Abstract
Free full text

Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression
Associated Data
Abstract
Skeletal muscle stem cells are regulated by Pax3/7. During development, Pax3 is required for the maintenance of these cells in the somite and their migration to sites of myogenesis; high levels of Pax3 interfere with muscle cell differentiation, both in the embryo and in the adult. Quantitative fine-tuning of Pax3 is critical, and microRNAs provide a potential mechanism. We identify microRNA-27b (miR-27b), which directly targets the 3′-UTR of Pax3 mRNA, as such a regulator. miR-27b is expressed in the differentiating skeletal muscle of the embryonic myotome and in activated satellite cells of adult muscle. In vivo overexpression of a miR-27b transgene in Pax3-positive cells in the embryo leads to down-regulation of Pax3, resulting in interference with progenitor cell migration and in premature differentiation. In a complementary experiment, miR-27b inhibitors were transfected into cultures of adult muscle satellite cells that normally express miR-27b at the onset of differentiation, when Pax3 protein levels undergo rapid down-regulation. Interference with miR-27b function results in continuing Pax3 expression leading to more proliferation and a delay in the onset of differentiation. Pax7 levels are not affected. Introduction of miR-27b antagomirs at a site of muscle injury in vivo also affects Pax3 expression and regeneration in vivo. We therefore conclude that miR-27b regulates Pax3 protein levels and this down-regulation ensures rapid and robust entry into the myogenic differentiation program.
The regulation of transcription factors that distinguish a progenitor cell from its tissue derivatives is an important facet of stem cell biology. Myogenesis provides a paradigm in which this level of control can be explored (1). Skeletal muscle progenitor cells are characterized by the expression of Pax3 and Pax7. These multipotent stem cells are present in the epithelial dermomyotome of the embryonic somite and subsequently enter the underlying myotome, the first differentiated muscle to form. Pax3/7-positive myogenic progenitors constitute a self-renewing population of cells that are essential for all subsequent skeletal muscle growth and contribute the satellite cells, required for regeneration of adult skeletal muscle. At the onset of myogenesis, Pax3-positive cells also migrate from the hypaxial domain of the dermomyotome to sites, such as those in the limbs, where skeletal muscles will form. Pax7 is subsequently up-regulated in these cells, which down-regulate the Pax genes before differentiation. The myogenic determination genes Myf5 (2) and MyoD (3) are direct targets of Pax3/7, which thus direct progenitors into the myogenic program. Such cells will proliferate as myoblasts and then differentiate after activation of Myogenin, another myogenic regulatory gene of the MyoD family. This is exemplified by the muscle satellite cell, which lies under the basal lamina of adult muscle fibers (4). On injury or in culture, before differentiation, these cells down-regulate Pax7 and Pax3, which is expressed in a subset of muscles at this stage (5). Low-level expression of Pax3/7 does not completely inhibit activation of Myogenin and muscle genes such as Troponin T (6), but the adverse effect of maintaining expression of these proteins is immediately evident when they are overexpressed, as reported for Pax7, which blocks differentiation (7). In addition to transcriptional repression of Pax3 and Pax7, the Pax3 protein is subject to a specific degradative pathway through monoubiquitination in satellite cells. When this is prevented, expression of differentiation markers is reduced (8).
In addition to regulation at the protein level, it is becoming increasingly evident that microRNA (miRNA) regulation of mRNAs also represents an effective way of rapidly interfering with function. This takes place when the miRNA, as part of the RNA-induced silencing complex (RISC), targets the 3′-UTR of transcripts by imperfect base pairing and functions to inhibit translation either at the initiation stage or at postinitiation steps. In some cases, degradation of the target mRNA is also a mechanism or consequence of miRNA-mediated suppression (9).
Although many miRNAs are expressed across multiple lineages or tissues (10, 11), some miRNAs are lineage restricted. This is the case for the muscle-specific “myomiRs,” which include miRNA-1 (miR-1) and its variant miR-206, along with miR-133, which is closely linked and coexpressed with both. Somitic expression of these miRNAs is restricted to the myotome, and this is consistent with their transcriptional regulation by MyoD and by MEF2, also implicated in skeletal myogenesis (12–14). Through the validation of some of the targets of these miRNAs, roles for them have been proposed in regulating the balance between proliferating and differentiating myoblasts by SRF/HDAC4 (15) or via an effect on DNA polymerase II (16). However, the potential role of miRNAs in repressing the expression of the key upstream regulators, Pax3 and Pax7, has not been addressed. In this article, we predict and validate miR-27 as a regulator of Pax3. This miRNA is expressed both in differentiating muscle masses, such as the myotome, in the embryo in differentiating satellite cells in the adult. It targets the 3′-UTR of Pax3 mRNA and affects Pax3 expression in vitro and in vivo, with consequences for the onset of skeletal muscle differentiation.
