Abstract
Free full text

“Co-transcriptionality” - the transcription elongation complex as a nexus for nuclear transactions
Abstract
Much of the complex process of RNP biogenesis takes place at the gene, co-transcriptionally. The target for RNA binding and processing factors is therefore not a solitary RNA molecule, but a transcription elongation complex (TEC) comprising the growing nascent RNA and RNA polymerase traversing a chromatin template with associated passenger proteins. RNA maturation factors are not the only nuclear machines whose work is organized co-transcriptionally around the TEC scaffold. In addition DNA repair, covalent chromatin modification, “gene gating” at the nuclear pore, Ig gene hypermutation, and sister chromosome cohesion have all been demonstrated or suggested to involve a co-transcriptional component. From this perspective, TEC’s can be viewed as potent “community organizers” within the nucleus.
What does it mean to be co-transcriptional?
The major steps in mRNA biogenesis; transcription, 5′ capping, splicing, and cleavage/polyadenylation can be re-constituted in vitro entirely independently of one another, yet in the nucleus they occur at the same time and place in intimate proximity (Bauren et al., 1998; Beyer and Osheim, 1988) (Rasmussen and Lis, 1993). The substrate for pre-mRNA processing is actually a transcript that is being extruded through an exit channel in the RNA polymerase. In bacteria, translation and ribosome assembly both occur co-transcriptionally suggesting that linking transcription to other steps in gene expression is an ancient invention. Co-transcriptional processes are often tailored to specific RNA polymerase. Ribosome assembly in E. coli requires transcription by the cell’s own polymerase and is impaired if the rDNA is transcribed by the bacteriophage T7 RNA polymerase (Lewicki et al., 1993). Some eukaryotic rRNA processing is also co-transcriptional and in yeast it can be disrupted by mutation of RNA polymerase I (pol I) (Schneider et al., 2007). By the same token, pre-mRNA capping, splicing and cleavage/polyadenylation require transcription by RNA pol II (Sisodia et al., 1987). RNA polymerases therefore appear to have been selected for the ability to support co-transcriptional events. RNA pol II in particular has acquired a unique C-terminal domain (CTD) comprising heptad repeats on its large subunit that enables efficient mRNA processing (Meinhart et al., 2005; Phatnani and Greenleaf, 2006)(Fig. 1A).
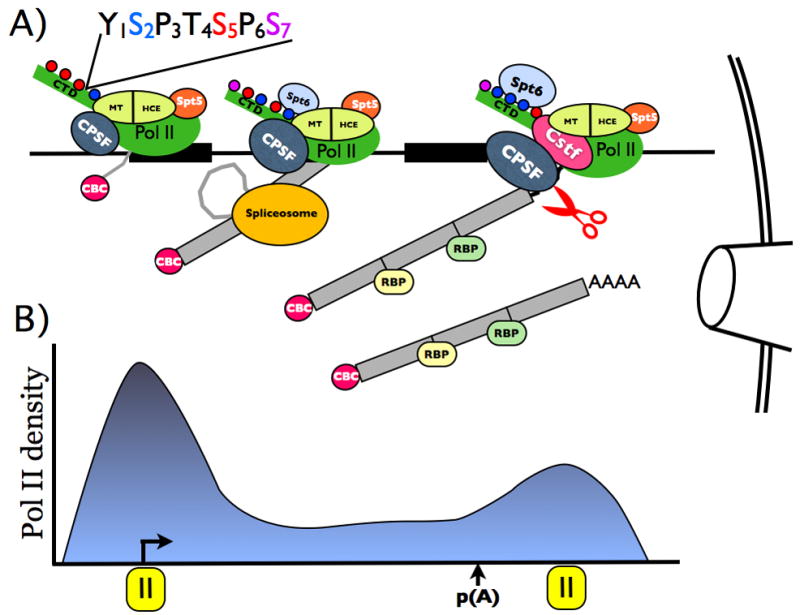
The conserved heptad repeat sequence with Serines 2, 5 and 7 that are differentially phosphorylated during the transcription cycle highlighted. The 5′-3′ distributions of CTD phosphorylations, processing factors and elongation factors are derived from ChIP studies (eg (Glover-Cutter et al., 2008) (Yoh et al., 2007)). Human capping enzyme (HCE), cap methyltransferase (MT) and the cleavage polyadenylation factors, CPSF, and CstF are shown. Elongation factors Spt5 that binds HCE and Spt6 that binds phospho-Ser2 are shown. Scissors depict RNA cleavage at the poly (A) site.
B). 5′-3′ distribution of RNA pol II density on a typical human gene showing pausing at the start site and downstream of the poly (A) site.
A number of general principles are emerging about the benefits of “co-transcriptionality” as a means of integrating diverse aspects of nuclear metabolism. In this review, we will attempt to highlight several of these principles and then discuss examples where they apply, focusing on mRNP biogenesis and modifications of the chromatin template. A number of excellent recent reviews also discuss aspects of this wide field (Iglesias and Stutz, 2008; Kornblihtt et al., 2004; Li and Manley, 2006; Schmid and Jensen, 2008) (Pandit et al., 2008).
Principles of “co-transcriptionality”
i. Co-transcriptionality permits “coupling” between different steps in mRNP biogenesis
When different steps in mRNA biogenesis occur at the same time and place there will be opportunities for “coupling” or cross-talk that makes those steps interdependent, thereby enhancing efficiency or accuracy. On the other hand, distinguishing between events that are simply “concurrent” from those that are functionally coupled is not always trivial (Lazarev and Manley, 2007). Coupling is implied when mutation of a protein that carries out one step, has an additional effect on a second step that occurs at the same time and place. For instance, coupling between pol II transcription and pre-mRNA processing is suggested by the fact that CTD deletion impairs processing (McCracken et al., 1997) in most cases studied so far but this effect can vary between genes (Ryman et al., 2007). The possibility of indirect secondary effects in genetic experiments must be kept in mind however and reconstituted in vitro systems can be helpful in ruling out such effects. Establishing functional coupling between transcription, processing and packaging of RNA in vitro is a major technical challenge, however some encouraging headway has been made (Das et al., 2007; Hicks et al., 2006; Rigo and Martinson, 2009b).
Coupling can work in different ways to link transcription with mRNA biogenesis and chromatin modification. The simplest form is a mass action effect resulting from co-localization of factors at the TEC thereby accelerating reactions that would otherwise be too slow. Localization is a major function of the pol II CTD that acts as a “landing pad”, binding directly to factors involved in pre-mRNA capping, 3′ end processing, transcription elongation, termination, and chromatin modification (Phatnani and Greenleaf, 2006)(Figs. 1A, ,2A).2A). A similar function is fulfilled by the unrelated CTD of RNA pol V, which binds an Argonaut protein that initiates gene silencing in plants (El-Shami et al., 2007). Direct interactions with the CTD have been characterized at the structural level for a capping enzyme Cgt1, a cleavage/polyadenylation factor, Pcf11, a histone methyltransferase, Set2, and a RNA binding termination factor, Nrd1 (Meinhart et al., 2005; Vasiljeva et al., 2008) (Figs. 1A, ,2A2A).

5′-3′ distributions of factors are based on ChIP studies as in Fig. 1A. Four factors that bind directly to the CTD are depicted: the cap methyltransferase (MT) and guanylyltransferase (GT), the termination factor Nrd1 and the cleavage/polyadenylation factor Pcf11. “ Gene gating” by putative interaction between the nuclear pore and the TEC is depicted by a yellow arrow. B). ChlP-Chip analysis of the 5′-3′ distribution of total pol II density (left) and Ser2 and Ser5 phosphorylated pol II (right) on a typical highly transcribed yeast gene, (RPL3). Results are from (P. Megee and D.B. unpublished).
A second coupling mechanism afforded by the meeting of factors at the TEC is allostery. An example is activation of the capping enzyme’s guanylyltransferase activity by the phosphorylated pol II CTD (Ho and Shuman, 1999). It seems likely that protein:protein interactions that first evolved to co-localize mRNA processing and transcription, may have subsequently acquired allosteric functions.
