Abstract
Free full text

Mutations in the inositol polyphosphate-5-phosphatase E gene link phosphatidyl inositol signaling to the ciliopathies
Abstract
Phosphotidylinositol (PtdIns) signaling is tightly regulated, both spatially and temporally, by subcellularly localized PtdIns kinases and phosphatases that dynamically alter downstream signaling events 1. Joubert Syndrome (JS) characterized by a specific midbrain-hindbrain malformation (“molar tooth sign”) and variably associated retinal dystrophy, nephronophthisis, liver fibrosis and polydactyly 2, and is included in the newly emerging group of “ciliopathies”. In patients linking to JBTS1, we identified mutations in the INPP5E gene, encoding inositol polyphosphate-5-phosphatase E, which hydrolyzes the 5-phosphate of PtdIns(3,4,5)P3 and PtdIns(4,5)P2. Mutations clustered in the phosphatase domain and impaired 5-phosphatase activity, resulting in altered cellular PtdIns ratios. INPP5E localized to cilia in major organs affected in JS, and mutations promoted premature destabilization of cilia in response to stimulation. Thus, these data links PtdIns signaling to the primary cilium, a cellular structure that is becoming increasingly appreciated for its role in mediating cell signals and neuronal function.
Joubert syndrome locus 1 (JBTS1, MIM%213300) was mapped to the distal q-arm of chromosome 9, between D9S1826-D9S1838 3, in two JS Emirate families variably associated with retinopathy, and a proven “molar tooth” sign (MTI-007 and MTI-008 previously called Family A and C, respectively, Fig. 1a, Supplemental Table 1) 3. To refine the candidate interval, we recruited two additional unaffected MTI-007 and two affected MTI-008 family members, and performed a 5K SNP analysis, confirming JBTS1 in MTI-007 (Supplemental Fig. 1). Family MTI-008 produced a linkage peak at JBTS1, among several other peaks of similar amplitude, with a maximum LOD score of about +1.94. As a result, these SNP scans failed to significantly narrow the candidate interval (Supplemental Table 2).
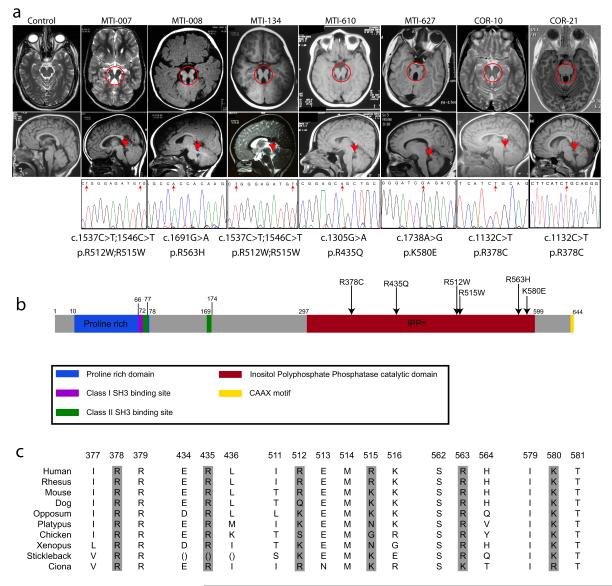
Missense mutations in the encoded enzymatic domain of INPP5E, (inositol polyphate-5-phosphatase E) in patients linked to the JBTS1 locus. (a) Top: axial brain MRIs showing molar tooth sign (red circle) from one affected from each linked family, representing four different countries of origin. Bottom: midline sagittal MRIs showing horizontally-oriented superior cerebellar peduncle (red arrow), not evident in control. Sequence chromatograms from one affected from each family, showing the nucleotide change (red arrow), with corresponding amino acid substitution listed below. Families MTI-007 and MTI-134 shared a common haplotype at the JBTS1 locus, as did COR-10 and COR-21, and they share common mutations. MTI-007 and MTI-134 have a compound homozygous mutation with a double R512W; R515W. (b) Predicted protein domains of INPP5E indicated by color. Each of the identified missense mutations (arrows) occurs in a basic residue within the catalytic domain and alters charge. (c) Evolutionary conservation of mutated amino acids.