Results
A single target site for miR-27 in the Pax3 3′-UTR is predicted by algorithms, including miRanda (17), TargetScan5.1 (18), and Diana-microT3.0 (19). In addition to a perfect 8-base-pair match in the seed region, there is an AU-rich flanking sequence and additional base pair matching that boost miRNA efficacy (Fig. 1A). Furthermore, secondary structure considerations (12) suggest the target site is accessible (Table S1). miR-27 includes miR-27a and miR-27b, which are transcribed from different chromosomes and differ by only one 3′ nucleotide. In the following experiments, similar results were obtained for miR-27a (Fig. S1). Functional tests in 293 cells, using a standard luciferase assay, demonstrate that miR-27b directly targets the Pax3 3′-UTR, specifically through the predicted site, as shown by mutation (Fig. 1B). We then verified the expression of miR-27b in the mouse embryo. At embryonic day 10.5 (E10.5) (Fig. 2A), it is expressed at a number of sites, including the somites. In immature, caudal somites, it is present in the ventral sclerotome, where Pax3 expression is down-regulated, but is excluded from the Pax3-positive dorsal dermomytome (Fig. 2B). In more mature interlimb somites, miR-27b has accumulated in the myotome (Fig. 2C), where skeletal muscle is forming, as indicated by Myogenin (MyoG) labeling (Fig. 2D).

The 3′-UTR of Pax3 is a target of miR-27. (A) The predicted interaction includes a perfect 8-base pair match to the seed region of the miRNA (dark gray). Additional features that would boost efficacy of the target interaction include a flanking AU-rich sequence (underlined). (B) Transfection of 293 cells, expressing the psicheck-2 luciferase vector containing the Pax3 3′-UTR downstream of the Renilla luciferase (R. luc) sequence, with miR-27b precursors, resulted in reduced R. luc activity that was lost on mutation of the miR-27 target site (Pax3–3′UTRmut). Conversely, transfection with a miR-27b inhibitor (anti-miR-27b), which blocks the activity of endogenous miR-27b in 293 cells (Fig. S2), resulted in increased R. luc activity. Reported results are normalized to firefly luciferase activity, which serves as an internal transfection control, and presented relative to R. luc activity after transfection with random, nontargeting, control oligonucleotides. Results are from 3 independent experiments.

Embryonic expression of miR-27b at E10.5. (A) Whole-mount in situ hybridization profile using an LNA probe specific to miR-27b indicates widespread expression in the limb buds (fl and hl) and in the tail bud, as well as the neural tube (nt) and heart (ht). It is also expressed in the somites (so) where myogenesis is initiated. (B and C) Transverse sections of an immature caudal (B) and more mature interlimb (C) somites from A show that miR-27b is not expressed in the dermomyotome (dm) where Pax3-positive myogenic cells are present. It is present in the sclerotome (sc) of the immature somite and has accumulated in the myotome (m) (distinguished from the dermomyotome by a white dotted line). (D) A transverse section at interlimb level for an E10.5 embryo, after immunohistochemistry with a Myogenin (MyoG) antibody that marks differentiating muscle cells and a Pax3 antibody, marking progenitor cells in the dermomyotome, strongly labeled for miR-27b in the epaxial (e) and hypaxial lips, as well as interspersed cells in the myotome.
