Abstract
Free full text

miR-145 and miR-143 Regulate Smooth Muscle Cell Fate Decisions
SUMMARY
microRNAs are regulators of myriad cellular events, but evidence for a single microRNA that can efficiently differentiate multipotent cells into a specific lineage or regulate direct reprogramming of cells into an alternate cell fate has been elusive. Here, we show that miR-145 and miR-143 are co-transcribed in multipotent cardiac progenitors before becoming localized to smooth muscle cells, including neural crest stem cell–derived vascular smooth muscle cells. miR-145 and miR-143 were direct transcriptional targets of serum response factor, myocardin and Nkx2.5, and were downregulated in injured or atherosclerotic vessels containing proliferating, less differentiated smooth muscle cells. miR-145 was necessary for myocardin-induced reprogramming of adult fibroblasts into smooth muscle cells and sufficient to induce differentiation of multipotent neural crest stem cells into vascular smooth muscle. Furthermore, miR-145 and miR-143 cooperatively targeted a network of transcription factors, including Klf4, myocardin, and Elk-1 to promote differentiation and repress proliferation of smooth muscle cells. These findings demonstrate that miR-145 can direct the smooth muscle fate and that miR-145 and miR-143 function to regulate the quiescent versus proliferative phenotype of smooth muscle cells.
MicroRNAs (miRNAs) represent a class of small (20–25 nucleotides), non-coding RNAs that are key regulators of many cellular events, including the balance between proliferation and differentiation during tumorigenesis and organ development1–3. miRNAs are initially transcribed as longer primary transcripts (pri-miRNAs) and processed first by the ribonuclease enzyme complex, Drosha/DGCR8, and then by Dicer, leading to incorporation of a single strand into the RNA-induced silencing complex (RISC). Each of the ~650 human miRNAs is predicted to interact with over one hundred target mRNAs in a sequence-specific fashion involving Watson-Crick base-pairing among nucleotides 2–8 of the miRNA4,5. miRNAs generally inhibit target mRNAs by repressing translation or reducing mRNA stability. miRNAs may also activate mRNA translation under certain cellular conditions6.
Regulation of cardiovascular cell fate decisions by miRNAs and control of proliferation and differentiation in cardiac progenitors has been reported, but remains inefficient7–12. A multipotent cardiac progenitor pool that can differentiate into cardiac myocytes, vascular smooth muscle cells (VSMCs) and endothelial cells exist13, as do multipotent neural crest stem cells that can also give rise to VSMCs, as well as melanocytes, chondrocytes and neurons14. Among these cell types, VSMCs are uniquely plastic, as they can oscillate between a proliferative or quiescent, more differentiated state15. This plasticity contributes to many human vascular diseases, including atherosclerosis16,17. The transcriptional control of this oscillation has been described16, but whether VSMC-enriched miRNAs exist or participate in this process is unknown. Here, we demonstrate that miR-145 and miR-143 are tightly integrated into a core transcriptional network involved in smooth muscle differentiation and proliferation, and that miR-145 functions as a critical switch in promoting smooth muscle differentiation.
miR-143 and miR-145 expression
We reported that miR-143 is the most enriched miRNA during differentiation of mouse embryonic stem (mES) cells into multipotent cardiac progenitors11. miR-143 is highly conserved and lies within 1.7 kilobases of another conserved miRNA, miR-145, on mouse chromosome 18 (Supplementary Fig. 1a, b). Both miRNAs are downregulated in various cancer cell lines, colon cancers, and lung cancers2. Given their genomic organization and proximity, miR-143 and miR-145 may be contained in a bicistronic primary transcript, but we were unable to amplify a common transcript from RNA, possibly because pri-miRNA transcripts are rapidly processed into their mature forms. DGCR8-null ES cells lack nuclear miRNA processing activity and have a defect in differentiation18, but can form mesoderm. Using primers for each miRNA and RNA from DGCR8-null embryoid bodies (EBs), we generated an amplicon that encompassed both miRNAs (Supplementary Fig. 1b), suggesting that miR-143 and miR-145 were transcribed as a bicistronic unit and therefore share common regulatory elements that control their expression.
To determine if these two miRNAs were also enriched in multipotent cardiac progenitors in vivo, we bred transgenic mice containing Cre recombinase in the Islet1 locus19 with Rosa26-EYFP mice20 and isolated YFP+ cardiac progenitor cells at E9.5 by fluorescence-activated cell sorting (FACS) (Supplementary Fig. 1c, d). The Islet1-Cre mice mark early multipotent cardiac progenitor cells that can differentiate into cardiac muscle, smooth muscle and endothelial cells21. Quantitative RT-PCR (qPCR) revealed that miR-143 and miR-145 were enriched in YFP+ cells (Supplementary Fig. 1e). qPCR with RNA from mouse hearts or whole embryos at varying stages of development also revealed enrichment of both miRNAs throughout cardiogenesis, before being downregulated in the adult heart (Supplementary Fig. 1f, g).
Transcriptional regulation of miR-143/145
To identify the tissue-specific expression and regulation of miR-143/miR-145 during mouse development, we searched for upstream regulatory regions. Comparison of genomic sequences across species revealed a 4.2-kilobase (kb) genomic region upstream of miR-143/miR-145 that was highly conserved between human and mouse (Supplementary Fig. 2a) and directed LacZ expression specifically in multipotent cardiac mesodermal progenitors of transgenic mice as early as embryonic day (E) 7.5 (Fig. 1a, b). By E9.5, LacZ expression was more robust and uniform in the heart and outflow tract and in cardiac progenitors of the pharyngeal mesoderm; expression was also present in the aorta just as smooth muscle differentiation began, but was absent in the cardinal vein (Fig. 1c, d). LacZ expression was robust in the endocardium and myocardium (Fig. 1d). During later cardiogenesis, expression became restricted to the ventricles and atria, but was notably absent in the aorta and pulmonary arteries (Fig. 1e). Postnatally, the pattern was reversed, with high transcript levels in smooth muscle of the aorta, pulmonary artery, and coronary vessels but undetectable levels in the ventricular myocardium (Fig. 1f–h). This enhancer was also active in the smooth muscle of the intestines (Supplementary Fig. 2b, c). The enhancer recapitulated the endogenous miR-145 expression, with transcripts in the smooth muscle of the adult aorta and coronary artery, but not ventricular myocardium, as shown by section in situ hybridization (Fig. 1i,j and Supplementary Fig. 2d).