An additional 25 consanguineous families with JS were analyzed, and three (MTI-134, MTI-610, and MTI-627, of Emirate, Turkish and Egyptian descent) showed at least one peak that overlapped with JBTS1. Two Italian families (COR-10 and COR-21) also linked to JBTS1 4. Haplotype analysis suggested that MTI-007 and MTI-134 were identical-by-descent, as were COR-10 and COR-21 (data not shown). Together these analyses defined a candidate interval of 3.5 MB in distal 9q34.2-tel, containing approximately 86 annotated candidate genes (Supplemental Table 3), 57 of which were screened for sequence changes, whereas the remaining 29 either lacked an open reading frame or were excluded due to lack of developmental expression. This extensive screening identified the INPP5E gene as mutated in each JBTS1-linked family (Fig. 1a-b).
The INPP5E mutations were notable for two reasons. First, all identified mutations were amino acid transversions that clustered within the enzymatically-active phosphatase domain (Fig. 1b). Second, each altered the charge of highly basic evolutionarily conserved amino acid residues (Fig. 1c), together suggesting that these mutations might alter enzymatic activity. None of these mutations were encountered among 188 control chromosomes from healthy ethnically matched individuals, and all segregated with the disease in each family. We modeled these mutations using the published crystal structure of synaptojanin5, the only inositol 5-phosphatase for which structural data is available. We found that each of the altered residues (with one exception, R563) was predicted to project its charged tail toward the presumed binding pocket of the PtdIns substrate (Supplemental Fig. 2), suggesting that these mutations might alter substrate specificity.
To test the effects of INPP5E mutations on PtdIns phosphatase activity, we immunoprecipated tagged wildtype and enzymatically null (D477N 6), together with each mutation separately from mammalian cells (resulting in similar protein levels, Supplemental Fig. 3) and then analyzed phosphatase activity against PtdIns(3,4,5)P3 and PtdIns(4,5)P2, the presumed cellular INPP5E substrates (Fig. 2a). We found that each of the JBTS1 missense mutations severely disrupted phosphatase activity towards PtdIns(3,4,5)P3 (Fig. 2b, p < 0.05 activity for each mutant, N = 3 independent experiments). Of note, since the R512W/R515W variants were observed on a single haplotype, we reasoned that probably only one was driving the defect in enzymatic activity, and under separate analysis, the R515W mutation was shown to be the major contributor to the defective enzymatic activity (data not shown). Comparison with phosphatase activity using PtdIns(4,5)P2 as substrate showed similar although not as severely compromised results as compared with wildtype (Fig. 2c), suggesting that PtdIns(3,4,5)P3 may be the relevant substrate in JBTS1. Because overexpression of INPP5E was previously shown to block Akt phosphorylation in response to PDGF stimulation in cultured cells (presumably by depleting levels of PtdIns(3,4,5)P3 and PtdIns(4,5)P2) 7, we next tested the effect of overexpression of each of the patient mutations in this published assay. Not only did patient mutations fail to block Akt signaling, but elevated basal levels of pAkt were also apparent in unstimulated cells (Supplemental Fig. 4). We conclude that the JBTS1-associated missense mutations impair phosphatase activity towards putative PtdIns substrates, which can alter downstream signaling events.