A third iteration is “kinetic coupling” that can facilitate mRNA biogenesis by optimising the timing of sequential events in this process. “Kinetic coupling” between transcript elongation and spliceosome assembly very likely regulates alternative splicing decisions (Kornblihtt et al., 2004).
ii. Co-transcriptionality can impose order or control over assembly of mRNPs and processing machines
Juxtaposition of proteins that have congregated at the TEC may permit assembly reactions, competitive interactions and handoffs that would not be possible post-transcriptionally without the TEC as a scaffold. Some protein complexes may be regulated by being assembled co-transcriptionally, rather than being loaded onto the TEC as fully pre-assembled units. A common relationship between TEC-associated factors is that of mutually exclusive protein:protein and protein:RNA interactions that replace one another in handoff reactions to establish an ordered sequence of events (Fig. 4B). One example is handoff of the yeast RNA helicase Sub2, from the THO complex, an elongation which rides with the TEC (Strasser et al., 2002), to the nascent transcript where it binds the mRNA export adaptor protein Yra1 (Iglesias and Stutz, 2008).
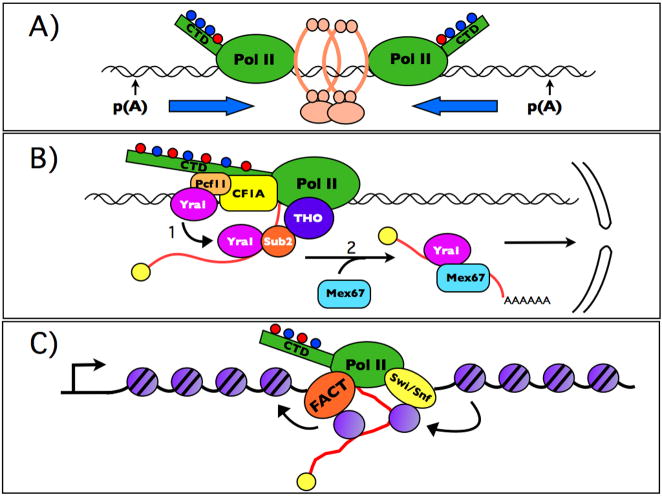
A) Possible mechanism of cohesin (pink rings) localization by convergent transcription in yeast (after (Lengronne et al., 2004)).
B) Mutually exclusive protein:protein and protein:RNA interactions and handoff reactions facilitate assembly of export-competent mRNP’s. In reaction 1 the yeast export adaptor Yra1 is handed off from Pcf11 to the RNA helicase Sub2. In reaction 2, Yra1 is handed off to the export receptor Mex67/Mtr2.
C) Mobilization of histones during pol II transcription elongation. FACT and Swi/Snf travel with the TEC to facilitate disassembly and reassembly of nucleosomes. Displaced histones could be handed off to the nascent RNA before they are re-deposited behind the polymerase.
A second way that order is imposed on co-transcriptional events is through directions emanating from phosphorylation of the pol II CTD heptad repeats (26 in yeast, 52 in mammals) with the consensus sequence YS2PTS5PS7. Phosphorylation of Ser5 by the TFIIH-associated kinase, Cdk7, or Kin 28 in yeast, occurs first, at initiation, whereas Ser2 phosphorylation by Cdk9/PTEFb (positive transcription elongation factor b) or Ctk1 in yeast, occurs later, during elongation (Komarnitsky et al., 2000; Phatnani and Greenleaf, 2006)(Fig. 2B). In addition, dephosphorylation at Ser5 and Ser2 by the Rtr1 and Fcp1 phosphatases, (Mosley et al., 2009; Phatnani and Greenleaf, 2006) helps define how the CTD is decorated as pol II transits from initiation, to elongation and termination of transcription. The combined action of kinases and phosphatases results in a characteristic switch from higher to lower ratios of Ser5:Ser2 CTD phosphorylation as pol II moves along a gene (Fig. 2B) (Komarnitsky et al., 2000). Ser7 residues of the CTD heptads are also phosphorylated within genes (Chapman et al., 2007) by Kin28/Cdk7 in yeast and mammalian cells (Akhtar et al., 2009). As a result of sequential phosphorylation, proteins that recognize Ser5 phosphorylated heptads, such as the yeast Nrd1 protein (Vasiljeva et al., 2008), will be recruited to the TEC earlier than those that recognize Ser2 phosphorylated heptads such as the 3′ processing factor, Pcf11 (Licatalosi et al., 2002).
iii. The TEC is a locator for nuclear machines
The impact of “co-transcriptionality” is not limited to mRNA production. The TEC is used to localize protein machines that carry out DNA repair, covalent DNA modification, and gene silencing to the places in the genome where they are required. An example of this locator function is transcription-coupled DNA repair where the nucleotide excision repair machine recognizes a stalled pol II TEC and specifically removes DNA lesions on the template strand (Lindsey-Boltz and Sancar, 2007). The TEC may also be an active participant in relocating chromatin associated proteins including histones and cohesins (Lengronne et al., 2004; Workman, 2006)(Fig. 4A, C).
The pol II CTD organizes a “mRNA factory”
The CTD functions as a flexible landing pad for pol II interacting proteins including pre-mRNA processing factors and chromatin modifiers (Phatnani and Greenleaf, 2006). The length of the fully extended CTD would be many times the diameter of core pol II (Meinhart et al., 2005) providing ample space in principle for binding to multiple partners. The heptad repeats are phosphorylated and dephosphorylated by kinases and phosphatases at the S2, S5, and S7 positions in a manner that is coordinated with the initiation, elongation and termination phases of the transcription cycle (reviewed in (Meinhart et al., 2005; Phatnani and Greenleaf, 2006)). CTD phosphorylation is potentially astronomically complex and elucidating how it controls loading and unloading of pol II passenger proteins is an important problem.
In vitro the CTD enhances capping, splicing and 3′ end formation independently of ongoing transcription, and CTD deletion disrupts these processing reactions in vivo (reviewed in (Hirose and Manley, 2000)). Although the CTD is necessary for efficient pre-mRNA processing, it is not sufficient. Simply pinning the CTD onto T7 RNA polymerase or pol I does not permit efficient processing (Natalizio et al., 2009).
A dynamic “mRNA factory” complex of pol II with associated proteins is probably responsible for simultaneous synthesis, processing, and packaging of the mRNP (Fig. 1A). The polymerase with nascent RNA and numerous passenger proteins would almost certainly exert too much viscous drag to move at high speed through nucleoplasm. The most likely solution to this hydrodynamic problem is that the chromatin template is threaded through a stationary “mRNA factory” (Jackson and Cook, 1995).
First, put your cap on
Pre-mRNAs are modified at the 5′ end by the addition of a 7-methyl G5′ppp5′N cap (reviewed in (Shuman, 2001) when the transcript is only 25–50 bases long. Capping is a three-step process that does not require specific sequences in the RNA. RNA triphosphatase removes the γ-phosphate of the first nucleotide, then GMP is added by RNA guanylyltransferase and finally the guanine is methylated at N7. In metazoans, the capping enzyme is a bifunctional polypeptide with triphosphatase and guanylyltransferase activities. The GMP transfer reaction is reversible, and capping is driven forward by the concerted action of both guanylyltransferase and methyltransferase, yet these two enzymes do not associate with one another (Shuman, 2001). The solution to this problem is that both the capping enzyme and the methyltransferase bind directly and specifically to the phosphorylated pol II CTD (Shuman, 2001). When transcription initiates, phosphorylation of the CTD on Ser5 residues permits loading of capping enzyme on to the TEC and allosteric activation of the guanylyltransferase (Ho and Shuman, 1999). The overall capping reaction is therefore facilitated by both co-localization of the capping enzymes on the phosphorylated CTD and allosteric activation.