(a–c) Whole mounts showing cardiac-specific β-gal activity in transgenic mouse embryos with 4.2 kb enhancer-lacZ construct (Supplementary Fig. 2a) at indicated time points. (d) Transverse section of (c) showing β-gal expression in pharyngeal mesoderm (pm), pharyngeal endoderm (pe), dorsal aorta (da), myocardium (mc), endocardium (ec). (e,f) β-gal expression in embryonic day (E) 15.5 (e) or post-natal (P21) (f) heart. Ao, aorta; pa, pulmonary artery. (g,h) Transverse sections of (f); co, coronary artery. (i,j) Section in-situ hybridization of miR-145 in P21 heart section. (h) and (j) represent higher magnification of boxed areas. (pcm, precardiac mesoderm; ht, heart; h, head; ot, outflow tract; rv, right ventricle; lv, left ventricle; cv, cardinal vein; ra, right atrium; la, left atrium).
Deletions of the 4.2-kb miR-143/145 enhancer revealed that a 0.9-kb region was sufficient for miR-143/145 cardiac and smooth muscle expression (Fig. 2a–e, Supplementary Fig. 3a). Within this regulatory region, we observed cis elements highly conserved between human, mouse, and zebrafish that represented potential binding sites for the essential cardiac transcription factors, serum response factor (SRF) and Nkx2.5 (Supplementary Fig. 3b). SRF plays a dual role in cardiac and SMC development, influencing both proliferation and differentiation depending on the types of co-activators or repressors present at specific developmental or cellular stages22. The potent SRF co-activator, myocardin (Myocd)23, is a component of a molecular switch for the VSMC fate24 and is sufficient to effect both structural and physiological attributes of this cell type25. SRF weakly activated the miR-143/145 enhancer upstream of a luciferase reporter, but co-transfection of Myocd synergistically and robustly activated luciferase activity in Cos cells (Fig. 2f). Mutation of the highly conserved CArG box in the SRF binding site decreased Myocd-dependent luciferase activity (Fig. 2f). Nkx2.5 could also independently activate this enhancer, and the combination of SRF, Myocd, and Nkx2.5, which also interacts with SRF26, had additive effects on luciferase activity. Mutation of each binding site progressively decreased luciferase activity (Fig. 2f).

(a–e) Lateral (a, b, c, e) or frontal (d) cardiac views of transgenic embryos containing indicated lacZ constructs and stained for β-Gal activity. (f) Fold-activation of luciferase activity directed by introduction of SRF, Myocd or Nkx2.5 expression vectors with the miR-143/145 enhancer in Cos cells. All changes were statistically significant (n=5). (g) miR-143 and miR-145 expression levels assessed by qPCR day 10 embyroid bodies (EBs) of indicated genotypes. (h) qPCR of miR-143 and miR-145 in Nkx2.5+/− and Nkx2.5−/− E9.5 hearts relative to WT. (i,j) qPCR of miRNAs in injured vessels (i) or atherosclerotic lesions (j) compared to normal arterial expression. Results shown in (f–j) are the average of three experiments. (ot, outflow tract; ra, right atrium; lv, left ventricle; rv, right ventricle; la, left atrium; dorsal aorta). *, p<0.05. Error bars indicate SD.
In vivo, mutation of the SRF binding site disrupted lacZ expression in the outflow tract and aorta, while disruption of the Nkx2.5 binding site diminished expression in the ventricles and atria (Fig. 2d, e), suggesting modular regulation by the enhancer. Mutation of both the SRF and Nkx2.5 binding sites abolished all activity of the enhancer within the heart (Fig. 2f). VSMC and atrial expression postnatally was also dependent upon the SRF-binding cis element (Supplementary Fig. 3c). Electromobility shift assay confirmed SRF could specifically bind to the putative binding site in the miR-143/145 enhancer (Supplementary Fig. 3d). Furthermore, miR-143 and miR-145 were each expressed at lower levels in SRF-null embryoid bodies (EBs) compared to wildtype EBs derived from the respective ES cells (Fig. 2g). The levels were also reduced in mesoderm-rescued SRF-null EBs11, confirming that the decreases did not reflect the absence of mesoderm (Fig. 2g). Similarly, miR-143 and miR-145 expression was also decreased in hearts of Nkx2.5 mutant mouse embryos in a dose-dependent fashion (Fig. 2h).
Dysregulation in vascular disease
The dynamic stage-dependent expression of miR-143 and miR-145 raised the possibility that their expression may also vary with the oscillation of VSMCs between differentiated and proliferative phenotypes. In a mouse model of this proliferative switch, ligation of the carotid arteries typically results in narrowing of the vascular lumen as a result of phenotypic modulation and proliferation of VSMCs. qPCR revealed marked decreases in miR-143 and miR-145 expression in injured carotid arteries compared to contralateral control vessels (Fig. 2i). miR-21 expression was increased as expected, and miR-16 was unchanged, demonstrating the presence of intact small RNAs27. In situ hybridization of injured and control carotid arteries also revealed marked downregulation of miR-143 and miR-145 expression in the thickened vascular wall, coincident with decreased expression of the differentiation marker, smooth muscle α-actin (Sm-α–actin) (Supplementary Fig. 4a). As a control, miR-143 and miR-145 levels were unchanged in cardiac muscle after injury (Supplementary Fig. 4b). Interestingly, transcripts of miR-145 were also downregulated to nearly undetectable levels in atherosclerotic lesions containing neointimal hyperplasia (Fig. 2j).