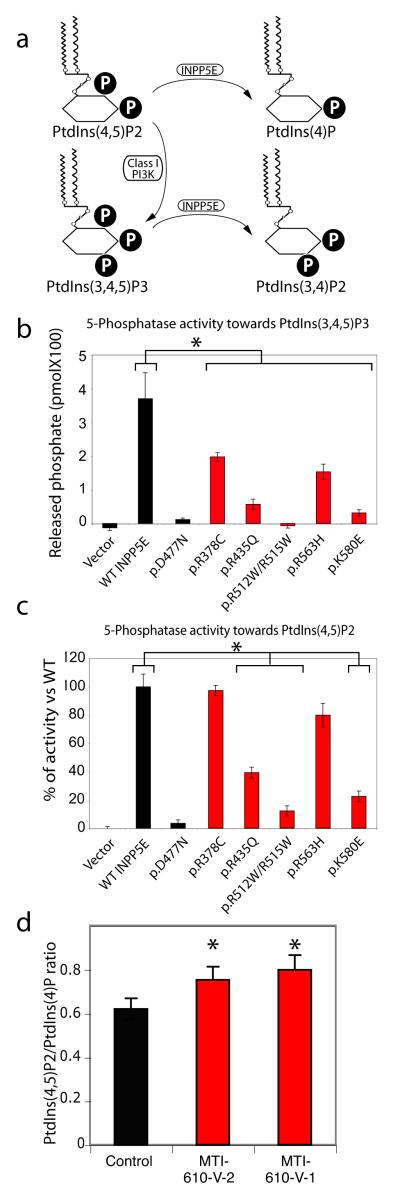
Impaired 5-phosphatase activity and altered ratio of PtdIns(4,5)P2 to PtdIns(4)P associated with JBTS1 INPP5E mutations. (a) Summary of PtdIns metabolism. P = phosphate. A block in INPP5E function is predicted to increase the PtdIns(4,5)P2:PtdIns(4)P ratio. (b-c) More severe reduction in 5-phosphatase activity of mutant INPP5E against PtdIns(3,4,5)P3 than PtdIns(4,5)P2 substrates. Note that activity was largely retained against PtdIns(4,5)2 for some mutations (mutants R435Q, R512W/R515W and K580E are severely defective, whereas R378C and R563H are only slightly diminished). D477N is known phosphatasedead, compared with each of the patient mutations. (N = 3 for each sample) (d) Elevated ratio of PtdIns(4,5)P2 to PtdIns(4)P in patient primary fibroblast lines MTI-610-V-2 and V-1, compared with control fibroblast. * represent p < 0.05 ANOVA two way corrected for multiple comparisons.
Next, we tested whether the absence of endogenous catalytic INPP5E activity results in an altered PtdIns profiles in cells. Although difficult to measure absolute levels PtdIns moieties in cells, the model predicted an increased ratio of PtdIns(4,5)P2/PtdIns(4)P due to a block in this enzymatic step (Fig. 2a), and thus skin fibroblasts from MTI-610 patient 1 and 2 (brother and sister) were assessed for PtdIns(4,5)P2/PtdIns(4)P ratio. In control unstimulated growth-arrested fibroblasts we found a PtdIns(4,5)P2/PtdIns(4)P ratio of about 0.63, whereas in both patient fibroblast samples the ratio was increased by about 20% (p = 0.05, N = 3 independent experiments, Fig. 2c). We found no differences in expression levels of INPP5E in this or other patient fibroblast samples with INPP5E mutations (Supplemental Fig. 5). We conclude that catalytic activity of INPP5E is required for maintenance of homeostatic ratios of PtdIns in cells.
In order to investigate a possible connection with ciliopathies, we examined INPP5E localization in cells with well-described cilia. The RPE-hTERT line (immortalized retinal pigment epithelia) display a primary cilia in >80% of growth-arrested cells 8,9. Coimmunostaining with pericentrin, a marker of the pericentriolar matrix, showed INPP5E immediately adjacent to the base of the cilium (Fig. 3a). This was confirmed by INPP5E coimmunostaining with acetylated tubulin, a marker of the ciliary axoneme, as well as with ARL13B, a cilia marker 10-12, which showed tight-colocalization of INPP5E to the ciliary axoneme, with only minimal INPP5E localized to non-ciliary locations (Fig. 3b). Transfection of wild-type GFP-tagged INPP5E into IMCD-3 cells under ciliary-permissive conditions demonstrated ciliary localization (Supplemental Fig. 6), and we found no defect in ciliary localization of any of the JBTS1-associated patient missense mutations when expressed in these cells (data not shown). By contrast, cells at mitotic stages (after cilium withdrawal) showed cytoplasmic localization (Supplemental Fig. 7). The data suggests that INPP5E localizes predominantly to the primary cilium during interphase in the cell lines tested.