Manipulation of miR-27b by using a lentivirus expression vector in somite explants leads to down-regulation of Pax3, but not Pax7, in transduced cells (Fig. 3A). When miR-27b is expressed in Pax3-positive cells as a conditional transgene in vivo, this resulted in a 2.5-fold increase in miR-27b expression, assessed in the limb buds by quantitative real-time PCR (qRT-PCR) (Fig. S3). Pax3 is reduced, exemplified by the reduction in Pax3-positive cells in forelimb buds (52 ± 9% of controls) (Fig. 3 B and C). Pax3 is required for the survival and migration of cells from the hypaxial dermomyotome to the limb buds (1, 2, 20). These results therefore suggest that Pax3 is affected by the presence of miR-27b. Lbx1, which also marks these migrating cells (21), was detectable in more cells (72 ± 9% of controls) than Pax3 (Fig. 3B), suggesting that in the transgenic embryos there is also ongoing down-regulation of Pax3 in the limb buds. We went on to analyze the balance between Pax3-positive progenitors and Myogenin-positive differentiating myogenic cells in limb buds. At E11.5, there is a striking shift toward differentiating cells (Fig. 3 D and E). In this situation, therefore, a reduction in Pax3 levels favors premature differentiation of those myogenic progenitor cells that have migrated into the limb bud. Promotion of differentiation is also detectable in the myotome of the somite at this stage (Fig. 3 D and E).
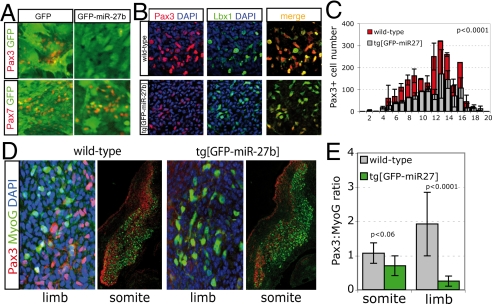
Overexpression of miR-27b in somite explant cultures and transgenic embryos reduces Pax3 protein levels and reduces the ratio of proliferating (Pax3+) to differentiating cells in the limb bud. (A) Newly formed somites in explant culture were transduced with lentivirus expressing GFP only (GFP) or GFP and miR-27b (GFP-miR-27b). After 3 days in culture, immunohistochemistry with Pax3 or Pax7 antibodies (red) indicates down-regulation of Pax3 for somites transduced with GFP-miR-27b, but not GFP only. Pax7, not a target of miR-27b, was not affected. (B) Immunohistochemistry with a Pax3 antibody (red), a Lbx1 antibody (green), and the two merged on serial sections through the complete forelimb bud of tg[miR-27b] embryos at E10.5 indicates a marked reduction in Pax3+ cells migrating to the limbs of transgenic, compared with wild-type embryos. (C) An example of Pax3+ cells per serial section spanning the limb bud is shown for control and transgenic embryos. (D) Immunohistochemistry with Pax3 (red) and Myogenin (green) antibodies on serial sections through the forelimb buds and interlimb somites of tg[miR-27b] and wild-type embryos at E11.5 shows a decrease in the ratio of Pax3/Myogenin (MyoG)-positive cells when miR-27b is overexpressed. Blue marks DAPI staining of nuclei. (E) Summary of results illustrated in D. Reduction in Pax3+ cells corresponds to an increase in myogenic differentiation, as indicated by the ratio of Pax3/MyoG expression.
We carried out the opposite experiment in adult muscle satellite cells by inhibiting miR-27b activity. Satellite cells from the diaphragm were isolated by flow cytometry from Pax3GFP/+ mice (5). In culture, these cells continue to express Pax3 transcripts, which are still present at lower levels after day 5, whereas Myogenin transcripts accumulate at this time (Fig. 4A). As cells prepare to differentiate by 3 days, miR-27b has increased. Similar results are shown for differentiating C2 myoblasts (Fig. S4). In contrast, miR-1 only accumulates later, indicative of its involvement in differentiated muscle (Fig. 4B). When an inhibitor to miR-27b is added to the culture, Pax3 protein increases, whereas Troponin T, a marker of differentiated muscle cells, decreases. Pax7, which is not a predicted target of miR-27b, was not affected (Fig. 4C). Pax3 transcript levels were not significantly affected (Fig. 4D). The effect of anti-miR-27b inhibitors on skeletal muscle differentiation is illustrated in Fig. 5 for Myogenin and Troponin T. After 4 days in culture, the levels of these differentiation markers are reduced and proliferation is higher, as indicated by the number of DAPI-positive nuclei (Fig. 5 A and B). In satellite cells, from Pax3GFP/flox null mice, no effect of miR-27b inhibition on differentiation is observed, showing that it acts primarily through Pax3 (Fig. 5C). After 5 days, in Pax3GFP/+cultures, Myogenin, which is expressed at the onset of differentiation, is now higher and Troponin T begins to be detectable, whereas in control satellite cell cultures large Troponin T-positive myotubes have formed in the control (Fig. 5 D and E). At this stage, the fusion index is reduced after transfection of anti-miR-27b inhibitors (Fig. 5F). These experiments show that, when Pax3 protein levels are maintained by preventing the effect of miR-27b on Pax3 mRNA, differentiation of satellite cells is delayed.