Regulation of cell fate and proliferation
The bimodal expression of miR-143 and miR-145 early during VSMC induction, and subsequently during the maturation into a non-proliferating, differentiated phenotype, led us to investigate their potential function in these settings. Since miR-145 and -143 expression was directly activated by SRF-Myocd, we first investigated whether either miRNA’s expression was necessary for Myocd-induced reprogramming of fibroblasts into VSMCs. Introduction of 1–2 ug of Myocd into fibroblasts reliably resulted in >50% conversion to VSMCs22. Inhibition of miR-145 using cholesterol-modified antisense oligonucleotides (antagomiRs)28 blocked Myocd’s ability to convert fibroblasts into VSMCs as illustrated by Sm-α-actin immunostaining and expression of multiple smooth muscle markers assessed by qPCR and western blot (Fig. 3a–c, e). The knockdown of miR-143 had little effect on Myocd-induced smooth muscle conversion (Fig. 3a, b and Supplementary Fig. 5a, b). Neither miRNA was sufficient to reprogram fibroblast cells. However, miR-145 potentiated Myocd’s reprogramming effects. Although 50 ng of Myocd was insufficient to induce VSMC gene expression, simultaneous addition of miR-145, but not miR-143, resulted in robust VSMC differentiation, equivalent to that observed with 1–2 ug of Myocd (Fig. 3b, d, e and Supplementary Fig. 5c). Thus, miR-145 activity was required for Myocd-dependent conversion of fibroblasts into VSMCs, and miR-145 robustly potentiated Myocd’s effects.
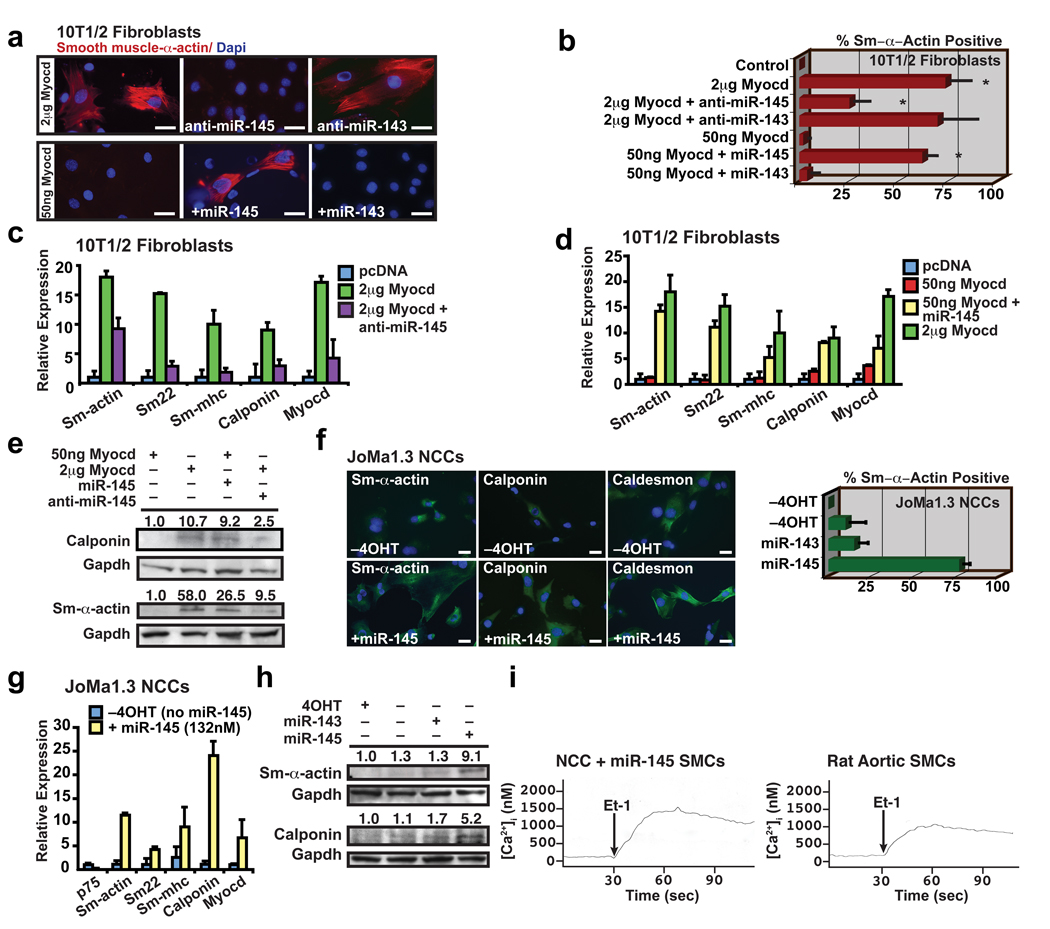
(a) Immunocytochemistry of 10T1/2 fibroblasts using smooth muscle (Sm) α-actin antibodies (red) under conditions indicated; nuclear stain, Dapi (blue). (b) Quantification of Sm-α-actin positive cells (n=6). (c) qPCR of Sm gene expression in fibroblasts transfected with Myocd with or without anti-miR-145 or (d) fibroblasts transfected with 50 ng Myocd with or without miR-145 (n=5). (e) Western blot of calponin and Sm-α-actin. (f) Immunocytochemistry of neural crest stem cells (Joma1.3 NCCs) with or without miR-145 using antibodies indicated (green); tamoxifen (4OHT) was removed to allow differentiation. Quantification of percent Sm-α-actin + cells relative to total Dapi+ nuclei (blue) (n=6). (g) qPCR of Sm gene expression in NCCs with miR-145 expression (n=5); p75 is a marker of undifferentiated neural crest cells. (h) Western blot of Sm-α-actin and calponin. (i) Calcium flux [Ca2+]i in SMCs derived from NCCs or rat aortic SMCs in response to endothelin-1 (Et-1) stimulation at 30 sec. Error bars indicating SD. *, p<0.05.
