Abstract
Free full text
Differential gene expression in LPS/IFNγ activated microglia and macrophages: in vitro versus in vivo
Abstract
Two different macrophage populations contribute to CNS neuroinflammation: CNS-resident microglia and CNS-infiltrating peripheral macrophages. Markers distinguishing these two populations in tissue sections have not been identified. Therefore, we compared gene expression between LPS (lipopolysaccharide)/interferon (IFN)γ-treated microglia from neonatal mixed glial cultures and similarly treated peritoneal macrophages. Fifteen molecules were identified by quantative PCR (qPCR) as being enriched from 2-fold to 250-fold in cultured neonatal microglia when compared with peritoneal macrophages. Only three of these molecules (C1qA, Trem2, and CXCL14) were found by qPCR to be also enriched in adult microglia isolated from LPS/IFNγ-injected CNS when compared with infiltrating peripheral macrophages from the same CNS. The discrepancy between the in vitro and in vivo qPCR data sets was primarily because of induced expression of the ‘microglial’ molecules (such as the tolerance associated transcript, Tmem176b) in CNS-infiltrating macrophages. Bioinformatic analysis of the ~19000 mRNAs detected by TOGA gene profiling confirmed that LPS/IFNγ-activated microglia isolated from adult CNS displayed greater similarity in total gene expression to CNS-infiltrating macrophages than to microglia isolated from unmanipulated healthy adult CNS. In situ hybridization analysis revealed that nearly all microglia expressed high levels of C1qA, while subsets of microglia expressed Trem2 and CXCL14. Expression of C1qA and Trem2 was limited to microglia, while large numbers of GABA+ neurons expressed CXCL14. These data suggest that (i) CNS-resident microglia are heterogeneous and thus a universal microglia-specific marker may not exist; (ii) the CNS micro-environment plays significant roles in determining the phenotypes of both CNS-resident microglia and CNS-infiltrating macrophages; (iii) the CNS microenvironment may contribute to immune privilege by inducing macrophage expression of anti-inflammatory molecules.
The CNS is supported and defended by two different macrophage populations: CNS-resident microglia and CNS-infiltrating macrophages (reviewed in Carson et al. 2007). In histological sections, these two cell types cannot be reliably distinguished from one another because CNS-resident microglia express most (if not all) common macrophage markers. In addition, both cell types can acquire either stellate or amoeboid morphologies depending on the CNS micro-environment. Despite this high degree of similarity, a large number of functional studies using flow cytometry and irradiation bone marrow chimeric mouse methodologies have convincingly demonstrated that these cells are functionally distinct (Hickey and Kimura 1988; Sedgwick et al. 1991; Vallieres and Sawchenko 2003; Byram et al. 2004; and reviewed in Ransohoff 2007).
Most tissue macrophages are relatively short-lived and are continually being replenished from bone marrow-derived cells (reviewed in Carson et al. 2006a,b; and Ransohoff 2007). By contrast, irradiation bone marrow chimeric mice reveal that CNS-resident microglia are largely self-renewing and rarely replenished from the bone marrow. Recent studies using non-irradiated parabiotic mice have confirmed these observations (Ajami et al. 2007). Irradiation bone marrow chimeric mice have also been used to selectively express major histocompatability complex (MHC) class II in either the radiation-insensitive (microglia) or radiation-sensitive compartment (peripheral CNS-infiltrating immune cells). These studies demonstrated that while peripheral immune cells were highly effective at initiating pro-inflammatory lymphocyte responses, CNS-resident microglia were not (Hickey and Kimura 1988; Greter et al. 2005). Rather CNS-resident microglia appeared to play an opposing role either in limiting pro-inflammatory lymphocyte responses or in initiating neuroprotective CD4+ T-cell responses (Byram et al. 2004; Takahasi et al. 2007).
Although CNS-resident microglia cannot be distinguished histologically from other macrophage populations, they can be distinguished using flow cytometry. Specifically, in CNS cell suspensions, CNS-resident microglia can be distinguished from acutely infiltrating peripheral macrophages by their lower levels of CD45 expression. Although the two levels of CD45 expression do slightly overlap, the bulk of each population can be reliably distinguished by this method (Sedgwick et al. 1991; Renno et al. 1995; Carson et al. 1998, 2006a,b; Fig. 1a). The stable differences in CD45 expression levels are also likely to have functional consequences. CD45 (also known as leukocyte common antigen) is a protein tyrosine phosphatase that has been implicated as an inhibitory receptor for a CD22; a ligand expressed by both CNS neurons and B cells (Mott et al. 2004). These data suggest that CNS neurons should be more effective at inhibiting infiltrating macrophages than CNS-resident microglia even when both are found in the same inflammatory foci! Considered together, these data demonstrate the continuing need to define the molecular differences between CNS-resident microglia and CNS-infiltrating macrophages.
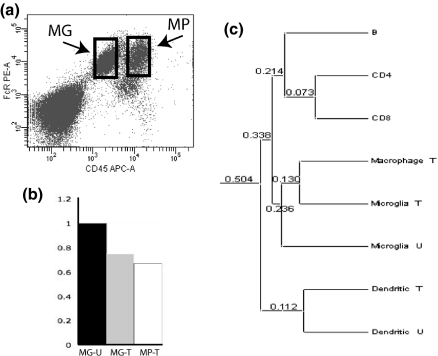
Microglia and macrophages isolated from the CNS show similar patterns of gene expression. (a): CNS-resident microglia (MG-boxed cells) and acutely infiltrating macrophages (MP-boxed cells) can be isolated from the same LPS/IFNγ-injected mouse brain and separated into two distinct cell populations using fluorescence-activated cell sorting. Microglia are defined as the Fc receptor positive, CD45lo cells (left box), and CNS-infiltrating macrophages are defined as the Fc receptor positive, CD45hi cells. (b) qPCR analysis of Tmem176b expression in microglia isolated from healthy unmanipulated adult CNS (MG-U), microglia isolated from LPS/IFNγ injected mouse CNS (MG-T) and acutely infiltrating macrophages isolated from the same injected CNS (MP-T). (c) Approximately 19000 different mRNAs were detected by TOGA gene expression profiling of lymphocytes (B cells, CD4+ and CD8+ T cells) isolated by flow cytometry from lymph nodes of naïve adult mice; unactivated CD45lo microglia from unmanipulated healthy adult mice, activated CD45lo microglia and CD45 high macrophages isolated by flow cytometry from LPS/IFNγ-injected adult murine; and in vitro unactivated and LPS-activated bone marrow-derived dendritic cells. Between each of the samples, pair-wise comparisons were made of the numbers of mRNAs expressed and the relative expression levels of the approximately total 19000 mRNAs detected by TOGA. From these comparisons, linear correlation coefficients were calculated in a pair-wise fashion for each of the samples. The difference index (numeric values listed at each branch in c) between each sample was calculated as 1 minus the correlation coefficient.