Inhibition of miR-27b activity in adult Pax3GFP/+ satellite cells. (A) qRT-PCR amplification of Pax3 and MyoG transcripts from sorted Pax3GFP/+ satellite cells after 3 (proliferating) and 5 (differentiating) days in culture. Results are normalized to GAPDH and relative to day 0. (B) qRT-PCR indicates mature miR-27b is up-regulated at 3 days, whereas miR-1 accumulates later at 5 days. (C) Protein levels of Pax3, Pax7, and Troponin T (TropT) were quantified by densitometry of Western blots after satellite cells were transfected with an anti miR-27b inhibitor (+) or control inhibitor (−) and cultured for 5 days. Pax3 levels increase in response to blocking miR-27b activity, confirming Pax3 as a direct target of the miRNA. Conversely, blocking the activity of miR-27b reduced the expression of Troponin T. Pax7, not predicted to be a target of miR-27, was unaffected. Results are indicated relative to β-tubulin expression and are from 2 independent Western blots. Representative immunoblots are shown. (D) Relative Pax3 transcript levels from cells transfected with control or anti-miR-27b inhibitors, normalized to GAPDH transcript levels.
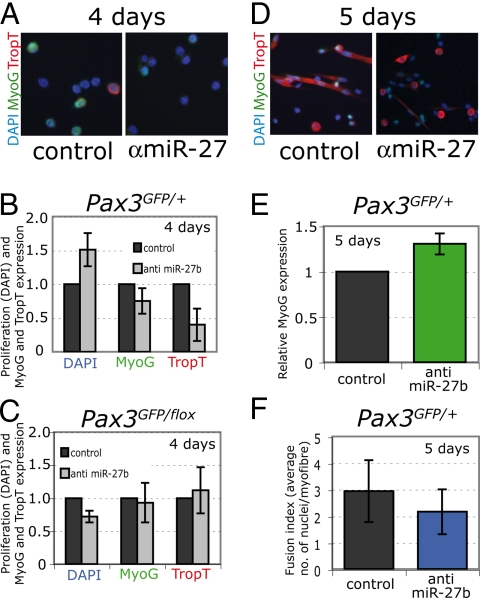
Inhibition of miR-27b activity in adult Pax3GFP/+ satellite cells delays differentiation. (A) Detection of Myogenin (MyoG) (green) and Troponin T (TropT) (red) by immunocytochemistry with the corresponding antibodies after satellite cells were transfected with an anti miR-27b inhibitor or control oligonucleotide and cultured for 4 days. DAPI staining of nuclei is shown in blue. Some MyoG-positive cells can be detected together with a low number of mononucleated TropT-positive cells. (B) The number of positive cells for these markers is reduced after transfection with anti miR-27b inhibitor, whereas the total number of DAPI-positive nuclei increases. (C) An equivalent series of results in satellite cells from Pax3GFP/flox mice that lack Pax3. (D) Immunocytochemistry as in A at 5 days shows that MyoG expression is normally reduced as cells differentiate into polynucleated, TropT-expressing myotubes. In contrast, cells transfected with anti-miR-27b inhibitor have higher MyoG expression and lower TropT expression (D), indicating that differentiation is delayed in response to inhibiting the activity of miR-27b. (E) Quantitation of these results as in B. At 5 days, only MyoG-positive nuclei were counted because TropT expression was found in the cytoplasm of polynucleated fibers at this stage. Results in B, C, and E are indicated relative to transfection with the negative control oligonucleotide and standard deviations are reported from 3 independent transfections, with P < 0.05 using paired Student's test. (F) Fusion index, estimated as the number of DAPI-positive nuclei per myofiber in control or anti-miR-27b inhibitor-treated satellite cell cultures. Results are averages with standard deviations from 3 independent transfections, with P < 0.05 using paired Student's test.