To test an alternative cell type in which miR-145 may be sufficient for VSMC differentiation, we used a multipotent neural crest stem cell line that can differentiate into numerous cell types (e.g., melanocytes, chondrocytes, neurons), including VSMCs, upon exposure to 5 days of TGF-β29. Remarkably, introduction of miR-145, but not miR-143, into neural crest stem cells was sufficient to guide ~75% of cells into the VSMC lineage within only twenty-four hours, as determined by immunocytochemistry with multiple markers (Fig. 3f). qPCR and western blot revealed upregulation of numerous markers of VSMC differentiation, including Sm-α-actin, Sm-22α, and smooth muscle myosin heavy chain (Sm-MHC) with miR-145 but not -143 (Fig. 3g, h; Supplementary Fig. 5d, e). The neural crest stem cell–derived SMCs exhibited calcium flux measurements similar to cultured VSMCs in response to endothelin-1 stimulation, indicating the differentiation of functionally mature smooth muscle (Fig. 3i, j). Thus, miR-145 was sufficient for directing the VSMC fate from multipotent neural crest stem cells that normally populate the aortic smooth muscle tissue, where miR-145 is expressed.
miRNA targets and mechanism
The mechanism by which this family of miRNAs regulates VSMCs is dependent upon their mRNA targets. A bioinformatics approach incorporating sequence matching and mRNA secondary structure to predict mRNA targets (K. Ivey & D. Srivastava, unpublished; also see Methods) revealed multiple highly conserved binding sites for miR-143 in the 3′ UTR of Elk-1 and for miR-145 in the 3′ UTR of Myocd (Supplementary Fig. 6a). Growth signals repress smooth muscle gene expression by displacing Myocd from SRF with Elk-1, a ternary complex factor that acts as a myogenic repressor and an activator of VSMC proliferation22. In this system, SRF serves as a platform for myogenic coactivators or corepressors that compete for a common docking site, thereby mediating VSMC phenotypic switching.
To determine whether Elk-1 and Myocd are direct targets of miR-143 or miR-145, respectively, we cloned the 3′ UTR of Elk-1 or Myocd into the 3′ UTR of a CMV-driven luciferase reporter. In the presence of the Elk-1 3′ UTR, miR-143 repressed luciferase activity; this repression was diminished upon mutation of one of the two miR-143 binding sites (Fig. 4a). The addition of an antagomiR to inhibit miR-143 in the A10 rat aortic VSMC line resulted in upregulation of Elk-1 protein, but not mRNA, consistent with translational repression of Elk-1 by miR-143 (Fig. 4b, Supplementary Fig. 6b). Furthermore, inhibition of miR-143 caused a doubling of the proliferative rate of VSMCs, demonstrating miR-143’s function in negatively regulating VSMC proliferation (Fig. 4c).

(a) Luciferase activity in Cos cells upon introduction of Elk-1 3′ UTR or mutant 3′ UTR (mut) downstream of a CMV-driven luciferase reporter with indicated miRNAs (n=5). (b) Elk-1 protein in cell lysates from A10 VSMCs transfected with a scrambled (scr) miRNA or anti-miR-143 or -miR-145 assessed by western blot. (c) Proliferation of VSMCs upon inhibition of miR-143 or miR-145 relative to control (5%FBS) (n=5). (d) Luciferase activity in Cos cells with Myocd 3′ UTR sequences with indicated miRNAs (n=5). The Myocd binding site (BS) was mutated in the context of a 4x concatemer. (e) Luciferase activity with wt or mutated Klf4-3′ UTR upon introduction of indicated miRNAs (n=5). (f) Analysis of Klf4 protein in cell lysates from A10 cells transfected with indicated anti-miRs by western blot. (g) Luciferase activity of wt or mutated CamkII-δ3′ UTR (n=5). (h) Western analysis for CamkII-δ protein in A10 cells transfected with scr miRNA or anti-miR-145. (i) Proliferation of VSMCs relative to control (n=5). Error bars indicate SD. Densitometry calculation performed by Image J. *, p<0.05.
The presence of putative miR-145 binding sites in the Myocd 3′ UTR seemed counter to the observed effects of miR-145 in potentiating Myocd’s reprogramming effects. When we cloned the Myocd 3′ UTR into a CMV-driven luciferase vector and introduced this into Cos cells, the constitutive luciferase activity decreased greater than 100-fold. Surprisingly, introduction of miR-145, but not miR-143, with the luciferase vector in Cos cells resulted in relief of the repression and an ~150-fold increase in luciferase activity compared to the CMV-luciferase-Myocd 3′ UTR-luciferase vector alone (Fig. 4d). The increase in luciferase activity was largely lost upon mutation of the miR-145 binding site in the Myocd 3′ UTR (Fig. 4d). In contrast, introduction of the same CMV-luciferase-Myocd 3′ UTR reporter did not cause a decrease in baseline luciferase activity in 293T cells. However, even in these cells, miR-145 consistently increased luciferase activity by ~1.5 fold (data not shown). Although antibodies to detect endogenous Myocd levels by western blot are not available, these findings are consistent with the recent observation that miRNAs can act as translational activators or repressors based upon the state of the cell cycle6. While the mechanism for this remains unclear, it will be interesting to determine if miR-145 prevents binding of a repressive RNA-binding protein enriched in Cos cells.