Using CD45 expression levels as a cell type specific maker, small numbers of microglia and CNS-infiltrating macrophages can be isolated from adult rodent CNS by flow cytometric cell sorting (Sedgwick et al. 1991; Renno et al. 1995; Carson et al. 1998). However, only a few hundred thousand cells per brain can be isolated by this method, which is insufficient for many functional and molecular assays (Carson et al. 1998, 2006a,b). We and others have therefore used primary microglia from neonatal mixed glial cultures and thioglycolate-elicted peritoneal macrophages as models for activated CNS-resident microglia and CNS-infiltrating macrophages, respectively (Carson et al. 1998, Papenfuss et al. 2007). Primary cultured microglia from mixed glial cultures have advantages over the common microglia cell lines as models for microglia in vivo. Primary cultured microglia are not oncogene immortalized and they are differentiated in the presence of astrocytes and oligodendrocytes. Thioglycolate-elicited peritoneal macrophages have advantages over splenic macrophages and oncogene-transformed macrophage cell lines in that they have been recently induced to infiltrate the peritoneum in response to pathogen recognition receptors. Here we test the predictive nature of these commonly used models to identify cell-type specific markers able to distinguish CNS-resident microglia from CNS-infiltrating macrophages.
Materials and methods
Microglia isolation from mixed glial cultures
Mixed glial cultures were prepared as previously described (Carson et al. 1998, 1999). Briefly, brains from postnatal day 1–3 C57Bl/6J mice were stripped of meninges, and the cortices mechanically dissociated, seeded into T-75 flasks, and maintained in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum and insulin (5 μg/mL). After 3 weeks, mixed glial cultures were trypsinized and incubated as a single cell suspension in Dulbecco’s modified Eagle’s media without phenol red for 30 min at 37°C to allow for the re-expression of trypsinized surface markers. Microglia were purified to > 98% by flow cytometry using phycoerythrin-conjugated antibodies directed against FcR/CD16/CD32 (Pharmingen, San Diego, CA, USA). Cytoplasmic mRNA was prepared from isolated cells immediately after isolation as previously described (Schmid et al. 2002).
Peritoneal macrophage and bone marrow-derived dendritic cell preparation
Peritoneal macrophages were prepared as previously described (Carson et al. 1998, 1999). Briefly, 3.0 mL of aged sterile thioglycolate broth were injected into the peritoneum of C57Bl/6J mice (Difco, Detroit, MI, USA). Mice were killed 3 days post-injection and peritoneal macrophages were harvested after killing by rinsing the peritoneal cavity with two 5 mL washes of OM5 media supplemented with 5.0 U/mL heparin (Sigma, St. Louis, MO, USA). Exudate cells were plated and allowed to adhere. Non-adherent cells were removed by rinsing the cultures. Greater than 90% of the remaining adherent cells were macrophages (CD11b+ cells). Bone marrow-derived dendritic cells were prepared by standard methods and purified by flow cytometry as described in Carson et al. (1999). Lymphocytes were isolated from lymph nodes of naïve mice by flow cytometry as previously described in Carson et al. 1999.
Microglia and macrophage isolation from healthy and inflamed adult CNS
Microglia were isolated as previously described (Carson et al. 1999) from the CNS of healthy adult (8–12 weeks old) C57Bl/6J mice or from mice injected intracerebrally with LPS (100 ng/mL) and interferon (IFNγ, 10 U/mL) in a total volume of 5 μL. In brief, mice were killed by halothane inhalation, and the brains of the mice rapidly removed and mechanically dissociated. The cell suspension was separated on a discontinuous 1.03/1.088 Percoll gradient and microglia/macrophages were collected from the interface as well as from the 1.03 Percoll fraction. Microglia and CNS-infiltrating macrophages were purified by flow cytometry using Allophycocyanin-conjugated antibodies against pan-CD45 and phycoerythrin-conjugated antibodies against FcR/CD16/CD32 (Pharmingen). Activated microglia were identified as FcR-positive, and CD45lo/intermediate and macrophages identified as FcR-positive, CD45hi.
RNA extraction and reverse transcription
RNA was extracted from isolated cells immediately after isolation as previously described (Schmid et al. 2002). RNA concentration was assessed by measuring the absorbance at 260 nm and was adjusted to 1 μg/μL. RNA integrity and absence of genomic DNA were verified by fractionating samples by electrophoresis in denaturing gels. Gels were stained with ethidium bromide and the ratio of 28S to 18S RNA was quantified. Two micrograms of RNA were RT using first strand cDNA synthesis kit according to manufacturer’s instructions (Amersham Biosciences, Buckinghamshire, UK).
Quantitative real-time PCR analysis
The oligonucleotide primers used are listed in Table S1. Primers were chosen to anneal with a DNA template at a temperature of 60°C and to encompass a coding fragment of 100–300 bp. qPCR was performed as previously detailed in Melchior et al. (2002). In brief, a constant amount of 200 ng cDNA from each RT or each dilution of the appropriate standard, was amplified in 25 μL of TaqMan PCR Core Reagent (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions. The reaction mixture consisted of 0.5 U of AmpliTaq Gold polymerase, each of the four dNTPs (0.2 mM), with dUTP replacing dTTP, each pair of primers (300 nM), and MgCl2 (3 mM final concentration) in the above-described Tris buffer. Amplifications were performed in an ABI Prism 7700 Sequence Detector System (Applied Biosystems). The reaction was started with a step of 10 min at 50°C for the removal of uracyl residues incorporated into the cDNAs with 1 U of uracyl-N-glycosylase (AmpErase reagent). This was followed by an incubation of 10 min at 95°C for AmpliTaq Gold activation, then 40 cycles each consisting of 15 s at 95°C and 1 min at 60°C. At the end of each experiment, amplification products were fractionated by gel electrophoresis to verify that they migrated as a single band with the expected size. Each sample was analyzed in duplicate. For the quantitative analysis, direct detection of PCR products was performed by measuring the progressive increase in fluorescence emitted by the binding of SYBR Green to double-stranded DNA.