The relationship between capping enzymes and pol II illustrates the two-way nature of communication between processing factors and the transcription machinery. Not only is capping enhanced by interaction of capping enzymes with the CTD but capping enzymes can also stimulate or inhibit transcription initiation and/or early elongation (Mandal et al., 2004; Myers et al., 2002; Schroeder et al., 2004). Polymerase complexes paused at 5′ ends are found on many metazoan genes and they are probably important for co-transcriptional capping. 5′ pausing is regulated by the pol II-associated elongation factor Spt5 which also allosterically activates the cap guanylyltransferase (Wen and Shatkin, 1999). Another regulator of elongation, the HIV Tat protein, also activates guanylyltransferase and enhances capping of viral transcripts (Chiu et al., 2002). Capping could be coupled to escape of paused pol II thereby ensuring that polymerases which enter productive elongation will have an appropriately modified 5′ end. Conversely pausing could promote capping perhaps by restricting the distance between the RNA 5′ end and capping enzymes sitting on the CTD. Capping is not usually regarded as a regulated step in gene expression however capping factor recruitment could in principle be regulated by Spt5 or CTD phosphorylation. A block to capping enzyme recruitment has been reported at yeast silent mating type loci (Gao and Gross, 2008). In future it will be of interest to investigate whether capping is affected by whether or not polymerase pauses at the transcription start site.
The influence of capping enzymes on mRNA production may not be limited to 5′ ends. Human capping enzymes are found at 5′ ends and throughout genes including 3′ flanking regions even more than a kilobase downstream of the poly (A) site (Glover-Cutter et al., 2008)(Fig. 1B). Capping factors therefore appear to linger on the pol II “landing pad” long after addition of the cap and they could therefore potentially influence elongation, termination, and 3′ end processing like the vaccinia virus capping enzyme. They might also cap the pervasive non-coding short transcripts detected within genes and at 5′ and 3′ ends (Kapranov et al., 2007). In addition, capping enzyme has been suggested to promote R-loop formation (see below) and could thereby affect transcription elongation (Kaneko et al., 2007).
Co-transcriptional splicing … some assembly required
Many but not all introns are removed co-transcriptionally rather than post-transcriptionally, as vividly shown by EM studies (Beyer and Osheim, 1988). Although not all splicing is completed co-transcriptionally, it is probable that assembly of most spliceosomes initiates on the nascent transcript. The spliceosome is one of the most elegant examples of ordered self-assembly. In vitro on a pre-made substrate without ongoing transcription, U1 snRNP first base pairs to the 5′ splice site and U2AF binds the 3′ splice site, followed by U2 snRNP base pairing the branch point. The tri snRNP U4-U6/U5 then engages and the complex rearranges to assume the catalytically active conformation, as U1 and U4 are discarded. ChIP analysis of co-transcriptional splicing on yeast genes supports a step-wise assembly process similar to that which occurs in vitro (Gornemann et al., 2005; Lacadie and Rosbash, 2005). On the other hand the possibility that pre-assembled higher order snRNP complexes are recruited co-transcriptionally cannot yet be excluded, especially in mammalian cells (Listerman et al., 2006). Little is known about spliceosome assembly at the TEC, but there are reasons for thinking it might differ from assembly that is uncoupled from transcription. Splicing of synthetic pre-mRNA’s in injected Xenopus ooctyes is less efficient than splicing coupled to transcription in the same cells consistent with stimulation of splicing in vitro by the phosphorylated CTD (Bird et al., 2004; Hirose and Manley, 2000). It has been suggested that co-transcriptional splicing might differ from splicing uncoupled from transcription because in the former case, exons are held in place by tethering to the polymerase (Dye et al., 2006). The functional significance of exon tethering for splicing remains to be determined however. Unlike the other mRNA processing steps, splicing is reiterated many times on most transcripts. How coupling with transcription might affect the re-cycling of spliceosome components for use on multiple introns within a transcript is an interesting open question.
A number of intriguing connections have been uncovered between the splicing machinery and TEC-associated proteins although the extent to which splicing is facilitated by direct protein:protein interactions between spliceosomes and pol II remains an open question. The yeast U1 snRNP protein Prp40, which bridges the 5′ splice site and branch point, can bind directly to the phospho-CTD (Phatnani and Greenleaf, 2006) but the functional significance of this interaction remains to be established. Human U1snRNP, but not other snRNP’s, also co-immunoprecipitates with pol II (Das et al., 2007). Furthermore U1snRNP at a 5′ splice site can activate recruitment of pol II and general transcription factors to the promoter, independent of splicing (Damgaard et al., 2008). The U1 snRNP may therefore have a special relationship with pol II TEC’s.
The SR family of splicing factors bind the nascent transcript at exonic splicing enhancer elements and regulate spliceosome assembly by contacting the U1 and U2 snRNPs (reviewed in (Long and Caceres, 2009)). SR proteins co-immunoprecipitate with pol II and the SC35 family member has been implicated in stimulating transcriptional elongation through its interaction with PTEFb (Lin et al., 2008). SR proteins other than SC35 appear to associate with TEC’s exclusively through the nascent RNA rather than through protein:protein contacts with pol II (Sapra et al., 2009). A functional link between SR’s and transcription is suggested by the finding that SRp20 regulation of alternative splicing requires the pol II CTD (de la Mata and Kornblihtt, 2006). In an in vitro system where pol II transcripts are selectively stabilized and spliced relative to T7 transcripts (Hicks et al., 2006). This channeling of pol II transcripts into a productive splicing pathway is probably due to facilitated binding of the nascent transcripts to RNA binding proteins (RBP’s) including SR proteins that protect them from degradation and enhance spliceosome assembly (Das et al., 2007).
In summary, while co-purification of splicing factors with pol II complexes is consistent with coupling between splicing and transcription, there is at present no compelling example of a functionally important direct interaction between a splicing factor and RNA pol II itself. It therefore remains possible that, in contrast to capping and 3′ end processing, all the major signals for loading of splicing factors onto the TEC lie in the nascent RNA, with the CTD and other transcriptional factors playing indirect roles. The extent to which co-transcriptional spliceosome assembly may vary between introns within a gene and between different genes remains an interesting open question.
Co-transcriptional splicing, elongation and RNA folding
RNA chain elongation is not a uniform monotonous process, but instead it is interrupted by numerous pauses dictated by the local sequence environment. Pol II elongation in live cells occurred at an average rate of 1.9 kb/min in one study (Boireau et al., 2007) and at a maximum rate of 4.3 kb/min in a second case (Darzacq et al., 2007). In addition to the many extrinsic factors that influence elongation, the intrinsic rate of elongation can be limited by the diffusion of NTP’s through the funnel domain to the active site. Conserved charged residues in the funnel hinder NTP diffusion to the active site potentially limiting the rate of transcription (Batada et al., 2004). Selective pressure acting on the funnel may therefore have tuned the transcription rate of RNA polymerase II so that it is within an optimal range that is compatible with co-transcriptional mRNA processing and packaging. One reason why transcription by T7 RNA polymerase does not support coupled pre-mRNA processing (Hicks et al., 2006; Natalizio et al., 2009) may be that it elongates several times faster than RNA pol II.
Nascent RNA is extruded through an exit channel in RNA polymerase that lies close to the attachment point of the CTD. Newly minted RNA sequences that exit the RNA polymerase are immediately available for interaction with RBP’s and base pairing with upstream RNA sequences. The folding pathway of a growing RNA chain differs fundamentally from the folding of a pre-made full-length transcript such as synthetic substrate RNA added to a processing reaction. Moreover the folding pathway adopted by a particular transcript can differ depending on its rate of growth and in particular on polymerase pausing (Pan and Sosnick, 2006)(Fig. 3B, C). Transcription by T7 RNA polymerase, which is 5–10 fold faster than E. coli polymerase, impairs co-transcriptional ribosome assembly (Lewicki et al., 1993) and correct folding of structured RNA’s in E. coli (Pan and Sosnick, 2006).
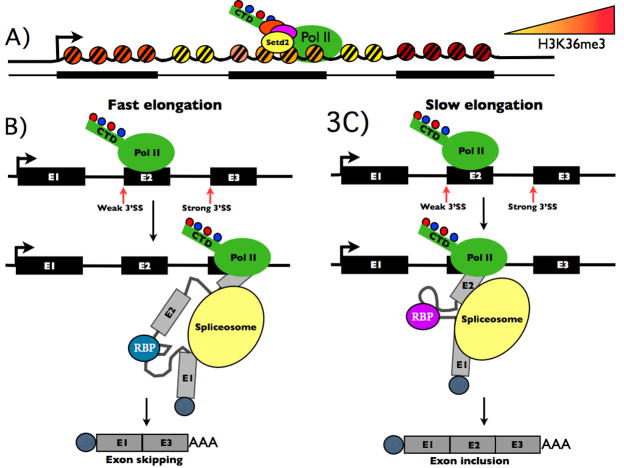
A) Differential histone H3 K36 trimethylation (H3K36me3) of worm and mouse exons and introns (Kolasinska-Zwierz et al., 2009) by Setd2 which binds the CTD (Yoh et al., 2008) is indicated. H3K36me3 increases 5′-3′ on most genes. B) A fast elongation rate and short “window of opportunity” favors exon skipping whereas slow elongation favors exon 2 (E2) inclusion (after (de la Mata et al., 2003). Elongation rate can also affect co-transcriptional RNA folding and potentially also binding of different RNA binding proteins (RBP).