While miR-145 may result in increased Myocd protein, its effects in potentiating Myocd-induced reprogramming of fibroblasts did not require presence of its binding site in Myocd’s 3′ UTR. miR-145’s potentiating effects could, however, be through effects on translation of endogenous Myocd mRNAs induced by the transfected Myocd protein; alternatively, miR-145 may promote differentiation through targets independent of Myocd. Indeed, our bioinformatics approach identified potential miR-145 binding sites in several other positive regulators of smooth muscle proliferation, including Kruppel-like factor 4 (Klf4) and Calmodulin kinase II-delta (CamkIIδ). Klf4 is a transcription factor involved in pluripotency30 that is also rapidly induced in post-injury proliferating VSMCs, where it interacts with enhancers in smooth muscle growth genes, inhibits smooth muscle differentiation genes, and represses Myocd expression31. The miR-145 binding site in the 3′ UTR of Klf4 specifically mediated miR-145-dependent repression in luciferase assays (Fig. 4e and Supplementary Fig. 6c). Furthermore, knockdown of miR-145 in rat A10 VSMCs resulted in an increase in Klf4 protein levels, but no change in Klf4 mRNA levels (Fig. 4f, Supplementary Fig. 6d). Similarly, a putative binding site in CamkII-δ, involved in multiple events including neointimal proliferation32,33, was validated as a miR-145-repressed target by luciferase and western analysis in VSMCs (Fig. 4g, h and Supplementary Fig. 6e). Numerous predicted targets for both miRNAs that were not validated in luciferase assays are shown (Supplementary Fig. 6f, g).
Consistent with miR-145 repression of genes involved in VSMC proliferation, introduction of miR-145 was sufficient to suppress the proliferative response normally induced by platelet-derived growth factor (Pdgf-β) in cultured VSMCs (Fig. 4i). In addition, lentiviral-mediated introduction of miR-145 into ligated mouse carotid arteries in vivo increased expression of markers of VSMC differentiation (e.g., Calponin and Sm-α-actin), as well as Myocd, compared to control-infected injured carotid arteries (Supplementary Fig. 7). These findings suggest that miR-145 may promote VSMC differentiation by directly repressing numerous transcription factors that promote the proliferative state while stabilizing factors that promote the differentiated state of VSMCs (Fig. 5).
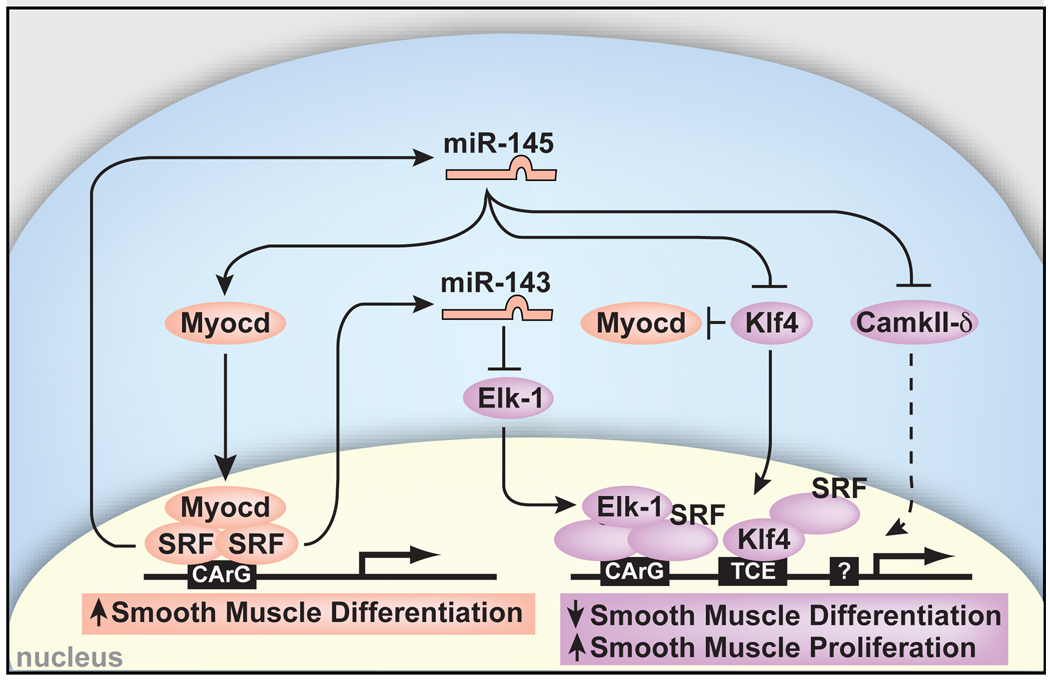
miR-143 and miR-145 are positively regulated by SRF and function to repress multiple factors that normally promote the less differentiated, more proliferative smooth muscle phenotype (purple). These include Klf4, which also represses Myocd. miR-145 has a positive effect on Myocd activity to concurrently promote the more differentiated smooth muscle phenotype (pink), thereby also functioning to reinforce its own expression. Effects of miR-145 and miR-143 converge on SRF-dependent transcription by regulation of co-activators and co-repressors to dictate the proliferative or differentiated phenotype of VSMCs. Positive regulation of Myocd by miR-145 results in reinforcement of miR-145 and miR-143 expression and the differentiated phenotype. Dashed lines indicate indirect effects.