Data were normalized to the endogenous reference hypoxanthine phosphoribosyl transferase (HPRT); then using the comparative cycle threshold (CT) method, the amount of RNA transcripts were expressed as a reference to a standardized unstimulated microglia sample. CT was defined as low as transcripts were abundant. For validation of the ddCt method, we also used standards for calibration of HPRT and Trem2 cDNA. Briefly, PCR fragments that served as standards for calibration of quantitative PCR were purified from preparative gels with an extraction kit (QiAquick; Qiagen Germany). Fragment concentrations were assessed by measuring their absorbance at 260 nm and according to their respective molecular weights; these standards were subsequently diluted to obtain 107, 106, 105, 104, 103, and 102 copies per well. The copy number was obtained by relating the fluorescence measured at the CT in unknown samples to the calibration curve derived from the standards. Using the standard curve, it was verified that the absolute copy number of HPRT transcripts was of the same order of magnitude in all samples analyzed by qPCR.
In situ hybridization
In situ hybridization was performed on free-floating cryosections sections as previously described (Schmid et al. 2002; Thrash et al. 2009, Papenfus et al. 2007). Briefly, coronal sections were hybridized at 55°C for 16 h with a 33P-labeled riboprobe (107 cpm/mL). Myeloid cells and blood vessels were identified by their ability to bind biotinylated tomato lectin (Vector Laboratories, Burlingame, CA, USA). Bound biotinylated tomato lectin was visualized by standard streptavidin, horseradish peroxidase-diaminobenzadine (DAB) methodology. Sections were mounted onto Fisherbrand (Fisher Scientific, Pittsburgh, PA, USA) superfrost/plus slides and dehydrated with ethanol and chloroform. Slides were exposed for 3 days to Kodak X-AR film and dipped in Ilford K-5 emulsion (Polysciences, Warrington, PA, USA). After 3 weeks, slides were developed with Kodak D19 developer, fixed, and counter-stained with Mayer’s hematoxylin. Riboprobes were prepared from antisense sequences corresponding to nucleotides 1282–1768 at the extreme 3′-end of the untranslated region of the large CXCL14 transcript to nucleotides 443–1005 within the open-reading frame of the published CXCL14 sequence (accession #: AF192557) and to the final 600 nucleotides within the 3′ untranslated region of C1qA (accession #: BC002086). Riboprobes for Trem2 were generated from reported plasmids as previously described (Schmid et al. 2002).
Northern blot
For Northern blots, 2 μg per lane of poly A+ or 10 μg per lane total RNA were resolved by electrophoresis in a 1.5% agarose/1.2 M formaldehyde gel, transferred to nylon membrane, and hybridized with 32P-radiolabeled probes (Schmid et al. 2002 and Thrash et al. 2009).
TOGA and bioinformatic comparison of microglial and macrophage transcriptomes
TOGA (Total Gene expression Analysis) analysis was performed as previously described (Schmid et al. 2002) on mRNA samples prepared from microglia and macrophages isolated from LPS/IFNγ-injected murine CNS. Briefly, RNA samples were converted to cDNA using a degenerate pool of biotinylated phasing primers that initiated synthesis at the beginning of the poly(A) tail on each mRNA. The cDNA collection was digested exhaustively with the restriction endonuclease MspI and the 3′ fragments were isolated by strepavidin-bead capture. These were released from the beads by cleavage with NotI, which recognized a site in the phasing primers. The captured fragments were modified at their 5′ ends to harbor a start site for in vitro transcription by ligation of an oligonucleotide adapter to the MspI overhang. Following incubation of the modified fragments with T3 polymerase, a collection of RNA fragments was produced corresponding to the 3′ portion of each starting mRNA, from its most 3′MspI recognition sequence to the beginning of its poly(A) tail, with each fragment flanked by linker tags of known sequence. For the initial PCR step, first-strand cDNA was prepared from the RNA pool by RT and used in four separate PCRs, in which a 5′ primer that extends by one of four possible nucleotides beyond the MspI site (N1 position) was paired with a universal 3′ primer to generate an N1-specific double-stranded DNA template. In the final step, 256 primers corresponding to all possible permutations of the four nucleotides immediately adjacent to the MspI recognition site (N1N2N3N4) were matched with the appropriate N1 template and utilized in individual robotically performed PCRs paired with a fluorescent primer to produce 256 non-overlapping pools of products, and these products were separated by electrophoresis. This process assigned each product a digital sequence tag address: an eight-nucleotide sequence (the four MspI recognition and the adjacent four parsing nucleotides) and a length, both of which were attributes of the individual mRNAs. The amplitudes of the fluorescent PCR products corresponded to the initial concentrations of their parent mRNAs. The amplitudes were automatically collated into a database, indexed by their digital sequence tag addresses that could be queried electronically to identify mRNAs, the concentration of which differed among the experimental samples.
Duplicate TOGA analysis of gene expression was performed on each of the cell population samples described in Fig. 1c and expression of 19023 different mRNAs was detected. Between each of the samples, pair-wise comparisons were made of the relative expression levels of ~19000 mRNAs detected by TOGA. From these comparisons, linear correlation coefficients were calculated in a pair-wise fashion for each of the samples. The difference index (DI) between each sample was simply calculated as 1 minus the correlation coefficient. The pairs of samples with the highest correlation coefficients (i.e., pairs with the least difference between samples) provided the first branching points. The peak data for these pairs were then averaged and the resulting data set was used to calculate new correlation coefficients with other combined data sets to identify the next branch points. This process was continued until all samples were connected in a complete tree as shown in Fig. 1c.