To look at effects on muscle satellite cell differentiation in vivo, the pectoralis muscle, which includes Pax7-positive satellite cells also expressing Pax3 (6), was injured by injection with cardiotoxin together with anti-miR-27b or control antagomirs. After 6 days, control injured muscle had regenerated to form new muscle fibers, with reconstitution of Pax7-positive satellite cells under the basal lamina (Fig. 6A). Some of these cells (10%) also express Pax3 (Fig. 6 B and C). In the presence of the antagomir, more satellite cells that coexpress Pax3 and Pax7 (40%) are present (Fig. 6 B and C), showing that Pax3 expression in activated satellite cells (8) is maintained. In addition, muscle injected with anti-miR-27b antagomirs had smaller fibers, indicative of a delay in regeneration (Fig. 6D). Fibers with more Pax3-positive cells are notably smaller in diameter, in keeping with a more immature state.
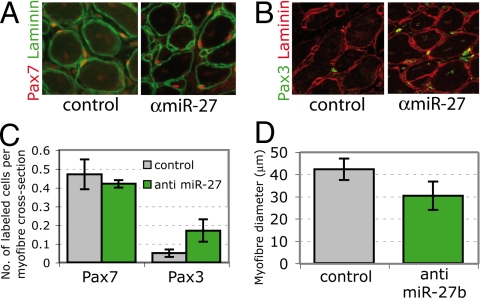
In vivo application of anti-miR-27b antagomir affects regeneration of injured muscle. (A) Immunohistochemistry of pectoralis muscle sections 6 days after cardiotoxin injury showing Pax7-positive satellite cells under the basal lamina of muscle fibers, outlined by laminin, with control or anti-miR-27b antagomirs. (B) As in A, using a Pax3 antibody to mark satellite cells. (C) Summary of the number of Pax3- or Pax7-positive cells per cross-section of a myofiber. (D) Average muscle fiber diameter in control and anti-miR-27b antagomir-treated regenerating muscle at 6 days.
Discussion
We show that the 3′-UTR of Pax3 mRNA is a target of miR-27. This is illustrated by standard luciferase assays in cell culture and by overexpression of miR-27 in somite explants or down-regulation of miR-27 in muscle satellite cells. In the context of skeletal myogenesis, in the embryo, this miRNA is notably absent from the epithelial dermomyotome where Pax3-positive myogenic stem cells are present. As the skeletal muscle of the myotome forms from Pax3-expressing cells, derived from the dermomyotome, miR-27 is up-regulated.
Overexpression of a miR-27 transgene in Pax3-expressing cells in the embryo results in a shift from Pax3/7-positive progenitor cells to Myogenin-positive cells that have entered the myogenic differentiation program. miRNA regulation of Pax3 mRNA is therefore another aspect of the control of this important equilibrium between self-renewing muscle stem cells and their tissue derivatives, also orchestrated by Pax3 targets such as components of the FGF signaling pathway (22), or by modulation of Notch (23, 24) or TGFβ/myostatin signaling (25). Manipulation of miR-27 expression in the embryo also affects Pax3 function before myogenesis; for example, at the level of migration of Pax3-positive progenitor cells from the somite to the limb bud. Here too, overexpression of miR-27 results in a specific down-regulation of Pax3, with less effect on Lbx1, also expressed in these cells (21). Lbx1 mRNA is not predicted to be a target of miR-27.
In adult muscle satellite cells, miR-27 is expressed, when Pax3 transcripts are still present, just before differentiation. In these myogenic progenitor cells, inhibitors to miR-27 delay the onset of skeletal muscle differentiation, in keeping with results that indicate an inhibitory effect of Pax3 on this process (8). These results underline a role for Pax3, as distinct from Pax7 that is not affected by miR-27, in adult skeletal muscle myogenesis. In vivo observations on injured muscle also support a role for Pax3 and show that interference with its down-regulation by miR-27 delays regeneration. miR-27 may well have other targets, some of which may be important in the myogenic context; however, altering miR-27 activity on different stages of myogenesis, in the embryo and in adult satellite cells, strongly suggests that the negative regulation of Pax3 protein production is an important aspect of its function. This is also indicated by the results on satellite cells for a Pax3GFP/flox-null mouse (26) where miR-27b inhibition no longer delays differentiation in the absence of Pax3.