The formation of productive versus non-productive processing complexes on nascent pre-mRNA’s is probably influenced by sequential folding of the RNA that exposes or sequesters splice sites and splicing enhancer and silencer sequences. Although the general significance of co-transcriptional RNA folding for splicing is not yet established, a growing body of evidence suggests that it can have important effects on alternative splicing (Shepard and Hertel, 2008). The practical significance of co-transcriptional folding in splicing is shown by the fact that the efficacy of therapeutic anti-sense oligonucleotides that induce exon skipping in the dystrophin mRNA can be predicted by taking into account the accessibility of target sequences during co-transcriptional folding (Wee et al., 2008).
A “window of opportunity” for alternative splicing
Alternative splicing affects the expression of most human genes and it is possible that the most important effect of transcription elongation on mammalian gene expression is mediated through effects on alternative splicing. The simplest way in which elongation rate can influence alternative splicing is by controlling the duration of the “window of opportunity” during which the upstream splice site can assemble a functional spliceosome before it has to compete with the downstream site (Fig. 3B, C). Hence, slowing elongation can enhance the use of poor upstream 3′ splice site relative to a better site downstream, and therefore favors inclusion of an alternative exon (Kornblihtt et al., 2004). It remains to be tested whether accelerated rates of transcription have the opposite effect, decreasing exon inclusion, as predicted by the “window of opportunity” model.
Given the intimate relationship between elongation and splicing, it is perhaps not surprising that numerous factors implicated in control of pol II pausing and processivity also affect constitutive or alternative splicing. These factors include the state of CTD phosphorylation, the elongation factor Spt5, promoter and enhancer-associated transcription factors and co-activators (reviewed in (Kornblihtt et al., 2004)), as well as covalent histone modifications (Kolasinska-Zwierz et al., 2009). The molecular basis for the connection between splicing and elongation is still unresolved however it is intriguing that the elongation factors PTEFb, CA150, and TAT-SF1 all associate directly or indirectly with spliceosomal U snRNP’s (Fong and Zhou, 2001; Lin et al., 2008; Pandit et al., 2008). PTEFb also interacts with SKIP, which is both a transcriptional co-activator and a subunit of U5 snRNP (Bres et al., 2005). Another connection is suggested by interaction of the U2 snRNP component SF3a with the chromatin remodeling ATPase Chd1 (Sims et al., 2007). It was recently discovered that elongation rates can be modulated by a physiological stimulus resulting in new alternative splice site choices. In response to UV-induced DNA damage, the CTD becomes hyperphosphorylated, transcription elongation slows down and alternative splice choices are switched in favor of the pro-apoptotic isoforms of Bcl-x and caspase 9 (Munoz et al., 2009).
An intriguing connection with chromatin is suggested by the discovery that the SWI/SNF and Chd1 chromatin remodeling ATPases influence splicing (Batsche et al., 2006; Sims et al., 2007). Batsche and colleagues suggested that alternative splicing decisions may be influenced by differences in elongation rates within constitutive versus alternatively spliced exons in a manner regulated by SWI/SNF and CTD phosphorylation (Batsche et al., 2006). Remarkably the chromatin of exons in worms and mice is enriched with histone H3 trimethylated on K36 and the amount of this modification correlates with the extent of splicing of alternative exons (Fig. 3A). Altered K36 methylation has also been correlated with regulation of NCAM alternative splicing following neuronal depolarization (Schor et al., 2009). Whether preferential H3K36 trimethylation in exons relative to introns affects splicing or vice versa is still unclear. H3K36 trimethylation by human HYPB/Setd2 enhances co-transcriptional loading of the mRNA export adaptor Aly by Iws1 a partner of Spt6, which binds to the CTD (Yoh et al., 2008). One possibility suggested by these results is that the level of K36 trimethylation could dictate differential elongation rates in exons and introns and by this means influences splicing and coupled export factor loading.
Co-transcriptional assembly of the 3′ processing machinery
3′ end processing of most mRNA’s is a two-step reaction comprising endonucleolytic cleavage shortly after the AAUAAA sequence, followed by polyadenylation of the exposed 3′ OH. Homologous multisubunit complexes including CstF (cleavage stimulation factor) and CPSF (cleavage polyadenylation specificity factor) in mammals and CF1A (cleavage factor 1A) and CPF (cleavage polyadenylation factor) in yeast perform coupled cleavage and polyadenylation. Some components including the endonuclease, CPSF73, are shared with the histone 3′ end processing complex that makes non-adenylated ends (reviewed in (Mandel et al., 2008)). Cleavage and early polyadenylation can occur at the site of transcription (Bauren et al., 1998) consistent with the fact that cleavage/polyadenylation factors are found at transcribed genes (Ahn et al., 2004; Gall et al., 1999; Glover-Cutter et al., 2008; Licatalosi et al., 2002). It has also been reported that poly (A) site cleavage can occur post-transcriptionally following polymerase release from the template (West et al., 2008)
The pol II CTD binds 3′ end processing factors and stimulates cleavage/polyadenylation in vivo and in vitro (Hirose and Manley, 1998; McCracken et al., 1997). The 50 kD subunit of CstF, the Pcf11 subunit of CF1A and the yeast termination factor Rtt103, all bind the CTD directly (Meinhart et al., 2005; Phatnani and Greenleaf, 2006) (Kim et al., 2004). Ser2 phosphorylation of the CTD is of special significance for 3′ end processing at least in part because the cleavage/polyadenylation factor Pcf11 preferentially binds to heptad repeats with this modification (Ahn et al., 2004; Licatalosi et al., 2002; Meinhart and Cramer, 2004). Modulation of CTD phosphorylation as polymerase traverses a gene therefore helps to coordinate the assembly of the 3′ end processing machinery at the site of transcription.
Unexpectedly, 3′ end processing factors are not confined to the 3′ ends of transcribed genes however. In fact human CPSF and yeast CF1A subunits (Dantonel et al., 1997; Glover-Cutter et al., 2008; Licatalosi et al., 2002) are found at 5′ ends, long before transcription of 3′ end processing signals. These factors are therefore probably recruited to the TEC initially by protein:protein interaction and subsequently handed off to the nascent RNA after the poly (A) site has been transcribed.
CPSF can bind both the body of pol II and CstF in a mutually exclusive way (Nag et al., 2007) suggesting that formation of a functional CPSF/CstF complex may be controlled by handoff of CPSF from pol II to CstF. A handoff reaction may also control assembly of the Pcf11 and Clp1 subunits of the yeast 3′ end processing complex, CF1A (Fig. 4B). Clp1 and the export adaptor, Yra1 (Johnson et al., 2009) bind the same short region of Pcf11 and are therefore likely to compete with one another. CF1A may therefore be recruited to the gene in a partially assembled form with Yra1 occupying the place of Clp1. At the poly (A) site Clp1 may displace Yra1, which is handed off to the RNA, thereby completing assembly of a functional 3′ processing complex (Saguez and Jensen, 2009).
Competition between mutually exclusive interactions at the TEC may be exploited for quality control of 3′ end processing. The yeast RBP’s Np13 and Rna15, a CF1A subunit, both load on to the TEC before it reaches the 3′ end of the gene. These two factors compete for binding to similar sites on the nascent RNA and it has been suggested this competition enhances the accuracy of 3′ processing by preventing Rna15 from recognizing cryptic poly (A) sites within genes (Bucheli et al., 2007).