Results
Functional differences between CNS-resident microglia and acutely infiltrating macrophages have been identified (reviewed in Carson et al. 2006a,b, 2007), but as yet markers able to distinguish these cell types in histological samples have not. We previously compared differences in gene expression between primary neonatal mixed glial culture microglia and peritoneal macrophages treated for 22 h with LPS (100 ng/mL) and IFNγ (10 U/mL) using the gene expression profiling method, TOGA (Schmid et al. 2002; Carson et al. 2004). Here, we used real-time qPCR and biological replicate samples of neonatal microglia and peritoneal macrophages to validate and quantify the degree of microglial-enriched expression of the 15 TOGA-identified molecules (Table 1, column 1). Enriched expression in neonatal microglia ranged from 2-fold for TCD1D16, Trem2, and Tmem176b (also known as TORID, LR8) to 100-fold for complement component C1qA, GPR84, and hypothetical protein LOC620695. The functions of many of these molecules are still unknown. However, consistent with identified neuroprotective functions of microglia, Trem2 and the tolerance-associated transcript, Tmem176b, each have demonstrated functions in decreasing adaptive immune responses when transfected into peripheral immune cells (Louvet et al. 2005; Takahasi et al. 2007).
Table 1
Comparison of differential gene expression in microglia and macrophages when stimulated in culture versus when stimulated within the intact adult murine CNS by LPS/IFN
| Gene | Relative expression in vitro: neonatal cultured microglia/peritoneal macrophages | Relative expression ex vivo: adult CNS microglia/CNS-infiltrating macrophage |
|---|---|---|
| LOC620695 | 250.00 | 0.72 |
| GPR84 | 100.00 | 0.76 |
| Sal3 | 100.00 | 0.56 |
| C1qA | 100.00 | 2.56 |
| NAV3 | 33.33 | 0.63 |
| USP2 | 20.00 | 0.68 |
| Tem7R/Plxdc2 | 14.29 | 1.14 |
| Hspa4l | 7.69 | 0.45 |
| CXCL14 | 6.67 | 5.56 |
| Rrbp1 | 6.67 | 1.49 |
| Trim47 | 5.00 | 0.79 |
| SPARC | 4.00 | 1.47 |
| Tmem176b | 2.78 | 1.10 |
| Trem2 | 2.44 | 9.09 |
| TDC1D16 | 2.04 | 0.56 |
Shaded rows indicate molecules showing enriched expression in LPS/IFNγ activated microglia in vitro and in vivo.
To test whether activated neonatal microglia successfully predicted microglial-enriched gene expression in the adult CNS, we quantified relative expression of the 15 TOGA identified molecules in CNS-resident microglia and CNS-infiltrating macrophages isolated from LPS/IFNγ-injected adult murine CNS. Intracerebral LPS/IFNγ injection caused a rapid, robust and transient, and self-resolving CNS inflammation. Specifically, peripheral macrophages (defined as CD45hi cells) comprised < 1% of the myeloid cells isolated from the unmanipulated healthy adult murine CNS, while all CNS-resident microglia (CD45lo cells) were nearly MHC class II negative (data not shown, Carson et al. 1998). By contrast, CD45hi macrophages comprised ~40% of the cells isolated and nearly all microglia expressed detectable levels of MHC class II, 22 h post-intracerebral LPS/IFNγ injection (data not shown; Carson et al. 1998). To provide polarized populations of ex vivo microglia and macrophages, only cells from the non-overlapping (boxed) regions of the CD45lo and CD45hi flow cytometry dot plot were collected by fluorescence activated cell sorting for subsequent RNA isolation (Fig. 1a).
Despite the high degree of enriched expression in activated neonatal microglia, only 3 of the 15 original TOGA-identified molecules were enriched in activated adult microglia when compared with acutely infiltrating macrophages isolated from LPS/IFNγ injected CNS: C1qA, Trem2, and CXCL14 (Table 1, column 3). The discrepancy between the in vitro and in vivo data sets was primarily because of increased expression of these molecules by CNS-infiltrating macrophages. For example, Tmem176b was nearly undetectable in peritoneal macrophages (unstimulated or stimulated), but was strongly induced in CD45hi macrophages infiltrating LPS/IFNγ-injected murine CNS (Fig. 1b). These data suggest that in vitro studies overestimated the molecular differences between microglia and macrophages because the peritoneal macrophages were not exposed to the regulatory effects of the CNS micro-environment. By contrast the cultured neonatal microglia isolated from mixed glia cultures were exposed to astrocytes and oligodendrocytes in vitro.
To determine the relative similarity of total gene expression between CNS-resident microglia and CNS-infiltrating macrophages, we compared the numbers and levels of total mRNA transcripts detected by TOGA and expressed in common between LPS/IFNγ stimulated microglia and macrophages. We previously reported comparative analysis of the ~15000 distinct mRNAs detected in TOGA profiling studies of neonatal microglia and peritoneal macrophages (Carson et al. 2004). In these in vitro studies, LPS/IFNγ-stimulated neonatal microglia showed greater similarity in total gene expression to unstimulated microglia (DI of 0.194) than to either unstimulated or LPS/IFNγ-stimulated peritoneal macrophages (DI between microglia and macrophages = 0.359) (Carson et al. 2004). By contrast, comparative analysis of the ~19000 mRNAs detected in the ex vivo TOGA profiling studies unexpectedly revealed that microglia isolated from LPS/IFNγ-injected mice showed greater similarity in total gene expression profiles to CNS-infiltrating macrophages (DI: 0.130) from LPS/IFNγ-injected mice than to microglia isolated from unmanipulated healthy adult murine CNS (Fig. 1c). As a control for this bioinformatic analysis, we compared the gene expression similarities of ex vivo lymph node B cells, CD4 T cells, CD8 T cells, and bone marrow-derived cultured dendritic cells profiled in the same TOGA experiment. The gene expression patterns of these cells grouped as predicted from known biology and previously published profiling experiments (Fig. 1c). Specifically, both types of T cells showed greater similarity to each other than to B cells and all lymphocytes displayed greater similarity with each other than with all ex vivo myeloid cells (microglia and macrophages) (Fig. 1c).