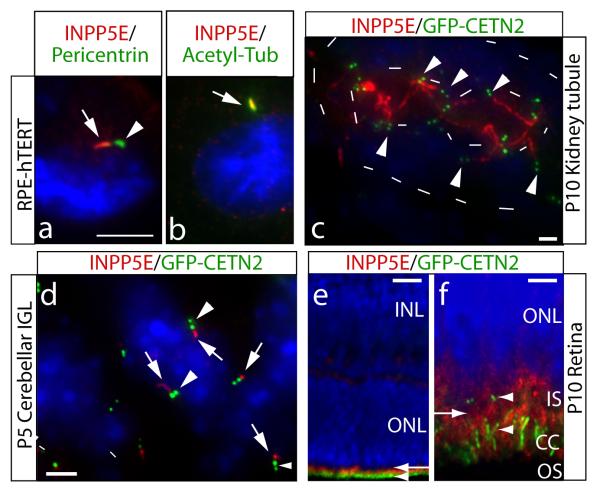
Ciliary axonemal localization of INPP5E and enhanced ciliary lability in INPP5E mutant cell line. (a) RPE-hTERT ciliated cells stained for INPP5E and cilia markers. INPP5E (red, arrow) localized adjacent to pericentrin-positive (green, arrowhead) pericentriolar matrix. (b) Colocalization of INPP5E (red, arrow) with ciliary axonemal acetylated tubulin (green). (c-f) show INPP5E (red) staining in GFP-CETN2 transgenic mouse (green pair of centrioles, arrowheads). (c) P10 kidney collecting tubule shows INPP5E-positive red cilia projecting into the renal tubule lumen (inner dashes), limits of tubule indicated by outer dashes. (d) P5 cerebellar internal granule layer with INPP5E (red, arrows) ciliary axoneme adjacent to the centrioles. (e) P10 retina, where GFP-CETN2 labels the basal body and connecting cilium (CC, green, arrowhead). INPP5E exclusively labels photoreceptor cells (red, arrow) (f) High-power view of (e) showing INPP5E staining (red, arrow) in the inner segment (IS), just adjacent to and above basal bodies and CC( arrowheads). Blue = Hoechst. Scale bar 5 um (a-b,d), 25 um (c), 50 um (e), 10 um (f).
We stained for INPP5E in organs affected in JS from the GFP-Centrin2 transgenic mouse (marks the base of the cilium) 13, and identified cilia localization in each. In renal collecting tubules, cilia were evident projecting from the epithelium into the lumen. In this mouse, many cells were marked by luminal localization of the pair of centrioles, and from this site, the majority of cells had an INPP5E-positive cilium projecting into the lumen (Fig. 3c). In the developing cerebellum, cilia have been identified on a population of neuroblasts in the cerebellar internal granule layer 14. We identified these ciliated cells based upon GFP signal at the centrioles, and identified INPP5E-positive cilia projecting into the parenchyma in the vast majority of these cells (Fig. 3d), and similar ciliary localization was observed in the hepatic bile ducts (not shown). We also observed that INPP5E localized to the region adjacent to the basal body and connecting cilium in retinal photoreceptor cells (Fig. 3e-f). Therefore, INPP5E is predominantly localized adjacent to or within the cilium in each of the major organs affected in JS.