3′ end processing and transcription elongation
Cleavage of the nascent transcripts at the poly (A) site probably occurs when pol II pauses downstream of the poly (A) site. Pausing in the 3′ flanking region approximately 1–2 kb downstream of the poly (A) site is a common feature of the pol II transcription cycle (Boireau et al., 2007; Darzacq et al., 2007; Glover-Cutter et al., 2008)(Fig. 1B). At this pause site, CTD Ser2 is highly phosphorylated (Fig. 1) and maximal levels of cleavage/polyadenylation factors are associated with the TEC (Glover-Cutter et al., 2008). Although the precise relationship between poly (A) site processing and subsequent transcription termination is unclear, it seems very likely that both events are coordinated with the downstream pause. Alternative poly (A) site choice has recently been recognized as a widespread phenomenon that determines the repertoire of 3′ UTR sequences (Licatalosi et al., 2008; Sandberg et al., 2008). How pausing in 3′ flanking regions is coordinated with alternative poly (A) site cleavage decisions is an interesting question for future investigation. While early recruitment of 3′ end processing factors appears to be a general phenomenon, it remains to be determined whether RNA cleavage at the poly (A) site always precedes transcription termination or whether the timing of cleavage and termination differs between genes.
Poly (A) site cleavage at the 3′ end may not be sufficient to cut the mRNA loose from the TEC. In mammalian cells, an additional release step requires the CTD (Custodio et al., 2007) and completion of splicing (Rigo and Martinson, 2009a). In yeast the THO complex, the RNA helicase Sub2, and the phosphatase Glc7 remodel the RNP at the 3′ end of the gene and pry it away from the cleavage/polyadenylation apparatus (Gilbert and Guthrie, 2004; Rougemaille et al., 2008).
Co-transcriptional recruitment of factors that dismantle the TEC
A major advantage of co-transcriptional over post-transcriptional recognition of 3′ end processing sites is that it permits coupling of transcription termination with recognition of the poly (A) signal that marks the end of the message (Rosonina et al., 2006). There are two main models for how cleavage/polyadenylation factors stimulate dissociation of the extraordinarily stable TEC. The “allosteric” model invokes a poly (A) site dependent conformational change in the TEC that reduces its processivity. The “torpedo” model, on the other hand, proposes that the cut site in the nascent RNA permits access to a 5′-3′ RNA exonuclease that degrades the nascent RNA tail and destabilizes the TEC. Evidence on these models is divided, but in both the “allosteric” and “torpedo” scenarios, it is clear that co-transcriptional loading of 3′ end processing factors including Pcf11 and the 5′-3′ RNA exonuclease Xrn2/Rat1 ultimately leads to dissociation of the TEC (reviewed in (Rosonina et al., 2006)).
A second mechanism of pol II termination is employed in yeast at non-coding genes that lack poly (A) sites. At these genes, a complex comprising Nrd1, Nab3, and the RNA helicase Sen1 is recruited to the TEC and terminates transcription independently of a 5′-3′ RNA exonuclease (Steinmetz et al., 2001) (Kim et al., 2006). The Nrd1 protein binds directly to Ser5 phosphorylated CTD heptads (Vasiljeva et al., 2008) as well as RNA. Position specific CTD phosphorylation on Ser5 at 5′ ends of genes ensures that this mechanism of termination only operates at short distances from the transcription start site (Gudipati et al., 2008). In summary pol II TEC’s, in a carefully regulated way, recruit the factors that ultimately lead to their dissociation from the template.
Specialized co-transcriptional RNA processing
In addition to capping, splicing and cleavage/polyadenylation, some transcripts are processed co-transcriptionally by A–I editing or miRNA excision from introns. In the nascent GluR-B pre-mRNA an ADAR (adenosine deaminase acting on RNA) recognizes RNA duplexes formed by intramolecular base pairing between exon 11 and intron 11 and converts an A to I thereby switching a Q codon to R. Editing in this case must occur before splicing and the pol II CTD is implicated in imposing this sequence of events (Ryman et al., 2007).
miRNA’s are released from pol II transcribed precursors by the microprocessor complex that includes the RNAse III family member Drosha which clips miRNA precursors at the base of a hairpin. Drosha is found at genes harboring intronic miRNA’s (Morlando et al., 2008) strongly suggesting that it works co-transcriptionally. Furthermore miRNA excision from introns is completely compatible with splicing (Kim and Kim, 2007). The microprocessor associates with spliceosomes (Kataoka et al., 2009) but it is not known whether, like RNA editing, the timing of miRNA excision relative to splicing is regulated by interactions with pol II.
Packaging, export, and the determination of RNA fate
As it is extruded from the polymerase, a nascent transcript encounters numerous RBP’s including cap binding complex (CBC) (Listerman et al., 2006; Visa et al., 1996), hnRNP’s (Daneholt, 2001), SR proteins (Long and Caceres, 2009), the exon-junction complex (EJC) (Custodio et al., 2004) and zipcode-binding proteins (ZBP) (Pan et al., 2007) some of which remain attached for much of the transcript’s lifetime. In this way co-transcriptional RBP loading governs the transport, translation, cytoplasmic localization, and lifespan of the mRNA (Daneholt, 2001; Glisovic et al., 2008).
In addition to providing early protection from nucleases, co-transcriptionality may impose order on RBP association with the nascent transcript as the mature mRNP assembles. For example CBC association with the 5′ cap early in transcript synthesis (Listerman et al., 2006; Visa et al., 1996) may direct subsequent interaction with the TREX complex (Cheng et al., 2006). TREX comprises a sub-complex called THO plus the export adaptor REF/Aly (Yra1 in yeast) and the RNA helicase UAP56 (Sub2 in yeast, Fig. 4B). TREX associates with the TEC and enhances elongation and mRNA export (Strasser et al., 2002).
A particularly critical aspect of co-transcriptional mRNP assembly is preparation for export. Nascent transcripts are packaged for export by loading of adaptor proteins that allow the mature mRNP to engage the export receptors Mex67/Mtr2 in yeast and TAP/p15 in metazoans (Iglesias and Stutz, 2008). Recruitment of the export adaptors Yra1 and REF/Aly to the nascent transcript is carefully monitored so that only perfectly formed fully processed mRNP’s become export competent (Lei et al., 2001; Schmid and Jensen, 2008). Yra1 binds the RNA helicase Sub2 that also associates with the THO complex on the TEC (Strasser et al., 2002). Yra1 recruitment requires interaction with a 3′ end processing factor, Pcf11 that binds the phosphorylated CTD (Johnson et al., 2009)(Fig. 4B). This mechanism therefore may ensure export adaptor loading will only occur if the machinery is in place to properly process the mRNA 3′ end. The mechanism of Yra1 loading also illustrates how the order of assembly of mRNP’s can be determined by the sequence of CTD phosphorylation. Ser2 phosphorylation specifically facilitates recruitment of Pcf11 and thereby indirectly specifies recruitment of the export factor Yra1 at later times in the transcription cycle. Following initial Yra1 loading by interaction with Pcf11, it is probably transferred to Sub2, whose helicase activity then facilitates a second transfer to the mRNA/Mex67 complex (Fig. 4B). Both handoff reactions most likely occur at the TEC since all players, Pcf11, Yra1, Sub2 and Mex67 localize to sites of transcription (reviewed in (Iglesias and Stutz, 2008)).
In mammalian cells, the Yra1 homologue Aly is loaded co-transcriptionally by interaction with Iws1, a partner of the elongation factor Spt6 that, like Pcf11, binds CTD heptads phosphorylated on Ser2 (Yoh et al., 2007). In summary, the sequence of CTD phosphorylations that accompanies the transcription cycle helps direct co-transcriptional mRNP assembly as well as processing of the nascent transcript.
Co-transcriptional Quality control
Quality control of mRNP’s is facilitated by their co-transcriptional assembly. This notion is suggested by the fact that slowing transcription elongation suppresses the growth defects of mutants that disrupt mRNP formation (Jensen et al., 2004). Yeast mRNP’s that are deemed not to be of “export quality” because they have improperly formed 3′ ends are detained close to the site of transcription in a process that requires the exosome (Hilleren et al., 2001), a complex of 3′-5′ exonucleases that degrades defective RNA’s. The exosome can be recruited to transcribed genes (Andrulis et al., 2002) and could therefore be positioned for degradation of defective mRNP’s when they are released from the template with exposed 3′ ends. A possible mechanism of exosome loading in yeast is through Nrd1 and Np13 which both bind the TEC and the exosome (Burkard and Butler, 2000; Vasiljeva and Buratowski, 2006).