C1qA and Trem2, two of the three molecules identified by qPCR as being enriched in adult microglia isolated from inflamed adult CNS (Table 1), are known to be expressed by microglia (Schmid et al. 2002; Takahasi et al. 2007; Farber et al. 2009; Thrash et al. 2009). Microglial expression of both C1qA (Fig. 2a and b) and Trem2 (Fig. 2c and d) was readily detected in both the healthy and inflamed adult CNS using in situ hybridization analysis. However, the in vivo expression patterns of the two molecules differed in microglia. In Fig. 2, we present the data from the healthy adult CNS, which lacks inflammatory macrophage infiltrates to clearly depict microglial (and not macrophage) expression of these molecules. While nearly all microglia expressed C1qA, the level of expression varied by twofold even in the healthy CNS when the number of emulsion grains per microglia was used as a measure of relative expression (Fig. 2a and b). By contrast, not all microglia expressed Trem2 in the adult CNS. In Fig. 2c and d, Trem2-positive- and Trem2-negative microglia are found in close proximity within the same brain region. These data demonstrate that while expression of these molecules was enriched in adult microglia, neither C1qA nor Trem2 is likely to be a useful universal marker distinguishing microglia from macrophages in CNS tissue sections. These data also demonstrate that microglias are not just heterogeneous between brain regions or during inflammatory responses but even within the same healthy brain region.
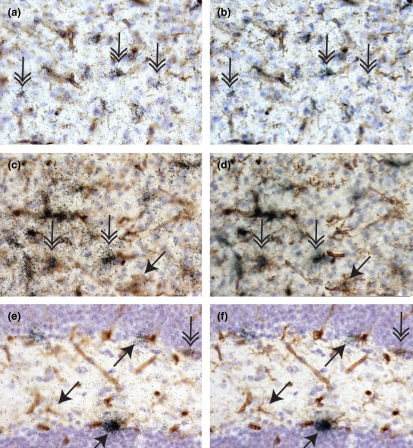
Heterogeneity of microglial gene expression in healthy murine CNS. All panels depict tissue sections prepared from a healthy unmanipulated adult mouse. In all panels, nuclei are labeled in blue with hematoxylin; microglia, macrophages, and blood vessels are labeled with tomato lectin and visualized in brown with DAB. C1qA (panels a, b), Trem2 (panels c, d), and CXCL14 (panels e, f) expression were detected using 33P-labeled gene-specific riboprobes. Expression is visualized by induction of black grains in the film emulsion coating each tissue section. In panels a, c, and e, the focal plane is on the level of emulsion. In panels b, d, and f, the focal plane is on level of immunohistochemistry. Downward pointing double arrows indicate examples of microglia positive for gene expression of C1qA (panels a, b), Trem2 (panels c, d), and CXCL14 (panels e, f). Angled downward single arrows indicate examples of microglia negative for gene expression. Upward angled single arrows indicate CXCL14 expression by lectin-negative cells. Panels a–d depict regions within the cortex, panels e and f depict regions within the hippocampus. DAB, diaminobenzadine.
CXCL14 was the third molecule identified as microglial-enriched from the qPCR screen and was also a demonstrated chemoattractant for B cells and monocytes. CXCL14 has been previously detected in CNS RNA but the brain regions and cells expressing CXCL14 had not been reported (Hromas et al. 1999; Cao et al. 2000; Frederick et al. 2000; Vadasz et al. 2007; Meuter and Moser 2008). Here, we examined the expression pattern of CXCL14 in unmanipulated healthy and LPS/IFNγ-injected adult murine CNS using two non-overlapping riboprobes that showed identical labeling in tissue sections (Figs. 2, 3 and Fig S1). CXCL14 expression was not regulated in vivo by LPS/IFNγ (Fig. 4e, and in situ hybridization data not shown). Therefore, CXCL14 expression is depicted only by the unmanipulated healthy adult CNS in Figs. 2e and f and 3 and Fig. S1. Surprisingly, the majority of CXCL14 expression was detected on lectin-negative cells (upward pointing arrows Fig. 2e and f). While most microglia were negative for CXCL14 expression (downward pointing single arrows Fig. 2e and f), low-level expression of CXCL14 was detected in rare microglia within the hippocampus, (downward pointing double arrow Fig. 2e and f) and in microglia located in the septum (Fig. 3a and b) and lateral striatum (Fig. S1 and data not shown). Within these last two regions, all cells including microglia expressed high levels of CXCL14 (Fig. 3a and b, Fig. S1). These data indicate that while CXCL14 mRNA is enriched in RNA pooled from all adult ex vivo microglia, CXCL14 expression is characteristic of only a very small subset of microglia in vivo.
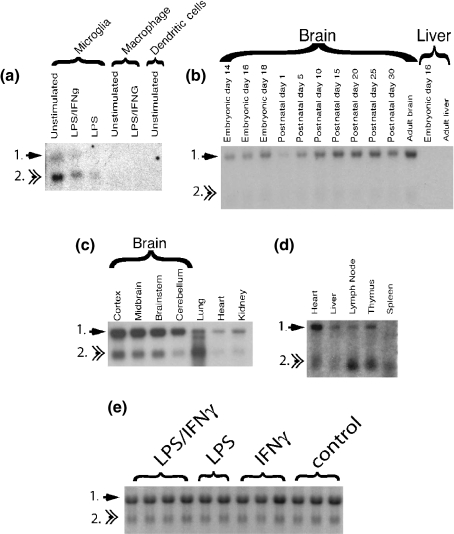
CXCL14 expression in cultured cells and murine tissues. Arrows point out the two known CXCL14 transcripts. (a) CXL14 expression in cultured cells. Cells were stimulated for 22 h with LPS (100 ng), IFNγ (10 U/mL) or LPS + IFNγ (10 U/mL, 100 ng/mL). (b) Developmental expression of CXCL14 in the CNS and liver. (c and d) Tissue distribution of CXCL14 expression. Northern blot in c was exposed overnight. The Northern blot in d was exposed for 2 weeks; e) each lane represents RNA isolated from a single mouse CNS that was unmanipulated or which received 5 μL intracerebral injections of IFNγ (10 U/mL), LPS (100 ng/mL) or IFNγ + LPS (10 U/mL, 100 ng/mL) 22 h prior to isolation.