Discussion
The ability of miR-145 to efficiently direct VSMC differentiation from multipotent stem cells is the first evidence, to our knowledge, of a miRNA capable of directing the VSMC fate. This is consistent with its early expression in the aorta of developing embryos as VSMC differentiation is initiated from neural crest and mesodermal progenitors. Once VSMC identity is established in the embryo, the downregulation of miR-143 and miR-145 in the developing embryo may allow necessary proliferation. Subsequent upregulation postnatally (coincident with the more differentiated VSMC state) and downregulation during neointimal hyperplasia indicate dynamic regulation that may contribute to the oscillating state of VSMCs. The observation that miR-145 is necessary and sufficient for VSMC differentiation raises the possibility that restoration of this miRNA could suppress the smooth muscle hyperplasia observed in vascular injury and atherosclerosis. Furthermore, the multiple targets we identified for miR-143 and miR-145 reveal an elegant mechanism by which this family of miRNAs promotes differentiation and simultaneously represses proliferation of VSMCs by converging on SRF-dependent co-activators and co-repressors (Fig. 5). Our findings suggest miR-145 may promote VSMC differentiation in part by increasing Myocd protein and functioning in a feed-forward reinforcement of its own expression by the SRF-Myocd complex, while miR-143 represses Myocd’s competitor, Elk-1. Given the potent effects on differentiation shown here, future studies will determine if restoration of normal levels of miR-143 and miR-145 holds therapeutic value in the setting of disease.
The downregulation of miR-145 in numerous cancers and our findings that it promotes differentiation raises the possibility that miR-145 functions as a pro-differentiation factor in a lineage-specific fashion depending on the cellular context. The targeting of Klf4 supports this notion, as Klf4 is expressed in undifferentiated embryonic stem cells and in other less differentiated cell types. Recent evidence indicates that miR-145 represses Klf4 in human embryonic stem cells as they begin to differentiate and is required for normal differentiation34. Since Klf4 is one of four factors described by Yamanaka and colleagues that together are sufficient to reprogram human fibroblasts into a pluripotent state (induced pluripotent stem (iPS) cells), it will be interesting to determine whether inhibition of miR-145 can enhance generation of iPS cells.
Methods Summary
Transgenic mice were generated by pronuclear injection and assayed by Bluo-gal (Invitrogen) staining as described7. Constructs contained promoter fragments cloned upstream of the pHsp68LacZ reporter. Embyronic hearts were collected at E9.5 from Islet1-cre mice19 crossed with Rosa26-YFP mice20, and miRNA microarray hybridizations and qPCR were performed from YFP+ cells. EMSA was performed as described35 using oligos corresponding to the conserved SRF-binding sites in the miR-143/145 enhancer. To identify and validate putative miRNA target genes, we used an in-house automated algorithm7,10 and cloned the 3′ UTR of each mRNA into the pMiR-Report luciferase reporter. Luciferase activity was measured using the Luciferase Dual-Reporter Kit. To assess miR-143/145 function, rat aortic A10 VSMCs or 10T1/2 fibroblasts were transfected with expression plasmids containing either pre-miR-145 or pre-miR-143, or inhibitors to miR-143 or -145, and then assayed for SM-marker gene expression by qPCR, western blot, and immunocytochemistry. Myogenic conversion assays were performed as described36 with 1–2 µg of Myocd. For multipotent stem cell studies, the JoMa1.3 neural crest cell line was maintained as reported29. To induce SMC differentiation, miR-145 was transiently transfected. A10 VSMCs proliferation studies were done as reported37 using the CellTiter 96TM assay and miRNAs were transfected at varying concentrations; 5ng/mL of PDGF-bb was added to appropriate wells post-growth arrest. Changes in intracellular calcium concentration ([Ca2+]i) were measured using a calcium fluorescent dye, Indo-1 AM, as described38 in A10 VSMCs or JoMa1.3 NCCs transfected with pre-miR-145 and exposed to 10 nM of the calcium agonist, Endothelin-1, for 30 seconds to stimulate calcium flux38. For assessment of miR-143 or -145 expression during vascular injury, ligated carotid arteries were collected from mice and sectioned, and aortic lesions from apolipoprotein E- null mice fed a Western diet were dissected; subsequently, expression was analyzed by qPCR and in-situ hybridization.
Methods
Transgenic Mice and Flow Cytometry
Transgenic mice were generated and Bluo-gal staining and histological analyses were performed as described7. For promoter analysis, fragments were subcloned into a pHsp68LacZ reporter vector and injected into pronuclei. The 4.2-kb regulatory element corresponds to mouse chr 18:61809195-61813466. Islet1-cre mice19 were crossed with Rosa26-YFP mice20, embryos were collected at E9.5, and heart and surrounding tissue was dissected, trypsinized, spun at 2000 rpm and the pellet was resuspended in PBS and filtered through a 40-µm Millipore membrane. Selection by FACs was based on expression of YFP. YFP+ cells were collected for RNA preparation.
miRNA Microarray and miRNA in Situ Hybridization
Total RNA was isolated (Trizol, Invitrogen) from mouse E9.5 embryonic hearts and used for miRNA microarray hybridizations (Exiqon) and quantitative real-time PCR. miRNA in situ hybridization analyses were performed as described39 with the following modifications: paraffin-embedded tissue sections or cryosections were treated for 15 minutes with Proteinase K, hybridized at 59°C (miR-145) or 42°C (miR-143), and final color development was performed with NBT/BCIP (Roche).