Invasive nascent transcripts and R-loops
Another important reason for having RNP’s assemble co-transcriptionally rather than post transcriptionally is to protect the DNA template from invasion by naked RNA. When normal co-transcriptional handling of nascent RNA by RBP’s is disrupted, R-loops can form by reannealing of the transcript with DNA behind the polymerase. The displaced single-stranded non-template DNA strands are highly recombinogenic causing genetic instability (Li and Manley, 2006). Similarly, in E. coli disruption of co-transcriptional translation may induce nascent mRNA, no longer engaged with ribosomes, to form R-loops (Gowrishankar and Harinarayanan, 2004). In yeast, R-loops form in mutants of the TREX complex (Huertas and Aguilera, 2003). R-loop formation and genomic instability also occur in mammalian cells when the SR proteins, ASF/SF2 or SC35 are depleted (Li and Manley, 2006). R-loops can obstruct elongating RNA polymerases if they are not removed by RNaseH and this mechanism may help explain the pile ups of pol II within transcribed genes when SC35 is depleted (Lin et al., 2008).
Nuclear pores and the TEC, a role in “gating”?
In addition to processing and export factors, the TEC can also be contacted by nuclear pore components, as predicted by the “gene gating” hypothesis which proposes that export is facilitated by localizing active genes at the pore (Akhtar and Gasser, 2007)(Fig. 2A). Coupling between transcription and mRNP exit from the nucleus is suggested by the observation that ongoing transcription facilitates interaction of Chironomus Balbiani Ring (BR) RNP’s with nuclear pores (Kylberg et al., 2008). Specific promoters, transcriptional activators, the co-activator SAGA, 3′ UTR’s, and the exosome have all been implicated in “gene gating” (reviewed in (Akhtar and Gasser, 2007)). TEC interaction with the pore is supported by the fact that when the THO complex is compromised, pore proteins accumulate at the 3′ ends of genes. This phenomenon may reflect a trapped intermediate in normal mRNP export (Rougemaille et al., 2008). TREX2 is a four-subunit complex that bridges the pore with the Mex67/Mtr2 and the THO complex (Fischer et al., 2002). Recent resolution of the TREX2 structure (Jani et al., 2009) gives cause for optimism that the connection between transcription and mRNP delivery to the nuclear pore will ultimately be elucidated at the atomic level.
Co-transcriptional looping?
Transcribed genes can adopt a looped conformation that was first graphically revealed in the lampbrush chromosomes of amphibian ooctyes. Pol II and mRNA processing factors are distributed around lampbrush loops and these structures critically depend on ongoing transcription (Gall et al., 1999). 3C studies which detect the spatial proximity of non-contiguous DNA regions in the nucleus (Dekker et al., 2002) show that loops also form between 5′ and 3′ ends of transcribed genes in somatic cells (O’Sullivan et al., 2004). Insertion of the finger domain of promoter-bound TFIIB into the RNA exit channel of pol II at both ends of the gene has been suggested as a mechanism for tying together the base of a loop (Singh and Hampsey, 2007). Co-transcriptional mRNA processing has also been proposed to promote looping of the HIV provirus (Perkins et al., 2008). Many questions remain about the role of co-transcriptional events in gene looping including: Are loops a result of threading a gene through a stationary ‘mRNA factory’ or a specific conformation designed to facilitate communication and polymerase recycling between the two ends of a gene? What fraction of the time does a transcribed gene adopt a looped conformation? And can a loop withstand the torsional stress exerted by multiple polymerases on the same gene?
The TEC is a locator for chromatin modifiers and silencers
The pol II TEC is targeted by diverse proteins that modify the chromatin template. One of the first co-transcriptional processes identified in eukaryotes was transcription-coupled DNA repair in which the TEC stalled at a DNA lesion attracts the nucleotide excision repair machinery. Indeed pol II itself might be the “the most specific damage recognition protein” (Lindsey-Boltz and Sancar, 2007). Another DNA modification that is probably guided by the pol II TEC is somatic hypermutation of immunoglobulin genes by deamination of C’s in the displaced non-template strand of transcribed DNA. This reaction is catalyzed by activation induced deaminase, AID, which is probably associated with pol II (Nambu et al., 2003).
Modification of histone N-terminal tails vastly enriches the chromatin landscape, and accurate deposition of these covalent marks is vital to many nuclear processes. The pol II TEC serves an important auxiliary function in guiding the placement of covalent histone modifications at specific regions of the genome. Some chromatin modifiers including the H3K4 and K36 methyltransferases Set1 and Set2 bind to the pol II CTD phosphorylated on Ser5 and Ser2 residues respectively (Phatnani and Greenleaf, 2006; Yoh et al., 2008). Transcription dependent H3K36 methylation by Set2 in yeast, recruits a histone deacetylase that helps shut down cryptic promoters (Workman, 2006). Other chromatin modifiers can latch on to the RNA component of the TEC. A Chironomus histone acetyltransferase is targeted to actively transcribed chromatin by interaction with a complex of the RBP hrp65 and actin that binds to nascent RNA (Sjolinder et al., 2005).
As a result of modifiers piggy-backing on the pol II TEC, the primary function of some transcription units is not to produce an RNA transcript, but to establish a chromatin domain marked by specific histone marks. In this way, one round of transcription can exert a profound influence on the future expression of that sequence. A recently identified antisense RNA of the yeast GAL10 gene is present in only one cell in fourteen, however the transient presence of the pol II TEC on the gene is sufficient to establish a stable domain of H3K36 trimethylation and H3 deacetylation (Houseley et al., 2008).
Paradoxically transcription by RNA pol II plays a central role in establishment of transcriptionally silent heterochromatin. This new paradigm is based on silencing of centromeric chromatin in fission yeast (reviewed in (Moazed, 2009)). Silencing is established by the RITS complex (RNA-induced initiation of transcriptional gene silencing) that recruits the methyltranferase CLRC which deposits the signature mark of heterochromatin, methylated H3K9. RITS is targeted to the TEC by binding of its Argonaut 1 subunit to short RNA duplexes formed between siRNAs and the nascent chains made as pol II transcribes the centromeric repeats. Transcription of the repeats is limited to a brief interval in S phase but this is sufficient to establish a heterochromatin state that persists throughout the cell cycle.
Silencing at fission yeast centromeres is specifically disrupted by mutations in the Rpb2 and Rpb7 subunits of pol II (Djupedal et al., 2005; Kato et al., 2005). Remarkably splicing factors, likely operating together with pol II, help to maintain centromeric silencing (Bayne et al., 2008). In plants, a separate non-essential nuclear RNA polymerase, pol V (IVb) forms the scaffold for co-transcriptional gene silencing. The CTD of the pol V large subunit has GW/WG containing repeats unrelated to the pol II CTD heptads, but like the pol II CTD, it is a landing pad for other proteins, notably Argonaut 4 (El-Shami et al., 2007). Argonaut 4 situated on the CTD is thought to initiate silencing by binding siRNA’s duplexed with nascent transcripts (Wierzbicki et al., 2008). In summary, like pre-mRNA processing, co-transcriptional gene silencing is specific to particular RNA polymerases.
The TEC as a machine that moves chromatin associated proteins
In addition to localizing histone modifiers, the pol II TEC probably also positions other chromatin proteins along the chromosome including histones. The act of transcription almost certainly influences nucleosome positioning by stimulating displacement in front of the TEC and replacement in its wake (Workman, 2006). This effect is mediated in part by remodelers and histone chaperones that travel with the TEC. In yeast, the remodeler SWI/SNF and the H2A/H2B histone chaperone, FACT travel with pol II (Workman, 2006) and facilitate transcription-coupled eviction and re-deposition of histones (Fig. 4C). Whether these factors bind directly to the TEC in addition to histones that are mobilized by passage of the TEC is not known. It is also possible that factor-independent effects of transcriptional elongation could influence the localization of chromatin-associated proteins as a result of torsional stress that accumulates within transcribed DNA. When a restrained template is transcribed, twin domains of supercoiling are established; positive in front, and negative behind the polymerase. Positive supercoiling ejects H2A/H2Bdimers from nucleosomes (Levchenko et al., 2005) in vitro consistent with their co-transcriptional displacement in vivo (Workman, 2006). Histones displaced from chromatin by T7 RNA polymerase transcription in vitro bind more tightly to the RNA transcript than to competitor DNA (Peng and Jackson, 1997). Displacement of histones by pol II passage could therefore precipitate their binding to the nascent transcript unless a mechanism is in place to avert this situation. Alternatively transient interaction with nascent RNA could serve to localize a pool of histones at the site of transcription ready for re-deposition (Fig. 4C).