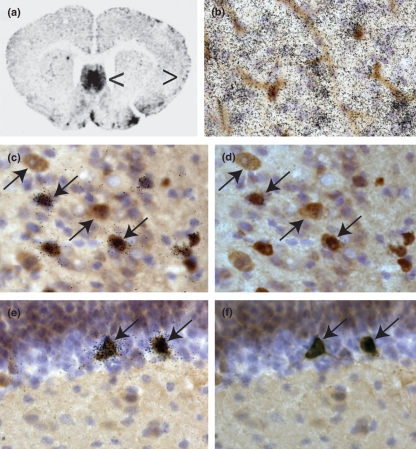
CXCL14 expression in healthy murine CNS in microglia and GABA+ neurons. Panel a is a single autoradiogram from the series depicted in Fig. S1. Left pointing arrow in panel a identifies region of septum depicted in panel b. Right pointing arrow in panel a depicts cortical area depicted in panels c and d. In panels b–f, nuclei are labeled in blue with hematoxylin. In panel b, microglia and blood vessels are labeled with tomato lectin and visualized brown with DAB. In panels c–f, GABA immunoreactivity is visualized in brown with DAB. CXCL14 expression was detected using 33P-labeled gene-specific riboprobes. Expression is visualized by induction of black grains in the film emulsion coating each tissue section. In panels b, c, and e, the focal plane is on the level of emulsion. In panels d and e, the focal plane is on level of immunohistochemistry. Downward-angled single arrows indicate examples of CXCL14+, GABA+ neurons in the cortex (panels c, d) and in the hippocampus (panels e, f). The upward pointing arrow indicates an example of CXCL14 negative, GABA+ neurons in the cortex (panels e, f). DAB, diaminobenzadine.
An overview of CXCL14 expression in the entire brain is depicted and described in Fig. S1. In brief, the highest levels of CXCL14 expression were observed in the septum, the islands of Calleja, the interpeduncular nucleus, the hippocampus, and the Purkinje layer of the cerebellum (Fig. S1). Cells expressing high levels of CXCL14 were found throughout the cerebral cortex but were most abundant in the cingulate cortex. In the striatum, CXCL14 expression delineated a lateral region of the striatum that has not been previously recognized as a defined striatal structure. Most CXCL14+ cells were identified as glial fibrillary acidic protein negative, 2′3′-cyclic nucleotide phosphodiesterase P-negative, and NeuN positive (data not shown). While nearly all CXCL14-positive cells displayed a GABAnergic neuronal phenotype (GABA-positive, large nucleus, and cell body: downward pointing arrows, Fig. 3b–d), not all GABAnergic neurons were CXCL14 positive (upward pointing arrows, Fig. 3b and c).
Finally, we examined CXCL14 expression in vitro and in vivo by Northern blot analysis (Fig. 4). CXCL14 is encoded by two mRNA transcripts with identical open reading frames (Hromas et al. 1999; Cao et al. 2000; Frederick et al. 2000; Vadasz et al. 2007; Meuter and Moser 2008). In cultured microglia (Fig. 4a), peritoneal macrophages (Fig. 4a), thymus (Fig. 4d), and secondary lymphoid structures (Fig. 4d; lymph node, spleen) the smaller CXCL14 transcript predominates (transcript 2, Fig. 4). By contrast, in non-immune organs including the CNS, the larger transcript predominates (transcript 1, Fig. 4a, c and e). Northern blot analysis confirmed enriched CXCL14 expression in cultured neonatal microglia when compared with peritoneal macrophages or bone marrow-derived dendritic cells (Fig. 4a). Because primary microglia from mixed glial cultures were derived from neonatal CNS cortices, we examined whether microglial expression of CXCL14 was more abundant in the embryonic or early postnatal CNS. By Northern blot analysis, the ratio of larger to smaller CXCL14 transcript did not change during development (Fig. 4b). By in situ hybridization analysis, the identical pattern of CXCL14 expression was observed at all ages examined from postnatal day 1 to 3 months (data not shown and Figs. 2 and 3, Fig. S1). These data indicated that detection of high levels of CXCL14 expression in neonatal microglia from mixed glial cultures was not a consequence of these cells reflecting an earlier stage of CNS development. Rather the robust expression of CXCL14 in cultured neonatal microglia was probably an artifact of culture in the absence of signals provided by an intact functional CNS.
Discussion
Several studies have identified functional differences in CNS-resident microglia and acutely infiltrating inflammatory macrophages (reviewed in Carson et al. 2007). However, as yet a universal marker that could be used to distinguish these functionally different cells in inflamed CNS tissue sections has not been identified. Here, we tested and confirmed by qPCR that a panel of 15 molecules previously identified by TOGA gene profiling were enriched in LPS/IFNγ-stimulated neonatal microglia when compared with similarly treated peritoneal macrophages.
Despite enriched expression from 2-fold to 250-fold in activated neonatal microglia in vitro, the majority of these TOGA-identified were not preferentially expressed in CNS-resident microglia when compared with CNS-infiltrating macrophages isolated from LPS/IFNγ-injected adult murine CNS. In large part, the failure to confirm preferential expression of 12 molecules in LPS/IFNγ-activated CNS-resident microglia was because of much higher expression of these molecules in CNS infiltrating macrophages (CD45hi cells) when compared with peritoneal macrophages (also CD45hi cells). Three molecules were confirmed by qPCR as enriched in adult microglia from LPS/IFNγ-injected adult CNS (C1qA, Trem2, CXCL14), but in situ hybridization analysis demonstrated that expression of these three molecules was heterogeneous in adult microglia in both the unmanipulated and inflamed CNS. Developmental regulation accounts for the observed heterogeneous pattern of microglia gene expression only in part. Before postnatal day 7, all microglia were Trem2-positive (Thrash et al. 2009). After eye opening in the pup, only subsets of microglia expressed varying levels of Trem2 (Schmid et al. 2002; Carson et al. 2006a,b; Thrash et al. 2009). By contrast, microglial expression of CXCL14 did not vary from postnatal day 1 adulthood.
The significance of CXL14 expression in the intact functional CNS is unknown. Because CXCL14 is a macrophage chemoattractant, it is tempting to speculate that it may contribute to organizing the close apposition of microglia to GABA-positive, CXCL14-positive neurons. However, as yet, microglia have not been formally demonstrated to respond to CXCL14 nor has the receptor for CXCL14 been identified. In addition, it is unknown why regions with high levels of CXCL14 expression do not show increased susceptibility to macrophage or B-cell infiltration. Is this an evidence of active immuno-suppression by the CNS environment or evidence of the failure to produce active bioavailable CXCL14 protein?