To assess the potential function of INPP5E at the primary cilium, we utilized primary fibroblasts from patient MTI-610-V-1. These cells displayed essentially normal growth characteristics, as well as percentage of ciliated cells compared with controls (81% vs. 76%, p > 0.05, Fig. 4a), suggesting that INPP5E enzymatic activity is not required for ciliogenesis or cilia maintenance, at least not in fibroblasts. However, the cilium is not a fixed structure, and is labile to the application of serum-derived factors such as PDGF 15. Since serum is a potent activator of the PtdIns pathway in cultured cells, we predicted that the altered PtdIns profile might render cilia in mutant cells more labile to the application of serum. We applied serum to control and MTI-610-V-1 cells and scored the percentage of cells retaining a cilium, based upon acetylated-tubulin and ARL13B staining at specific time intervals. By 4 hr after serum application, wild-type cells had an 8% cilia withdrawal rate whereas INPP5E mutant cells had a 33% cilia withdrawal rate, (73% vs. 43%, p < 0.01, N = 3 independent experiments). At the 8 hr and 24 hr timepoints, the percentage of cells with visible cilia had equalized to about 40% in the two genotypes, suggesting that INPP5E controls timing of cilia disassembly during early response to serum.
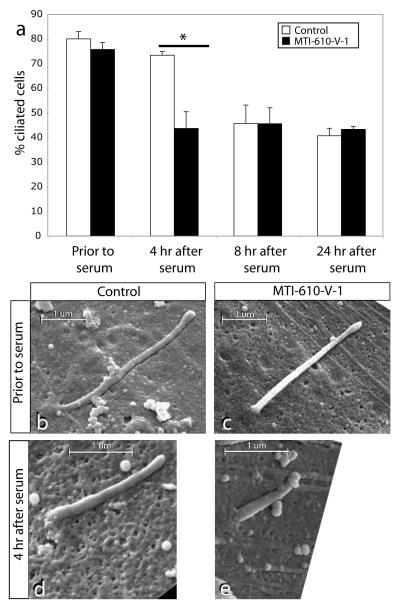
(a) Lability of cilia in patient primary fibroblasts with mutant INPP5E. Prior to serum stimulation, serum-starved control and patient fibroblasts displayed comparable percentage of ciliated cells. By 4 hr after serum stimulation, the ciliated cells dropped to 43% in patient fibroblast as compared to 73% in control (* = p < 0.01, N =3, total cells counted >250 for each condition). By 8 hr or 24 hr after stimulation, ciliated cells were again comparable between patient and control samples. (b-e) Scanning electron micrographs of control vs. patient primary fibroblast with mutant INPP5E. Prior to serum stimulation, ciliary axonemal length is comparable, although slightly shorter in patient sample. By 4 hr after stimulation, axonemal length is shorter in patient sample. Scale bar indicated in um.
To determine if the reduced number of ciliated mutant cells is due to an enhanced rate of cilia withdrawal in the absence of functional INPP5E, we performed scanning electron microscopy. Prior to the addition of serum, we found that the length of the primary cilium was approximately the same in control vs. MTI-610-V-1 serum-starved fibroblasts, measuring ~3 um in length (Fig. 4b-e). At the 4 hr timepoint (where we saw the maximal difference in cilia number), however, cilium length of the MTI-610 fibroblasts was about half of control. These data were quantified in x-y sections from cells with remaining visible cilia as evidenced by immunostaining for ARL13B prior to, and then at 4 hr and 8 hr after serum addition. We found a significant decrease in average cilia length at 4 hr after serum stimulation in mutant vs. controls (2.73 vs. 1.28 um, wildtype vs. MTI-610, p < 0.01, n > 30 cells for each condition), whereas no difference was apparent either before serum or after 8 hrs (Supplemental Fig. 8). Thus ciliary disassembly is more rapid in the absence of full INPP5E activity compared with wildtype.
