this report, we sequenced all coding exons of TET2 to define the spectrum of somatic TET2 mutations in myeloid malignancies. The broad range of myeloid disorders linked to mutations in TET2 suggests that mutations in TET2 have a pleiotropic role in myeloid transformation. Although our data suggest that TET2 mutations may hold prognostic significance in AML, larger clinical correlative studies will be needed to more accurately assess the effect of TET2 mutations on prognosis, diagnosis, and therapeutic relevance to myeloid malignancies. Whether TET2 mutations dysregulate pathways already known to contribute to hematopoietic transformation, or represent a novel pathway, remains to be elucidated, and the role of TET family alterations in neoplasms other than myeloid malignancies is not yet known.
Acknowledgments
The authors thank the patients who have contributed to our understanding of these disorders.
This work was supported by the National Institutes of Health (grant CA10863101, G.G.-M., H.M.K., and D.G.G.; HL082677, R.L.L.), the Howard Hughes Medical Institute, the Starr Cancer Consortium (New York, NY), the Myeloproliferative Disorders Foundation (Chicago, IL), the Doris Duke Charitable Foundation (New York, NY), and the ECOG Leukemia Tissue Bank (Boston, MA; CA021115 and CA114737). O.A.-W. is supported by a Dana Fellowship from the Memorial Sloan Kettering Clinical Scholars Program. O.K. is supported by a grant from the Academy of Finland (Helsinki, Finland). D.G.G. is an Investigator of the Howard Hughes Medical Institute and is a Doris Duke Charitable Foundation Distinguished Clinical Scientist. R.L.L. is an Early Career Award recipient of the Howard Hughes Medical Institute, a Clinical Scientist Development Award recipient of the Doris Duke Charitable Foundation, and the Geoffrey Beene Junior Chair at Memorial Sloan Kettering Cancer Center. J.D.C. is a Scholar of the Leukemia & Lymphoma Society (White Plains, NY).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: O.A.-W., A.M., J.P., D.G.G., J.D.C., and R.L.L. designed the study; C.H., G.G.-M., M.W., S.M., J.Y., R.B., E.P., M.M.L.B., M.B., M.S.T., H.M.K., and R.L.L. collected and processed samples and provided genetic and clinical annotation; O.A.-W., J.P., K.H., S.T., I.D., A.H., and R.L.L. performed sequence analysis, analyzed sequence traces, and validated mutations; O.A.-W., B.L.E., R.M.S., and R.L.L. acquired and analyzed SNP array data; O.A.-W., J.P., and O.K. performed methylation-specific polymerase chain reaction; O.A.-W. and R.L.L. wrote the manuscript with assistance from A.M., C.H., and J.D.C.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ross L. Levine, Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, 1275 York Ave, Box 20, New York, NY 10065; e-mail: gro.ccksm@renivel.
References
Articles from Blood are provided here courtesy of The American Society of Hematology
Full text links
Read article at publisher's site: https://doi.org/10.1182/blood-2009-03-210039
Read article for free, from open access legal sources, via Unpaywall:
https://ashpublications.org/blood/article-pdf/114/1/144/1315723/zh802709000144.pdf
Free after 12 months at www.bloodjournal.org
http://www.bloodjournal.org/cgi/content/full/114/1/144
Free to read at www.bloodjournal.org
http://www.bloodjournal.org/cgi/content/abstract/114/1/144
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1182/blood-2009-03-210039
Article citations
Cell-Free DNA Hydroxymethylation in Cancer: Current and Emerging Detection Methods and Clinical Applications.
Genes (Basel), 15(9):1160, 03 Sep 2024
Cited by: 0 articles | PMID: 39336751 | PMCID: PMC11430939
Review Free full text in Europe PMC
Oncogenic Enhancers in Leukemia.
Blood Cancer Discov, 5(5):303-317, 01 Sep 2024
Cited by: 0 articles | PMID: 39093124
Review
TET3 downregulation and low 5-hydroxymethylcytosine are epigenetic signatures of head and neck carcinoma.
Mol Biol Rep, 51(1):877, 31 Jul 2024
Cited by: 0 articles | PMID: 39083093
Ultrasensitive detection of 5-hydroxymethylcytosine in genomic DNA using a graphene-based sensor modified with biotin and gold nanoparticles.
Mater Today Bio, 27:101123, 11 Jun 2024
Cited by: 0 articles | PMID: 38988817 | PMCID: PMC11234158
DNA methylation in human diseases.
Heliyon, 10(11):e32366, 04 Jun 2024
Cited by: 0 articles | PMID: 38933971 | PMCID: PMC11200359
Review Free full text in Europe PMC
Go to all (478) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mutation in TET2 in myeloid cancers.
N Engl J Med, 360(22):2289-2301, 01 May 2009
Cited by: 1170 articles | PMID: 19474426
TET2 deletions are a recurrent but rare phenomenon in myeloid malignancies and are frequently accompanied by TET2 mutations on the remaining allele.
Br J Haematol, 156(1):67-75, 24 Oct 2011
Cited by: 16 articles | PMID: 22017486
TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia.
Haematologica, 94(12):1676-1681, 01 Oct 2009
Cited by: 163 articles | PMID: 19797729 | PMCID: PMC2791942
Mutations of the TET2 and CBL genes: novel molecular markers in myeloid malignancies.
Ann Hematol, 89(7):643-652, 02 Mar 2010
Cited by: 39 articles | PMID: 20195608
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (5)
Grant ID: U10 CA021115
Grant ID: U24 CA114737
Grant ID: R01 CA101774-07
Grant ID: CA10863101
Grant ID: R01 CA101774
NHLBI NIH HHS (2)
Grant ID: HL082677
Grant ID: K08 HL082677
 1,2
1,2