In aggregate, these data contribute to the growing literature demonstrating that microglia cultured in the absence of signals provided by an intact functional CNS do not fully mimic microglia located within the intact adult CNS (reviewed in Carson et al. 2007). Here, we show that cultured microglia expressed high levels of molecules such as CXCL14 that were rarely expressed by these cells in vivo. In addition, these data confirmed that microglial gene expression was not homogeneous throughout the CNS. Indeed, microglial phenotype, both in vivo and in vitro appeared to be determined by local environmental cues. Furthermore, the continuing failure to identify a definitive microglial-specific marker suggests that any molecule expressed by all microglia subsets in all micro-environments will also be expressed by nearly all inflammatory macrophages. Finally, induced expression of ‘microglial-specific’ molecules with immunosuppressive functions by CNS-infiltrating macrophages adds to the growing literature that the CNS actively regulates microglia and macrophage phenotypes via a wide array of neuronal and glial-expressed molecules (including CD22, CD200, fractalkine, and vasoactive intestinal protein) (Getts et al. 2008 and reviewed in Carson et al. 2007). The existence of active regulation of CNS inflammation by the CNS itself has consequences for the aging and diseased CNS. Failures in CNS immunoregulation have the potential to contribute to the onset and progression of chronic neuroinflammation characteristic of many neurodegenerative disorders.
Acknowledgments
This work was supported by grants from the NIH (NS39508; NS045735), Dana Foundation, UCR Biomedical Sciences PIC Program (to MJC) and a NMSS post-doctoral fellowship (BM).
Glossary
Abbreviations used
| CT | cycle threshold |
| DI | difference index |
| HPRT | hypoxanthine phosphoribosyl transferase |
| IFN | interferon |
| LPS | lipopolysaccharide |
| MHC | major histocompatability complex |
| TOGA | Total Gene expression Analysis |
Conflicts of interest
MJC has received a grant from BiogenIdec on research topics unrelated to the current study; the authors declare no conflicts of interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1 qPCR primer pairs.
Figure S1 CXCL14 expression in unmanipulated adult murine CNS.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ajami B, Bennett JL, Kreiger C, Tetzlaff W, Rossi RM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007;10:1538–1543. [Abstract] [Google Scholar]
- Byram SC, Carson MJ, DeBoy CA, Serpe CJ, Sanders VM, Jones KJ. CD4-positive T cell-mediated neuroprotection requires dual compartment antigen presentation. J. Neurosci. 2004;24:4333–4339. [Europe PMC free article] [Abstract] [Google Scholar]
- Cao X, Zhang W, Wan T, He L, Chen T, Yuan Z, Ma S, Yu Y, Chen G. Molecular cloning and characterization of a novel CXC chemokine macrophage inflammatory protein-2gamma chemoattractant for human neutrophils and dendritic cells. J. Immunol. 2000;165:2588–2595. [Abstract] [Google Scholar]
- Carson MJ, Reilly CR, Sutcliffe JG, Lo D. Mature microglia resemble immature antigen-presenting cells. Glia. 1998;22:72–85. [Abstract] [Google Scholar]
- Carson MJ, Reilly CR, Sutcliffe JG, Lo D. Disproportionate recruitment of CD8+ T cells into the CNS by professional antigen-presenting cells. Am. J. Pathol. 1999;154:481–494. [Europe PMC free article] [Abstract] [Google Scholar]
- Carson MJ, Thrash JC, Lo D. Analysis of microglial gene expression: identifying targets for CNS neurodegenerative and autoimmune disease. Am. J. Pharmacogenomics. 2004;4:321–330. [Abstract] [Google Scholar]
- Carson MJ, Thrash JC, Walters B. The cellular response in neuroinflammation: the role of leukocytes, microglia and astrocytes in neuronal death and survival. Clin. Neurosci. Res. 2006a;6:237–245. [Europe PMC free article] [Abstract] [Google Scholar]
- Carson MJ, Doose JM, Melchior B, Schmid CD, Ploix C. CNS immune privilege: hiding in plain sight. Immunol. Rev. 2006b;213:48–65. [Europe PMC free article] [Abstract] [Google Scholar]
- Carson MJ, Bilousova TV, Puntambekar SS, Melchior B, Dose JM, Ethell IM. A rose by any other name: the potential consequences of microglial heterogeneity during CNS health and disease. Neurotherapeutics. 2007;4:571–579. [Europe PMC free article] [Abstract] [Google Scholar]
- Farber K, Chenung G, Mitchell D, Wallis R, Weihe E, Schwaeble W, Kettemann H. C1q, the recognition subcomponent of the classical pathway of complement drives microglial activation. J. Neurosci. Res. 2009;87:644–652. [Europe PMC free article] [Abstract] [Google Scholar]
- Frederick MJ, Henderson Y, Xu X, Deavers MT, Sahin AA, Wu H, Lewis DE, El-Naggar AK, Clayman GL. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am. J. Pathol. 2000;156:1937–1950. [Europe PMC free article] [Abstract] [Google Scholar]
- Getts DR, Terry RL, Getts MT, Muller M, Rana S, Shrestha B, Radford J, Van Rooijen N, Campbell IL, King NJ. Ly6c+ “inflammatory monocytes are microglial precursors recruited in a pathogenic manner in West Nile virus encephalitis. J. Exp. Med. 2008;205:2319–2337. [Europe PMC free article] [Abstract] [Google Scholar]
- Greter M, Heppner FL, Lemos MP, Odermatt BM, Goebels N, Laufer T, Noelle RJ, Becher B. Dendritic cells permit immune invasion of the CNS in an animal model of MS. Nat. Med. 2005;11:328–334. [Abstract] [Google Scholar]
- Hickey WF, Kimura H. Perivascular microglial cells of the CNS are bone marrow-derived and present antigen in vivo. Science. 1988;239:290–292. [Abstract] [Google Scholar]