Another example of positioning by the pol II TEC has been proposed for cohesins that are thought to encircle pairs of sister chromatids and maintain their cohesion during interphase. Cohesins show a remarkable tendency to congregate between convergent genes in yeast (Lengronne et al., 2004) where they may facilitate transcriptional termination (Gullerova and Proudfoot, 2008). Preventing transcription of one member of a pair of convergent genes can abolish the build-up of cohesin in the intergenic region suggesting that the pol II TEC pushes cohesins into place (Lengronne et al., 2004)(Fig. 4A). It remains to be demonstrated directly however, that the TEC can take on this “bulldozer” –like function.
Future Directions
To understand the role of the TEC as an integrator and organizer of co-transcriptional activities in detail will require answers to a number of important questions including:
How do the right factors associate with the pol II TEC at the right time on a particular gene? To what extent does a code of CTD phosphorylation provide the necessary information as opposed to specific signals in the sequence of the nascent transcript?
What is the relationship between co-transcriptional pre-mRNA processing and chromatin? How does chromatin structure affect co-transcriptional processing? Conversely, can processing, particularly splicing, feed back on chromatin modification?
How does the rate of growth of the RNA chain affect mRNP assembly and processing, particularly alternative splicing? How is spliceosome assembly affected by being coupled to transcription and how does co-transcriptional spliceosome assembly differ between different constitutive and alternative introns.
What is the structural basis for targeting of the TEC by processing, packaging, and chromatin modifying machines? In a few cases, structures of these complexes have been elucidated, but in many other cases, the nature of contacts with the TEC are still undefined.
Acknowledgments
We apologize to colleagues whose work could not be referenced because of space limitations. Work in the authors’ lab is supported by NIH grants GM58163 and GM063873. We thank S. Johnson, S. Kim, R. Allshire, A. Kornblihtt, M. Neuberger, D. Black and P. Megee for helpful comments and B. Erickson for help preparing the figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SH, Kim M, Buratowski S. Phosphorylation of Serine 2 within the RNA Polymerase II C-Terminal Domain Couples Transcription and 3′ End Processing. Mol Cell. 2004;13:67–76. [Abstract] [Google Scholar]
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. [Abstract] [Google Scholar]
- Akhtar M, Heidemann M, Tietjen J, Zhang D, Chapman RD, Eick D, Ansari A. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. [Europe PMC free article] [Abstract] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. [Abstract] [Google Scholar]
- Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc Natl Acad Sci U S A. 2004;101:17361–17364. [Europe PMC free article] [Abstract] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. [Abstract] [Google Scholar]
- Bauren G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 1998;12:2759–2769. [Europe PMC free article] [Abstract] [Google Scholar]
- Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. Splicing factors facilitate RNAi- directed silencing in fission yeast. Science. 2008;322:602–606. [Europe PMC free article] [Abstract] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. [Abstract] [Google Scholar]
- Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy- terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol. 2004;24:8963–8969. [Europe PMC free article] [Abstract] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol. 2007;179:291–304. [Europe PMC free article] [Abstract] [Google Scholar]
- Bres V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19:1211–1226. [Europe PMC free article] [Abstract] [Google Scholar]
- Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Np13 and polyadenylation factor CFI. RNA. 2007;13:1756–1764. [Europe PMC free article] [Abstract] [Google Scholar]
- Burkard KT, Butler JS. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Np13p. Mol Cell Biol. 2000;20:604–616. [Europe PMC free article] [Abstract] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. [Abstract] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. [Abstract] [Google Scholar]
- Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell. 2002;10:585–597. [Abstract] [Google Scholar]
- Custodio N, Carvalho C, Condado I, Antoniou M, Blencowe BJ, Carmo-Fonseca M. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. Rna. 2004;10:622–633. [Europe PMC free article] [Abstract] [Google Scholar]
- Custodio N, Vivo M, Antoniou M, Carmo-Fonseca M. Splicing- and cleavage-independent requirement of RNA polymerase II CTD for mRNA release from the transcription site. J Cell Biol. 2007;179:199–207. [Europe PMC free article] [Abstract] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29:271–278. [Abstract] [Google Scholar]
- Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci U S A. 2001;98:7012–7017. [Europe PMC free article] [Abstract] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. [Abstract] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. [Abstract] [Google Scholar]
- Das R, Yu I, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. [Abstract] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. [Abstract] [Google Scholar]
- de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. [Abstract] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. [Abstract] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. [Europe PMC free article] [Abstract] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. [Abstract] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. [Europe PMC free article] [Abstract] [Google Scholar]
- Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. [Europe PMC free article] [Abstract] [Google Scholar]
- Fong Y, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–932. [Abstract] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. [Europe PMC free article] [Abstract] [Google Scholar]
- Gao L, Gross DS. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol Cell Biol. 2008;28:3979–3994. [Europe PMC free article] [Abstract] [Google Scholar]
- Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–212. [Abstract] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. [Europe PMC free article] [Abstract] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. [Europe PMC free article] [Abstract] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. [Abstract] [Google Scholar]
- Gowrishankar J, Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol Microbiol. 2004;54:598–603. [Abstract] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–794. [Abstract] [Google Scholar]
- Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. [Abstract] [Google Scholar]
- Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. [Abstract] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. [Abstract] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. [Abstract] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [Abstract] [Google Scholar]
- Ho C, Shuman S. Distinct Effector Roles for Ser2 and Ser5 Phosphorylation of the RNA polymerase II CTD in the Recruitment and Allosteric Activation of Mammalian Capping Enzyme. Mol Cell. 1999;3:405–411. [Abstract] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. [Abstract] [Google Scholar]
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. [Abstract] [Google Scholar]
- Iglesias N, Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 2008;582:1987–1996. [Abstract] [Google Scholar]
- Jackson DA, Cook PR. The structural basis of nuclear function. Int Rev Cytol. 1995;162a:125–149. [Abstract] [Google Scholar]
- Jani D, Lutz S, Marshall N, Fischer T, Kohler A, Ellisdon A, Hurt E, Stewart M. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell. 2009;33:727–737. [Europe PMC free article] [Abstract] [Google Scholar]
- Jensen TH, Boulay J, Olesen JR, Colin J, Weyler M, Libri D. Modulation of transcription affects mRNP quality. Mol Cell. 2004;16:235–244. [Abstract] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell. 2009;33:215–226. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaneko S, Chu C, Shatkin AJ, Manley JL. Human capping enzyme promotes formation of transcriptional R loops in vitro. Proc Natl Acad Sci U S A. 2007;104:17620–17625. [Europe PMC free article] [Abstract] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. [Abstract] [Google Scholar]
- Kataoka N, Fujita M, Ohno M. Functional association of the Microprocessor complex with spliceosome. Mol Cell Biol 2009 [Europe PMC free article] [Abstract] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. [Abstract] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rail exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. [Abstract] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct Pathways for snoRNA and mRNA Termination. Mol Cell. 2006;24:723–734. [Abstract] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. [Europe PMC free article] [Abstract] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. [Europe PMC free article] [Abstract] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. [Europe PMC free article] [Abstract] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. [Europe PMC free article] [Abstract] [Google Scholar]
- Kylberg K, Bjorkroth B, Ivarsson B, Fomproix N, Daneholt B. Close coupling between transcription and exit of mRNP from the cell nucleus. Exp Cell Res. 2008;314:1708–1720. [Abstract] [Google Scholar]
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell. 2005;19:65–75. [Abstract] [Google Scholar]
- Lazarev D, Manley JL. Concurrent splicing and transcription are not sufficient to enhance splicing efficiency. RNA. 2007;13:1546–1557. [Europe PMC free article] [Abstract] [Google Scholar]