Other miRNAs, miR-1/miR-206 and miR-133, that show skeletal muscle specificity, have been proposed to interfere with proliferation/differentiation of myogenic cells through effects on SRF/HDAC4 (15) or DNA polymerase II (16). There is a potential target site in the 3′-UTR of Pax3 mRNA for miR-1/miR-206. However, in our hands, miR-1 did not have as robust an effect as miR-27 in the standard luciferase assay. In addition, as we show, miR-1 accumulates after miR-27b, in differentiating satellite cells. Our results identify miR-27 as an important player in skeletal myogenesis, through its control of the 3′-UTR of Pax3 mRNA.
Materials and Methods
DNA Constructs.
The 3′-UTR of the Pax3 transcript was amplified by PCR by using forward 5′-AAAAGCGGCCGCGTTTAAACGAGAAGATTCTTCATGTCTA-3′ and reverse 5′-AAAACTCGAGGTTTAAACTAGGGGGTAGTTCCTCCT-3′ primers, cloned into the pGEMT-Easy vector (Promega), and the sequence was verified. The predicted miR-27b “seed” site was mutated with mutagenic primers 5′-GCTGATCTTGCCCCTAACCCCGAATTGTCCCAGCATGACC-3′ and 5′-GATCTTGCCCCTAACCCCCTTTTGTCCCAGCATGACC-3′ by using the QuickChange Multisite mutagenesis kit (Stratagene); mutations were confirmed by sequence analysis. The wild-type and mutated 3′-UTRs were subcloned into the NotI–XhoI site of the psicheck-2 vector (Promega). The miR-27b precursor sequence was amplified from genomic DNA by PCR using forward 5′-AAAAGTCGACCGAAGATGCTCACCAGCCCTT-3′ and reverse 5′-AAAAGTCGACGGCAGTGGCCTCTGCCTGGC-3′ primers and cloned into the pGEMT-Easy vector. The verified sequence was subcloned into either the SalI site of pLenti-PGK-GFP (for explant transduction) or blunt-end ligated into the EcoRV site of pBSKIsceIGFP vector (for transgenesis), such that the stem-loop precursor sequence was located in the 3′-UTR of GFP (GFP-miR-27b), as described for enhanced expression of RNAi in refs. 27 and 28. Insertion of EGFP into pBSKIsceI (29) was by HindIII cloning of an amplified EGFP PCR product from pIRES2-EGFP (Clontech) using forward 5′-AAAAAGCTTCACAACGATGGTGAGCAAGGG-3′ and reverse 5′-AAAAAAGCTTTTACTTGTACAGCTCGTCCAT-3′ primers. Sequence and orientation were confirmed.
Cell Culture.
293 cells were maintained in DMEM supplemented with 10% FBS and antibiotics. Pax3GFP/null mice were obtained by crossing Pax3GFP/+ mice with mice bearing the Pax3 locus floxed (Pax3fl/) (26) together with an inducible Cre recombinase (Rosa26CreERT2) (30). The conditional Pax3 mutant mice (Pax3GFP/fl: Rosa26CreERT2) were injected with tamoxifen to induce deletion of the Pax3 allele. Satellite cells were isolated from the diaphragms of 5- to 6-week-old Pax3GFP/+ or Pax3GFP/flox mice after tamoxifen induction, sorted by flow cytometry, and grown as described in ref. 6. Before Western blot analysis, satellite cell cultures were supplemented with the proteasome inhibitor MG132 (Sigma) for 4 h.
Transfections.
293 cells were seeded at 1.0 × 104 cells per well of a 12-well plate and grown for 24 h. A total of 50 ng of the appropriate psicheck-2 luciferase reporter construct was cotransfected with 20 nM final concentration of either double-stranded RNA oligonucleotides designed to mimic miR-27b, anti-miR-27b molecules, or their corresponding negative controls (Ambion) using Lipofectamine 2000 (Invitrogen). Twelve hours after transfection, the medium was changed and cells were grown for an additional 24 h before assay. Pax3GFP/+ sorted cells were reverse transfected with the oligonucleotides by using RNAiMax (Invitrogen). Twelve hours after transfection, the medium was changed and cells were grown for the indicated periods before assay. Oligonucleotides were transfected efficiently and were stable throughout the course of the experiments (Fig. S5).
Luciferase Assay.
Luminescent signals arising from psicheck-2 transfected cells were quantified by using the Dual Luciferase assay system (Promega) with a GloMax luminometer (Promega). All values are given relative to transfections with the appropriate negative control oligonucleotide.
In Situ Hybridization.