- Hromas R, Broxmeyer HE, Kim C, Nakshatri H, Christopherson K, II, Azam M, Hou YH. Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem. Biophys. Res. Commun. 1999;255:703–706. [Abstract] [Google Scholar]
- Louvet C, Chiffoleau E, Heslan M, et al. Identification of a new member of the CD20/FcepsilonRIbeta family overexpressed in tolerated allografts. Am. J. Transplant. 2005;5:2143–2153. [Abstract] [Google Scholar]
- Melchior B, Remy S, Nerriere-Daguin B, Heslan JM, Soulilou JP, Brachet P. Temporal analysis of cytokine gene expression during infiltration of porcine neuronal grafts implanted into the rat brain. J. Neurosci. Res. 2002;68:284–292. [Abstract] [Google Scholar]
- Meuter S, Moser B. Constitutive expression of CXCL14 in healthy human and murine epithelial tissues. Cytokine. 2008;44:248–255. [Abstract] [Google Scholar]
- Mott RT, Ait-Ghezala G, Town T, Mori T, Vendrame M, Zeng J, Ehrhart J, Mullan M, Tan J. Neuronal expression of CD22: novel mechanism for inhibiting microglial proinflammatory cytokine production. GLIA. 2004;46:369–379. [Abstract] [Google Scholar]
- Papenfuss TL, Thrash JC, Danielson PE, Foye PE, Hilbush BS, Sutcliffe JG, Whitacre CC, Carson MJ. Induction of golli-MBP expression in CNS macrophages during acute LPS-induced CNS inflammation and experimental autoimmune encephalomyelitis (EAE) ScientificWorldJournal. 2007;7:112–120. [Europe PMC free article] [Abstract] [Google Scholar]
- Ransohoff RM. Microgliosis: the questions shape the answers. Nat. Neurosci. 2007;10:1507–1509. [Abstract] [Google Scholar]
- Renno T, Krakowski M, Piccirillo C, Lin JY, Owens T. TNF-alpha expression by resident microglia and infiltrating leukocytes in the central nervous system of mice with experimental allergic encephalomyelitis. Regulation by Th1 cytokines. J. Immunol. 1995;154:944–953. [Abstract] [Google Scholar]
- Schmid CD, Sautkulis LN, Danielson PE, Cooper J, Hasel KW, Hilbush BS, Sutcliffe JG, Carson MJ. Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 (TREM-2) on adult murine microglia. J. Neurochem. 2002;83:1309–1320. [Europe PMC free article] [Abstract] [Google Scholar]
- Sedgwick JD, Schwender S, Imrich H, Dorries R, Butcher GW, ter Meulen V. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. PNAS. 1991;88:7438–7442. [Europe PMC free article] [Abstract] [Google Scholar]
- Takahasi K, Prinz M, Stagi M, Chechneva O, Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007;4:e124. [Europe PMC free article] [Abstract] [Google Scholar]
- Thrash JC, Torbett BE, Carson MJ. Developmental regulation of TREM2 and DAP12 in the murine CNS: implications for Nasu-Hakola disease. Neurochem. Res. 2009;34:38–45. [Europe PMC free article] [Abstract] [Google Scholar]
- Vadasz C, Smiley JF, Figarsky K, Saito M, Toth R, Gyetvai BM, Oros M, Kovacs KK, Mohan P, Wang R. Mesencephalic dopamine neuron number and tyrosine hydroxylase content: genetic control and candidate genes. Neuroscience. 2007;149:561–572. [Europe PMC free article] [Abstract] [Google Scholar]
- Vallieres L, Sawchenko PE. Bone marrow-derived cells that populate the mouse brain preserve their hematopoietic identity. J. Neurosci. 2003;23:5197–5207. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1111/j.1471-4159.2009.05984.x
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/j.1471-4159.2009.05984.x
Citations & impact
Impact metrics
Citations of article over time
Article citations
Microglia contribute to neuronal synchrony despite endogenous ATP-related phenotypic transformation in acute mouse brain slices.
Nat Commun, 15(1):5402, 26 Jun 2024
Cited by: 0 articles | PMID: 38926390 | PMCID: PMC11208608
Astrocyte-Derived Exosomal miR-148a-3p Suppresses Neuroinflammation and Restores Neurological Function in Traumatic Brain Injury by Regulating the Microglial Phenotype.
eNeuro, 11(2):ENEURO.0336-23.2024, 08 Feb 2024
Cited by: 1 article | PMID: 38272675 | PMCID: PMC10860656
Novel image analysis tool for rapid screening of cell morphology in preclinical animal models of disease.
Heliyon, 9(2):e13449, 08 Feb 2023
Cited by: 1 article | PMID: 36873154 | PMCID: PMC9975095
Editorial: The role of microglia in the pathogenesis of neurodegenerative diseases.
Front Aging Neurosci, 14:1105896, 06 Jan 2023
Cited by: 1 article | PMID: 36688165 | PMCID: PMC9853986
Microglial polarization in TBI: Signaling pathways and influencing pharmaceuticals.
Front Aging Neurosci, 14:901117, 01 Aug 2022
Cited by: 12 articles | PMID: 35978950 | PMCID: PMC9376354
Review Free full text in Europe PMC
Go to all (88) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (1 citation) ENA - AF192557
- (1 citation) ENA - BC002086
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification of new CNS-resident macrophage subpopulation molecular markers for the discrimination with murine systemic macrophages.
J Neuroimmunol, 169(1-2):39-49, 16 Sep 2005
Cited by: 12 articles | PMID: 16169092
Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice: implications for vaccine-based therapies for Alzheimer's disease.
ASN Neuro, 2(3):e00037, 12 Jul 2010
Cited by: 86 articles | PMID: 20640189 | PMCID: PMC2905103
RNA-Seq transcriptomic profiling of primary murine microglia treated with LPS or LPS + IFNγ.
Sci Rep, 8(1):16096, 31 Oct 2018
Cited by: 29 articles | PMID: 30382133 | PMCID: PMC6208373
Exploiting dynamic enhancer landscapes to decode macrophage and microglia phenotypes in health and disease.
Mol Cell, 81(19):3888-3903, 30 Aug 2021
Cited by: 20 articles | PMID: 34464593 | PMCID: PMC8500948
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NINDS NIH HHS (5)
Grant ID: R01 NS039508
Grant ID: NS39508
Grant ID: R01 NS039508-06
Grant ID: NS045735
Grant ID: R01 NS045735