Electromobility Shift Assay (EMSA)
Oligoribonucleotides corresponding to the conserved SRF-binding sites in the miR-143/145 enhancer were synthesized (Integrated DNA Technologies) as follows:
SRF binding site: GGGAGCAGCCTTGCCATATAAGGGCAGG; SRF mutant binding site: GGGAGCAGCCTTGCTACCGAAGGGCAGG. EMSA was performed as described35.
miRNA Target Prediction
Putative miRNA target genes were identified using an in-house automated algorithm based on empirical miRNA:mRNA interaction data7,10 that qualifies mRNAs based on 1) complementarity between the seed region of the miRNA and the mRNA 3′ UTR as annotated in RefSeq; 2) identification of an extended binding site; 3) favorable binding affinity between the miRNA and mRNA target site as calculated by RNAhybrid40; 4) high free energy in the regions flanking the putative binding site as determined by mFold41; 5) absence of stabilizing elements in the binding site; 6) presence of destabilizing elements in the region surrounding and including the possible binding site; and 7) conservation over a number of species.
miR-143 and miR-145 Target Analyses and Expression
A 250-bp fragment encompassing miR-145 was ligated into pSilencer 4.1-CMV (Ambion). A 250-bp fragment containing miR-143 was ligated into pEF-Dest-51 (Invitrogen). The entire 3′ UTR of each mRNA containing predicted miR-143 and/or miR-145 binding sites was cloned into the pMiR-Report luciferase reporter (Applied Biosystems). All assays were performed in quadruplicate in 12-well plates of Cos cells and transfected with siPort XP-1 (Ambion). After 24 hours, cells were harvested and luciferase activity was measured with the Luciferase Dual-Reporter Kit (Promega). Renilla assays were performed in parallel to normalize for transfection efficiency.
Embryonic stem (ES) cells or embryoid bodies (EBs), A10 cells or differentiated 10T1/2 fibroblasts were harvested in Trizol (Invitrogen) for total RNA isolation. Total RNA (2 µg) from each sample was reversed transcribed with Superscript III (Invitrogen). Taqman primers were used to amplify genes (ABI; primer sequences available upon request). The primers to detect the 1.7 kb miR-143/145 primary transcript were as follows: Forward: GCATCTCTGGTCAGTTGGG, Reverse: GACCTCAAGAACAGTAT. GAPDH was used as a control. DGCR8null EBs (Day 8, D10 EBs) were a gift from R. Blelloch18. miRNA qPCR on β-MHC-GFP control EBs, SRFnull EBs, or SRFnull EBs expressing miR-1 or miR-133 was performed as described above; miR-16 was used as the endogenous control. Each qPCR was performed at least three times; representative results are shown as fold expression relative to undifferentiated ES cells.
Tissue Culture
10T1/2 fibroblasts were maintained at low density (~30% confluence) in DMEM with 10% FBS and were transfected with Lipofectomine 2000 (Invitrogen) and 1–2 µg of full length or smooth muscle isoform of Myocd36. Pre-miR-145 sequence containing ~250 bp amplified from genomic DNA was cloned into pSilencer 4.1-CMV vector (Ambion) and pre-miR-143 was cloned into the pEF-Dest-51 vector (Invitrogen). Two days after transfection, media was replaced with differentiation medium (DMEM, 2% horse serum). 4–5 days later, further analyses, including immunocytochemistry, Western blot, and RT-PCR were performed.
A10 SMCs and Cos cells were maintained in DMEM with 10% FBS. A10 cells were transfected with BlockIT Fluorescent oligo (Invitrogen), miR-143 or miR-145 inhibitor (Dharmacon) or antagomiR (IDT Technologies), or miR-143, 145 mimic (Dharmacon). 24–48 hours later, western blot or RT-PCR was performed.
The Joma1.3 neural crest cell line was maintained as reported29. NCCs were plated (~7.5 X105 cell density) on plastic culture dishes coated with fibronectin, and kept in an undifferentiated state by the addition of 200 nm 4-OHT (Tamoxifen) every 24 hrs. For differentiation into SMCs, TGF-β was added 24 hrs after last 4-OHT treatment, which was stopped to allow differentiation to take place within 4–6 days. Pre-miR-145 or -miR-143 was transfected in 6-well culture dishes using 10 µl lipofectamine (Ambion) at concentrations ranging from 66 nm to 132 nm to induce SMC differentiation 24 hrs after removal of Tamoxifen.
Proliferation assays
Rat aortic A10 VSMCs proliferation studies were done as reported37. Briefly, cells were plated at a density of 5,000 cells/well in 96-well plates containing 5% FBS/DMEM. After plating, miRNAs were transfected at varying concentrations ranging from 20 nM to 240 nM. Twelve hours later, media was washed 3 times and changed to serum free DMEM with antibiotics. Serum free conditions were maintained for 48 hours to allow growth arrest. The medium was then changed to 5% FBS/DMEM and 5ng/mL of PDGF-bb (R&D Systems) was added to appropriate wells. After 24 hours, rates of proliferation were determined using the CellTiter 96TM assay (Promega). Proliferation was measured by the amount of 490 nm absorbance and is directly proportional to the number of living cells. Proliferation was subsequently expressed as absorbance of cells with treatment compared to cells without treatment. Each experiment was done in quintuplicate.
Immunohistochemistry and Western Blot Analysis
Immunostaining was performed using pre-ready mouse anti-smooth muscle actin (Dako, 1A4), 1:500 mouse anti-caldesmon (Abcam, 12B5), and 1:50 rabbit anti-calponin (Chemicon) antibodies and 1:400 Tritc- or Fitc-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Jackson ImmunoResearch). Myogenic conversion assays were performed as described, and protein lysates collected36. Rat aortic A10 cells were collected and assayed using Elk-1, Klf-4, and CamKII-δ (Cell Signaling) antibodies.