Embryos were harvested and fixed for 24 h in 4% paraformaldehyde, washed 3 times in PBS, and stored in methanol for a minimum of 24 h. Endogenous miR-27b was detected by using a locked nucleic acid (LNA) probe against miR-27b (31). Prehybridization and hybridization was with 60% formamide, at 57 °C. After washes, embryos were incubated with anti-digoxigenin (DIG) antibody (Roche) overnight at 4 °C and washed extensively for 5 days before staining with BM Purple AP substrate (Roche). For optimal signal-to-background ratios, DIG-labeled LNA probes were recycled 2 additional times.
RNA and Protein Analysis.
Total RNA was harvested from cells by using TRIzol reagent (Invitrogen). Detection of miRNAs from samples by quantitative RT-PCR was performed by using the miRcury LNA MicroRNA PCR system (Exiqon) according to the manufacturer's instructions. Amplification of U6 RNA was used to normalize the data. For detection of mRNAs, total RNA was reverse transcribed with a random primer (Ambion) by using SuperScript II reverse transcriptase (Invitrogen). Primers were forward 5′-TCCATCCGACCTGGTGCCAT-3′ and reverse 5′-TTCTCCACGTCAGGCGTTG-3′, Pax3; forward 5′-CAACCAGGAGGAGCGCGATCTCCG-3′ and reverse 5′-AGGCGCTGTGGGAGTTGCATTCACT-3′, Myogenin; and forward 5′-CACCATCTTCCAGGAGCGAG-3′ and reverse 5′-CCTTCTCCATGGTGGTGAAGA-3′, GAPDH. PCRs were performed with Power SYBR Green PCR Master Mix (Applied Biosystems). For protein analysis, cell lysates were prepared with 50 μL of lysis buffer (1% Nonidet P-40, 0.15 M NaCl, 50 mM Tris) supplemented with Complete protease inhibitor mixture (Roche). Twenty micrograms of total protein were loaded into wells of NuPAGE NOVEX Bis-Tris Gels (Invitrogen). Western blots were performed with monoclonal anti-Pax3 (DSHB), anti-Pax7 (DSHB), anti-Troponin T (Sigma), and anti-β-tubulin (Chemicon) antibodies. Detection was by ECL Plus Western Blotting Detection System (GE Healthcare), and blots were directly processed on a Storm 840 Optical scanner (Amersham) with the exception of anti-Pax3 Western blots, which were exposed to Hyperfilm MP (Amersham). Densitometry was performed with ImageQuaNT software (Molecular Dynamics).
Lentiviral Production.
293T cells were transfected with the pLentiPGKGFP constructs indicated above, pREV, VSVG, Δ874 (kind gifts from G. Cossu, DIBIT Universita Vita-Salute San Raffaele, Milan), by using CaCl2/Hepes-buffered saline precipitation. Twenty-four hours later, the medium was changed and collected after a further 48 h, filtrated through a 44-μm filter, and concentrated by using Vivaspin 20 concentrators with a 50,000 molecular weight cutoff (Sartorius Biotech). Titers were calculated by GFP FACS analysis of transduced 293 cells.
Somite Explant Culture and Transduction.
Mouse embryos were taken at E9.5 and treated with 0.1% trypsin, 0.25% pancreatin for 5 min at 4 °C, as described in ref. 32. The three most recently formed somites were transferred to a 200-μL drop of medium (40% DMEM, 40% F12, 20% FBS). After 10 min, when somites had attached, plates were carefully transferred to the incubator. Twelve to 16 h later, somites were transduced with lentivirus expressing either GFP or GFP-miR-27b at a concentration of 1.0 × 104 virus particles per μL. Twenty-four hours after transduction, the lentivirus-containing medium was removed and replaced with 1 mL of fresh medium. Somites were cultured for a total of 3 days.
Immunofluorescence.
Fixed somite explants or cultured Pax3GFP/+ cells were permeabilized with 0.2% Triton, 50 mM NH4Cl, and blocked by using 0.2% gelatin in PBS. Frozen transverse sections were treated as described in ref. 33. Primary antibodies used were monoclonal anti-Pax3 (DSHB), monoclonal anti-Pax7 (DSHB), monoclonal anti-Troponin T (Sigma), polyclonal anti-Myogenin (Santa Cruz), polyclonal Lbx1 (a kind gift from C. Birchmeier, Max Delbrück Center, Berlin-Buch, Germany), polyclonal anti-laminin (Sigma), and polyclonal anti-GFP (MBL). Fluorochrome conjugated secondary antibodies (Molecular Probes) were used for visualization and DAPI was included in the washing steps at 1:1,000 before mounting with Gel-Mount (Electron Microscopy Sciences).