Following ascertainment of fibroblast samples from additional patients with INPP5E mutations, we noted some alterations in cell proliferation rates in culture, and thus tested quiescent cells for ability to reenter the cell cycle. While MTI-610 fibroblasts had no apparent alteration in cell cycle reentry (based upon Ki67 staining or BrdU incorporation), MTI-134 and MTI-627 exhibited a reduction in the percentage of mitotically active cells following serum addition (Supplemental Fig. 9). As MTI-134 and MTI-627 had more severe reduction in enzymatic activity than MTI-610 in the in vitro assay (0% and 8.5% vs.15%, presented in Fig. 2), the data suggests that residual but not full function of INPP5E may be required for this serum-based cell-cycle reentry. MTI-610 fibroblasts exhibited a normal percentage of mitotically active cells, however, a greater percentage of these mitotically synchronized cells reentered the cell cycle initially (Ki67 staining between 18-24hrs) and progressed through the cell cycle more rapidly (reduction of BduU incorporation from 24hrs to 30hrs) than control fibroblasts, suggesting that INPP5E enzymatic activity may also influence the rate of cell cycle progression. Not all serum-based responses are abnormal in these patient samples, however. For instance, there were no alterations in low-dose serum-dependent scratch-wound assay healing (Supplemental Fig. 10), which we designed to be migration-dependent but largely proliferation-independent 16. We conclude that INPP5E may play specific serum-dependent cellular roles, mediating both ciliary stability as well as cell cycle dynamics.
Our data demonstrates that hypomorphic mutations in INPP5E lead to JS, providing the first evidence of a link between PtdIns signaling and the ciliopathies. It will be important to determine if the developmental defects observed in JS are the result of defects in cilia maintenance or due to a more broadly defined disruption in PtdIns signaling. Since PDGFαα receptor has been localized to primary cilia 17, and since PDGF is one of the key serum-derived factors that mediates cilia withdrawal concurrent with cell cycle reentry 18 it is possible that PDGF and other factors that utilize cilia as signaling centers have downstream pathways that are regulated through specific localization and function of INPP5E.
Previous studies have hinted at possible connections between PtdIns signaling and the cilium. Tubby mutant mice have cilia-like phenotypes and the protein encoded from the mutant gene binds specific PtdIns including PtdIns3,4,5(P3) 19. Furthermore, Bardet-Biedl syndrome shares similar ciliopathy features to JS, and the BBS5 protein can bind PtdIns 9. However, our findings are the first, to our knowledge, to directly implicate defective enzymatic conversion within the PtdIns class. The fact that INPP5E displays enrichment within the ciliary axoneme makes it tempting to speculate that PtdIns signaling may not just function to regulate cilia stability, but also that this signaling may occur within the cilium. An alternative model is that INPP5E might be sequestered within the cilium, but exert its activity within the cytoplasm, similar to the models being proposed for Sonic hedgehog signaling 20. The identification of a key regulatory enzyme in this process highlights an important area of research with clinical relevance.
Methods
Patients and families
Twenty-seven consanguineous families from the Middle East, Turkey and Europe were included in this study based upon the following criteria: 1) at least one individual with a neuroradiographically proven “molar tooth sign” associated with any JS or related disorder phenotype; 2) evidence for linkage to the JBTS1 locus; 3) exclusion of linkage to any other of the known JBTS loci (JBTS2-9). Whenever possible, patients underwent a full diagnostic protocol as previously reported 21. Brain MRI analysis was completed during the clinical assessment using standard sequence protocols. Parental written informed consent was obtained from all families, and the study was approved by the HRPP Committees of UCSD (La Jolla) and CSS-Mendel Institute (Rome).
Genome-wide screen and fine mapping of the JBTS1 locus
A 5K whole genome linkage scan was performed in all informative families using the Illumina Linkage IVb mapping panel 22, and analyzed with easyLINKAGE-Plus software 23, which runs Allegro version 1.2c in a PC Windows interface to calculate multipoint LOD scores. Parameters were set to autosomal recessive with full penetrance, and disease allele frequency of 0.001. Fine mapping on selected individuals was performed with the Affymetrix 250K Nsp1 SNP array and results analyzed using pedigree-free identity-by-descent mapping. Short tandem repeat polymorphic markers at the JBTS1 locus were used as previously published 4.
Mutation screening
Mutations were screened using a combination of single stranded conformational polymorphism analysis and direct sequencing as described 24,25.