Calcium Flux Assays
Calcium studies were performed as described38. In brief, rat aortic smooth muscle cells or JoMa1.3 NCCs transfected with pre-miR-145 were grown to 95% confluency, and resuspended in media containing 1 mg/ml BSA and 10 mM HEPES to make 1×107 cells/ml. Then the cells were loaded with cell permeant Indo-1 AM (Invitrogen) for 40 minutes at 37°C, and subsequently washed and resuspended with Hank’s Buffered Saline Solution containing 1 mg/ml BSA and 10 mM HEPES. Indo-1 was excited at 350 nm and measured at 410 and 490 nm. The fluorescence intensity ratio (F410nm/F490nm) was calculated using a Hitachi F-2000 fluorescent spectrophotometer and Intracellular Cation Measurement System software, version 1.03 (Hitachi, Tokyo, Japan). Cells were exposed to 10 nM of the calcium agonist, Endothelin-1 (Sigma) at 30 seconds to stimulate calcium flux38.
Mouse Vascular Injury and Atherosclerosis Models
Mice that had their left carotid artery ligated were sacrificed 21-days post ligation, fixed and sectioned to obtain cross-sections of the left carotid artery as described42; the contralateral right carotid artery was used for control. Lentivirus harboring GFP + miR-145 or GFP alone (1×10e7 ifu, System Bio) was mixed in 20% pluronic gel and kept cold before application to 12-week old FVB/NJ mice. Lentiviral-pluronic gel was immediately applied to the external surface of the ligated vessel following arterial injury. Nine days post-ligation, the proximal portion of the injured carotid arty was rapidly removed for total RNA isolation (Trizol) and qPCR (BioRad, MylQ) analysis. 12-week-old apolipoprotein (Apo) E- null mice were fed a Western diet for 4 weeks, and aortic lesions were dissected and collected for RNA analysis.
Statistical analysis
The two-tailed student’s t-test, type II, was used for data analysis. P<0.05 was considered significant.
Acknowledgments
The authors thank R. Blelloch for DGCR8-null EBs; R.J. Schwartz for SRF-null ES cells; I. Charo and N. Saederup for RNA from atherosclerotic tissue; J. Maurer for JoMa neural crest cell line; L. Qian and Y. Huang for generously providing mouse cardiac infarct RNA; C. Tsou for help with calcium flux assays; E.N. Olson for the myocardin expression plasmid; P. Swinton for generation of transgenic mice; J. Fish and C. Miller for histopathology support; S. Ordway and G. Howard for scientific editing; B. Taylor for manuscript preparation. We also thank members of the Srivastava lab for helpful discussion. J.M.M. was supported by HL62572 and HL091168 from NHLBI/NIH. D.S. was supported by grants from the NHLBI/NIH and the California Institute for Regenerative Medicine (C.I.R.M.) and was an Established Investigator of the American Heart Association. This work was also supported by NIH/NCRR grant (C06 RR018928) to the Gladstone Institutes.
Footnotes
Author contributions
K.R.C. and D.S. designed the study and K.R.C. executed or oversaw execution of all experiments; N.T.S. and E.B. performed the NCC studies; M.W. and K.N.I. performed some expression studies and K.N.I. helped supervise the project; A.N.M. provided technical support; T.L. and J.M.M. performed carotid artery ligation studies; S.U.M. isolated YFP+ progenitor cells and performed some expression studies; J.M.M. assisted K.R.C and D.S. in editing the manuscript; K.R.C. and D.S. wrote the manuscript and D.S. supervised all aspects of the project.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature08195
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2769203
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature08195
Article citations
New insights on genetic background of major diabetic vascular complications.
Diabetol Metab Syndr, 16(1):243, 07 Oct 2024
Cited by: 0 articles | PMID: 39375805 | PMCID: PMC11457557
Regulation of idiopathic pulmonary fibrosis: a cross-talk between TGF-<i>β</i> signaling and MicroRNAs.
Front Med (Lausanne), 11:1415278, 25 Sep 2024
Cited by: 0 articles | PMID: 39386739 | PMCID: PMC11461268
Review Free full text in Europe PMC
Vascular smooth muscle cell phenotypic switching in atherosclerosis.
Heliyon, 10(18):e37727, 10 Sep 2024
Cited by: 0 articles | PMID: 39309965 | PMCID: PMC11416558
Review Free full text in Europe PMC
Hormonal influence: unraveling the impact of sex hormones on vascular smooth muscle cells.
Biol Res, 57(1):61, 04 Sep 2024
Cited by: 0 articles | PMID: 39227995 | PMCID: PMC11373308
Review Free full text in Europe PMC
The role of microRNAs in pregnancies complicated by maternal diabetes.
Clin Sci (Lond), 138(18):1179-1207, 01 Sep 2024
Cited by: 0 articles | PMID: 39289953 | PMCID: PMC11409017
Review Free full text in Europe PMC
Go to all (1,034) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells.
Atherosclerosis, 219(2):468-474, 09 Sep 2011
Cited by: 44 articles | PMID: 21945499
MicroRNA MiR-199a-5p regulates smooth muscle cell proliferation and morphology by targeting WNT2 signaling pathway.
J Biol Chem, 290(11):7067-7086, 16 Jan 2015
Cited by: 40 articles | PMID: 25596533 | PMCID: PMC4358129
Serotonin augments smooth muscle differentiation of bone marrow stromal cells.
Stem Cell Res, 12(3):599-609, 13 Feb 2014
Cited by: 2 articles | PMID: 24595007
The mechanism of stem cell differentiation into smooth muscle cells.
Thromb Haemost, 104(3):440-448, 10 Jun 2010
Cited by: 22 articles | PMID: 20539914
Review
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: C06 RR018928
NHLBI NIH HHS (7)
Grant ID: HL62572
Grant ID: R01 HL091168-01A1
Grant ID: R01 HL132574
Grant ID: R01 HL062572-12
Grant ID: R01 HL091168
Grant ID: R01 HL062572
Grant ID: HL091168