Transgenesis.
The transgenic plasmids indicated above were digested with IsceI, agarose gel-purified, and transgenic animals were obtained as described in ref. 34. Three transgenic animals were obtained with the miR-27b transgene. Mice were crossed with the Pax3Cre/+ line (35) to activate miR-27b expression in Pax3-positive cells. This is under the control of the CAG promoter and contains an EGFP reporter to verify activation after Cre-mediated recombination. Embryos were dated taking the day after mating as E0.5.
Muscle Regeneration.
Adult 8-week-old male Swiss mice were anesthetized by i.p. injection of a mixture of Imalgene 1,000 (ketamine, 100 mg/mL, 10 mg/kg) and Rompun (xylazine, 2%, 0.06 mL/kg). Breast skin was opened in anesthetized mice, and pectoralis muscles were exposed and injected with a 100-μL solution containing 10 μM cardiotoxin and 10 μg of either anti-miR-27 or negative control antagomirs (Dharmacon). Six days after injection, pectoralis muscles were dissected and immediately frozen in isopentane cooled in liquid nitrogen and stored at −80 °C until analysis. Care and handling of animals were in accordance with the regulations of the French Ministry of Agriculture and Fisheries practiced by the Institut Pasteur animal facility.
Acknowledgments.
We thank C. Bodin for technical assistance, E. Perret for advice on image acquisition, and B. Gayraud-Morel for assistance in lentivirus production. This work was supported by the Institut Pasteur and the Centre National de la Recherche Scientifique as well as by grants from the Association Francaise Contre les Myopathies and from the FP7 EuroSyStem Integrated Project and the FP6 MYORES Network of Excellence of the European Union. C.G.C. was supported by MYORES and latterly by EuroSyStem.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900210106/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0900210106
Read article for free, from open access legal sources, via Unpaywall:
http://www.pnas.org/content/106/32/13383.full.pdf
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/106/32/13383
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/106/32/13383
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/106/32/13383.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/158437848
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0900210106
Article citations
Elucidation of how the Mir-23-27-24 cluster regulates development and aging.
Exp Mol Med, 56(6):1263-1271, 14 Jun 2024
Cited by: 1 article | PMID: 38871817 | PMCID: PMC11263685
Review Free full text in Europe PMC
The Role of MicroRNA in the Pathogenesis of Duchenne Muscular Dystrophy.
Int J Mol Sci, 25(11):6108, 01 Jun 2024
Cited by: 2 articles | PMID: 38892293 | PMCID: PMC11172814
Review Free full text in Europe PMC
Post-transcriptional regulation of myogenic transcription factors during muscle development and pathogenesis.
J Muscle Res Cell Motil, 45(1):21-39, 11 Jan 2024
Cited by: 2 articles | PMID: 38206489
Review
The roles of miRNAs in adult skeletal muscle satellite cells.
Free Radic Biol Med, 209(pt 2):228-238, 24 Oct 2023
Cited by: 3 articles | PMID: 37879420
Review
Oncomeric Profiles of microRNAs as New Therapeutic Targets for Treatment of Ewing's Sarcoma: A Composite Review.
Genes (Basel), 14(10):1849, 23 Sep 2023
Cited by: 0 articles | PMID: 37895198 | PMCID: PMC10606885
Review Free full text in Europe PMC
Go to all (200) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
miR-27b regulates myogenic proliferation and differentiation by targeting Pax3 in goat.
Sci Rep, 8(1):3909, 02 Mar 2018
Cited by: 19 articles | PMID: 29500394 | PMCID: PMC5834623
microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7.
J Cell Biol, 190(5):867-879, 01 Sep 2010
Cited by: 381 articles | PMID: 20819939 | PMCID: PMC2935565
A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis.
PLoS Genet, 6(4):e1000897, 01 Apr 2010
Cited by: 55 articles | PMID: 20368965 | PMCID: PMC2848553
Skeletal muscle progenitor cells and the role of Pax genes.
C R Biol, 330(6-7):530-533, 13 Jun 2007
Cited by: 98 articles | PMID: 17631448
Review