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. [Europe PMC free article] [Abstract] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. [Europe PMC free article] [Abstract] [Google Scholar]
- Levchenko V, Jackson B, Jackson V. Histone release during transcription: displacement of the two H2A–H2B dimers in the nucleosome is dependent on different levels of transcription-induced positive stress. Biochemistry. 2005;44:5357–5372. [Abstract] [Google Scholar]
- Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. JMol Biol. 1993;231:581–593. [Abstract] [Google Scholar]
- Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. [Abstract] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. [Abstract] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. [Europe PMC free article] [Abstract] [Google Scholar]
- Lindsey-Boltz LA, Sancar A. RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Proc Natl Acad Sci U S A. 2007;104:13213–13214. [Europe PMC free article] [Abstract] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. [Abstract] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. [Abstract] [Google Scholar]
- Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7572–7577. [Europe PMC free article] [Abstract] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. [Europe PMC free article] [Abstract] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan GH, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-Terminal Domain Of RNA Polymerase II Couples Messenger RNA Processing to Transcription. Nature. 1997;385:357–361. [Abstract] [Google Scholar]
- Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy- terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. [Abstract] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. [Abstract] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. [Europe PMC free article] [Abstract] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. [Abstract] [Google Scholar]
- Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. Rtrl is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell. 2009;34:168–178. [Europe PMC free article] [Abstract] [Google Scholar]
- Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano II, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. [Abstract] [Google Scholar]
- Myers LC, Lacomis L, Erdjument-Bromage H, Tempst P. The yeast capping enzyme represses RNA polymerase II transcription. Mol Cell. 2002;10:883–894. [Abstract] [Google Scholar]
- Nag A, Narsinh K, Martinson HG. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol. 2007;14:662–669. [Abstract] [Google Scholar]
- Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. [Abstract] [Google Scholar]
- Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA. The carboxyl-terminal domain of RNA polymerase II is not sufficient to enhance the efficiency of pre-mRNA capping or splicing in the context of a different polymerase. J Biol Chem 2009 [Europe PMC free article] [Abstract] [Google Scholar]
- O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. [Abstract] [Google Scholar]
- Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol Cell Biol. 2007;27:8340–8351. [Europe PMC free article] [Abstract] [Google Scholar]
- Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. [Abstract] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. [Europe PMC free article] [Abstract] [Google Scholar]
- Pandya-Jones A, Black D. Co-transcriptional Splicing of Constitutive and Alternative Exons. RNA. 2009 in press. [Europe PMC free article] [Abstract] [Google Scholar]
- Peng HF, Jackson V. Measurement of the frequency of histone displacement during the in vitro transcription of nucleosomes: RNA is a competitor for these histones. Biochemistry. 1997;36:12371–12382. [Abstract] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. [Europe PMC free article] [Abstract] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. [Abstract] [Google Scholar]
- Rasmussen EB, Lis JT. In-vivo transcriptional pausing and cap formation on 3 drosophila heat-shock genes. Proc Natl Acad Sci USA. 1993;90:7923–7927. [Europe PMC free article] [Abstract] [Google Scholar]
- Rigo F, Martinson HG. Polyadenylation releases mRNA from RNA polymerase II in a process that is licensed by splicing. RNA 2009a [Europe PMC free article] [Abstract] [Google Scholar]
- Rigo F, Martinson HG. Polyadenylation releases mRNA from RNA polymerase II in a process that is licensed by splicing. RNA. 2009b;15:823–836. [Europe PMC free article] [Abstract] [Google Scholar]
- Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. [Abstract] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, Libri D. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. [Abstract] [Google Scholar]
- Ryman K, Fong N, Bratt E, Bentley DL, Ohman M. The C- terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. [Europe PMC free article] [Abstract] [Google Scholar]
- Saguez C, Jensen TH. Assembly of export-competent mRNP: it’s all about being connected. Mol Cell. 2009;33:139–140. [Abstract] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. [Europe PMC free article] [Abstract] [Google Scholar]
- Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. [Abstract] [Google Scholar]
- Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117:419–429. [Abstract] [Google Scholar]
- Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell. 2007;26:217–229. [Europe PMC free article] [Abstract] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106:4325–4330. [Europe PMC free article] [Abstract] [Google Scholar]
- Schroeder S, Zorio D, Schwer B, Shuman S, Bentley D. A Function of Yeast mRNA Cap Methyltransferase, Abd1, in transcription by RNA Polymerase II. Mol Cell. 2004;13:377–387. [Abstract] [Google Scholar]
- Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–1469. [Europe PMC free article] [Abstract] [Google Scholar]
- Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. [Abstract] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. [Europe PMC free article] [Abstract] [Google Scholar]
- Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. [Abstract] [Google Scholar]
- Sisodia SS, Sollner WB, Cleveland DW. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol Cell Biol. 1987;7:3602–3612. [Europe PMC free article] [Abstract] [Google Scholar]
- Sjolinder M, Bjork P, Soderberg E, Sabri N, Farrants AK, Visa N. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev. 2005;19:1871–1884. [Europe PMC free article] [Abstract] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. [Abstract] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez- Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. [Abstract] [Google Scholar]
- Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. [Abstract] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5- phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. [Europe PMC free article] [Abstract] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. [Europe PMC free article] [Abstract] [Google Scholar]
- Wee KB, Pramono ZA, Wang JL, MacDorman KF, Lai PS, Yee WC. Dynamics of co-transcriptional pre-mRNA folding influences the induction of dystrophin exon skipping by antisense oligonucleotides. PLoS ONE. 2008;3:e1844. [Europe PMC free article] [Abstract] [Google Scholar]
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. [Europe PMC free article] [Abstract] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. [Europe PMC free article] [Abstract] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. [Europe PMC free article] [Abstract] [Google Scholar]
- Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. [Abstract] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. [Europe PMC free article] [Abstract] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2009.09.018
Read article for free, from open access legal sources, via Unpaywall:
https://www.cell.com/article/S1097276509006765/pdf
Subscription required at www.molecule.org
http://www.molecule.org/cgi/content/reprint/36/2/178
Citations & impact
Impact metrics
Citations of article over time
Article citations
Splicing regulation through biomolecular condensates and membraneless organelles.
Nat Rev Mol Cell Biol, 25(9):683-700, 21 May 2024
Cited by: 2 articles | PMID: 38773325
Review
The Nuclear Cap-Binding Complex, a multitasking binding partner of RNA polymerase II transcripts.
J Biochem, 175(1):9-15, 01 Dec 2023
Cited by: 2 articles | PMID: 37830942 | PMCID: PMC10771035
Review Free full text in Europe PMC
Regulation of the dynamic RNA Pol II elongation rate in Drosophila embryos.
Cell Rep, 42(10):113225, 12 Oct 2023
Cited by: 1 article | PMID: 37837623 | PMCID: PMC10842316
Control of transcription elongation and DNA repair by alarmone ppGpp.
Nat Struct Mol Biol, 30(5):600-607, 30 Mar 2023
Cited by: 4 articles | PMID: 36997761 | PMCID: PMC10191844
The Role of RNA-Binding Proteins in Hematological Malignancies.
Int J Mol Sci, 23(17):9552, 23 Aug 2022
Cited by: 10 articles | PMID: 36076951 | PMCID: PMC9455611
Review Free full text in Europe PMC
Go to all (243) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing.
J Mol Biol, 428(12):2623-2635, 20 Apr 2016
Cited by: 159 articles | PMID: 27107644 | PMCID: PMC4893998
Review Free full text in Europe PMC
Cotranscriptional RNA checkpoints.
Epigenomics, 2(3):449-455, 01 Jun 2010
Cited by: 5 articles | PMID: 22121903
Review
THO/Sub2p functions to coordinate 3'-end processing with gene-nuclear pore association.
Cell, 135(2):308-321, 01 Oct 2008
Cited by: 103 articles | PMID: 18957205
The link between mRNA processing and transcription: communication works both ways.
Exp Cell Res, 296(1):91-97, 01 May 2004
Cited by: 85 articles | PMID: 15120999
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (6)
Grant ID: R01 GM063873-06
Grant ID: R01 GM058613-11
Grant ID: GM063873
Grant ID: R01 GM063873
Grant ID: GM58163
Grant ID: R01 GM058613