Bioinformatics
Protein evolutionary conservation was determined by aligning amino acids from the Human Genome Browser (http://www.genome.ucsc.edu). Ciliary proteome was searched using web-based tools 26,27. Protein folding and intermolecular interactions were predicted using Swiss-Model with crystallized PtdIns 5-phosphatase domain of synaptojanin bound to Ca(2+) and PtdIns(1,4)P2 5. The resulting INPP5E structure was manipulated with PyMOL software by importing amino acid sequence for wildtype and mutant and comparing the likely docking site for PtdIns.
Biochemical assays
INPP5E cDNA 7 was cloned in frame with EGFP into pcDNA3.0 for expression. Patient mutations were individually engineered into the INPP5E cDNA in a Myc-tagged vector, stably transfected into 293T cells using the T-Rex tetracycline-regulated system (Invitogen)7, immunoprecipitated using Myc-tag, confirmed by Western to each express comparable amounts (Supplemental Fig. 3), and then used in a modified PtdIns(4,5)P2 hydrolysis assay as described 28. PtdIns(3,4,5)P3 hydrolysis was measured by adding purified substrate to the immunoprecipitates at 37°C for 10 min, followed by malachite green for an additional 10 min 29. Released inorganic phosphate was measured at 650 nm, compared against a standard curve, and performed in triplicate. Assessment of PtdIns(4,5)P2/PtdIns(4)P ratio from patient primary cells were metabolically labeled and performed as described 30. Stably transfected 293T cells were stimulated with 50ng/ml of PDGF and analyzed for Akt signaling as described7.
Histology
Mouse tissues from wildtype or GFP-CETN2 mice were immunostained with an affinity purified peptide-specific INPP5E rabbit antisera that reacts with human and mouse as reported 31 at 1:200 and images acquired on a DeltaVision imaging system with 5-10 cycles of deconvolution.
Fibroblasts assay
Primary skin fibroblasts from patient and unaffected passage-matched control were grown at similar confluence for 3 days in the absence of serum. Ciliary disassembly and wound healing was assessed as described 18,32. Cell-cycle re-entry was interrogated as described 33 with 20% serum addition to sub-confluent fibroblasts. BrdU (10 uM) was applied for 1 hr, and antigens detected with antibodies to Ki67(1:1000) and BrdU (1:1500) (Abcam # ab66155 and ab6326).
Scanning Electron microscopy
Fibroblasts were fixed in 2.5% glutaraldehyde/0.1 M cacodylate buffer (CB) (pH 7.4) for 30 min at 25C, rinsed (0.1 M CB), postfixed (1% OsO4/0.1 M CB) for 30 min at 4C, dehydrated with a graded alcohol series, which was displaced with hexamethyldisilazane, and evaporated overnight before sputter coating samples with palladium and imaged on XL30 ESEM-FEG.
Cilia length quantification
Cilia length was correlated in the x-y-z vs. x-y using the Volocity rendering software. 3D reconstruction of cilia using DeltaVision imaging system with 100X objective with 0.1 um z-steps allowed for length assessment irrespective of angle of orientation. Cilia length was also measured in the same cells using x-y only projections, and measurements were compared for five cells. No observable difference in cilia length measurements were detected, and thus subsequent lengths were measured from x-y projections.
Acknowledgements
We thank the Marshfield Clinic Research Foundation, Center for Inherited Disease Research and UCLA Microarray Core (supported by NHBLI and NIH) for genotyping support. Jennifer Meerloo at the UCSD Neuroscience Microscopy Imaging Core provided imaging support. Ryan Anderson of the UCSD Material Sciences provided electron microscopy support. We thank the Dixon lab (UCSD) for suggestions and help with protein modeling, the Mitchell lab (Monash University) for reagents, and the Schumans lab (IBMM) for help with the phosphatase assay and for communicating results prior to publication. This work was supported by the UCSD Neuroplasticity of Aging Training Grant (to SLB.), Italian Ministry of Health (RC2008 to BD, Ricerca Finalizzata 2006 to EMV), Telethon Foundation Italy (GGP08145 to EB/EMV) NIH HL 16634 (PWM and MVK), American Heart Association Grant 0730350N (MVK), NINDS, Burroughs Welcome Fund, the March of Dimes, and the Howard Hughes Medical Institute (to JGG).
Footnotes
GenBank accession numbers. Human INPP5E is assigned GenBank number NM_019892, and the encoded protein NP_063945. There is one minor splice variant annotated in human (U45974), due to a deletion of the 3′ stop codon, resulting in an additional 50 amino acids of the C-terminal protein. This stretch has no similarity in the database and is of unknown function.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ng.423
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2746682?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/ng.423
Article citations
The Knowns and Unknowns of Membrane Features and Changes During Autophagosome-Lysosome/Vacuole Fusion.
Int J Mol Sci, 25(20):11160, 17 Oct 2024
Cited by: 0 articles | PMID: 39456939 | PMCID: PMC11508585
Review Free full text in Europe PMC
A class I PI3K signalling network regulates primary cilia disassembly in normal physiology and disease.
Nat Commun, 15(1):7181, 21 Aug 2024
Cited by: 0 articles | PMID: 39168978 | PMCID: PMC11339396
Molecular and structural perspectives on protein trafficking to the primary cilium membrane.
Biochem Soc Trans, 52(3):1473-1487, 01 Jun 2024
Cited by: 0 articles | PMID: 38864436 | PMCID: PMC11346432
Review Free full text in Europe PMC
Parkinsonism Sac domain mutation in Synaptojanin-1 affects ciliary properties in iPSC-derived dopaminergic neurons.
Proc Natl Acad Sci U S A, 121(17):e2318943121, 18 Apr 2024
Cited by: 1 article | PMID: 38635628 | PMCID: PMC11047088
Molecular Mechanism of Autophagosome-Lysosome Fusion in Mammalian Cells.
Cells, 13(6):500, 13 Mar 2024
Cited by: 2 articles | PMID: 38534345 | PMCID: PMC10968809
Review Free full text in Europe PMC
Go to all (268) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases
- (1 citation) OMIM - 213300
Nucleotide Sequences
- (1 citation) ENA - U45974
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NM_019892
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse.
Nat Genet, 41(9):1027-1031, 09 Aug 2009
Cited by: 240 articles | PMID: 19668215
A compartmentalized phosphoinositide signaling axis at cilia is regulated by INPP5E to maintain cilia and promote Sonic Hedgehog medulloblastoma.
Oncogene, 36(43):5969-5984, 26 Jun 2017
Cited by: 30 articles | PMID: 28650469
The Joubert Syndrome Protein Inpp5e Controls Ciliogenesis by Regulating Phosphoinositides at the Apical Membrane.
J Am Soc Nephrol, 28(1):118-129, 08 Jul 2016
Cited by: 30 articles | PMID: 27401686 | PMCID: PMC5198270
Regulation of PtdIns(3,4,5)P3/Akt signalling by inositol polyphosphate 5-phosphatases.
Biochem Soc Trans, 44(1):240-252, 01 Feb 2016
Cited by: 36 articles | PMID: 26862211
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (3)
Grant ID: P30 CA023100
Grant ID: R01 CA138676
Grant ID: R01 CA138676-01
NCRR NIH HHS (2)
Grant ID: P41 RR011823
Grant ID: P41 RR011823-135880
NHLBI NIH HHS (2)
Grant ID: R01 HL016634
Grant ID: HL 16634
NIH HHS (2)
Grant ID: DP2 OD004265-01
Grant ID: DP2 OD004265
NINDS NIH HHS (7)
Grant ID: P30 NS047101
Grant ID: R13 NS053097
Grant ID: R01 NS048453
Grant ID: R13 NS053097-01
Grant ID: R01 NS052455
Grant ID: P30 NS047101-03
Grant ID: R01 NS048453-01
Telethon (1)
CLINICAL AND MOLECULAR CHARACTERIZATION OF JOUBERT SYNDROME AND RELATED DISORDERS
Prof. Enrico Silvio Bertini, Ospedale Pediatrico Bambino Gesù - IRCCS, Area di Ricerca Genetica e Malattie Rare
Grant ID: GGP08145





