Abstract
Free full text

Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Abstract
Flaviviruses are a group of human pathogens causing severe encephalitic or hemorrhagic diseases that include West Nile, dengue and yellow fever viruses. Here, using X-ray crystallography we have defined the structure of the flavivirus cross-reactive antibody E53 that engages the highly conserved fusion loop of the West Nile virus envelope glycoprotein. Using cryo-electron microscopy, we also determined that E53 Fab binds preferentially to spikes in noninfectious, immature flavivirions but is unable to bind significantly to mature virions, consistent with the limited solvent exposure of the epitope. We conclude that the neutralizing impact of E53 and likely similar fusion-loop-specific antibodies depends on its binding to the frequently observed immature component of flavivirus particles. Our results elucidate how fusion-loop antibodies, which comprise a significant fraction of the humoral response against flaviviruses, can function to control infection without appreciably recognizing mature virions. As these highly cross-reactive antibodies are often weakly neutralizing they also may contribute to antibody-dependent enhancement and flavi virus pathogenesis thereby complicating development of safe and effective vaccines.
Introduction
Flaviviruses are a group of human pathogenic, enveloped RNA viruses, which include dengue (DENV), yellow fever and West Nile viruses (WNV), and cause severe hemorrhagic or encephalitic disease of global impact. They are composed of an outer protein shell with 180 copies of the envelope (E) protein and membrane (M) protein, a lipid bilayer and an inner core that contains the infectious RNA genome and capsid (C) protein. X-ray crystal structures (Rey et al, 1995; Modis et al, 2003; Zhang et al, 2004; Nybakken et al, 2006) have established that the E glycoprotein of flaviviruses has three domains, DI, DII and DIII, with a flexible ‘hinge' between DI and DII (Zhang et al, 2007). At the tip of domain DII is the conserved ‘fusion loop', which is required for acid-catalysed type II fusion with the host endosomal membrane (Allison et al, 2001; Bressanelli et al, 2004; Modis et al, 2004).
In the flavivirus life cycle, formation of immature virus occurs in the endoplasmic reticulum. The virus matures while being transported to the plasma membrane through the trans-Golgi network (TGN). The low pH environment of the TGN causes the heterodimeric complex of E with the premembrane protein (prM) on the viral surface to reorganize into E homodimers (Wengler and Wengler, 1989; Li et al, 2008; Yu et al, 2008). This structural rearrangement leads to the exposure of a furin cleavage site on the prM molecule. After cleavage, the pr molecules remain associated with the hydrophobic fusion loop of the E proteins while still in a low pH environment thus preventing fusion of the virus with membranes in the host cell. As the virus is released into the neutral pH extracellular environment, the pr peptides disassociate from the viral surface. Subsequently, after virus attachment and internalization through poorly characterized cellular receptors, a series of structural transitions, triggered by the low pH of the endosomes, convert the mature virion into a fusion-competent particle that promotes the release of the viral genome into the cytoplasm of the host cell.
In the immature virion, the E and the prM glycoproteins form heterodimers grouped into 60 trimeric spikes (Zhang et al, 2003) (Figure 1A). In the mature virus, the E glycoprotein associates into 90 homodimers, forming a smooth viral surface (Kuhn et al, 2002) (Figure 1B). Both immature and mature viruses have icosahedral symmetry with an external diameter of about 600 and 500 Å, respectively.
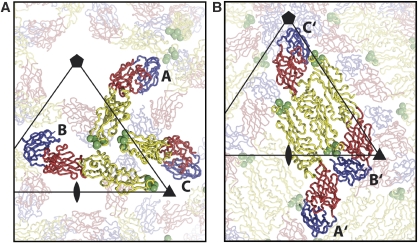
Organization of the E glycoprotein on the surface of (A) immature and (B) mature virus. E monomers are shown as a Cα backbone with DI, DII and DIII in red, yellow and blue, respectively. The black triangle represents an icosahedral asymmetric unit. Green spheres mark the E53 epitope residues (75, 76, 99, 106 and 107) determined by yeast surface display.
Although human vaccines are available for yellow fever virus, tick-borne encephalitis virus and Japanese encephalitis virus, at present none are approved for use against DENV or WNV (Widman et al, 2008). Vaccine development for DENV, which has four serologically distinct serotypes, has been particularly problematic because of the possibility of antibody-dependent enhancement (ADE) in myeloid cells expressing activating Fc-γ receptors (Brandt, 1982). Poorly neutralizing cross-reactive antibodies that are generated during primary infection can enhance replication of a heterologous DENV strain during a subsequent secondary challenge. The majority of these antibodies are generated against the E glycoprotein, which is the major antigenic and immunodominant protein on the virus surface.
Antibodies that recognize the conserved flavivirus fusion loop are generally cross-reactive and comprise a significant fraction of the humoral response to WNV in humans (Oliphant et al, 2007; Roberson et al, 2007), although they have weak neutralizing activity (Crill and Chang, 2004; Oliphant et al, 2006, 2007; Stiasny et al, 2006). Mapping studies have shown that the monoclonal antibody (MAb) E53 recognizes an epitope within the fusion loop of the WNV E glycoprotein. Initial functional studies showed that MAb E53 protects mice from lethal WNV infection when administered prophylactically (Oliphant et al, 2006) and blocks Vero cell infection primarily by inhibiting cellular attachment (Nybakken et al, 2005). Subsequent investigations showed that E53 had decreased inhibitory activity when pr was completely cleaved off the virus particle, showing that the neutralizing activity is dependent on the maturation state of the virion (Nelson et al, 2008). The E53 epitope was mapped by yeast surface display to include residues in the fusion loop and adjacent bc-loop in domain II of E (Oliphant et al, 2006) (Figure 1).
Here, using X-ray crystallography of the E53–WNV E glycoprotein complex and cryo-electron microscopic (cryoEM) reconstructions of WNV and DENV complexed with Fab fragments, we show that the E53 epitope is largely inaccessible in mature virions and the antibody binds preferentially to the spike in immature virions. The structural description of an antibody that neutralizes infectivity by binding to an immature virus particle strongly supports the earlier hypothesis (Nelson et al, 2008) that hybrid mature/immature particles contribute to virus infectivity and pathogenesis.
Results
Crystal structure of the E53 Fab fragment in complex with WNV E glycoprotein
The structure of E53 Fab in complex with the WNV E ectodomain was determined crystallographically to 3.0 Å resolution (see Materials and methods and Supplementary Table I), showing that E53 engages 12 residues within the fusion loop (E residues G104, C105, G106, L107, G109 and K110) and the disulphide-linked bc-loop (E residues C74, P75, T76, M77, G78 and E79) (Figure 2). Yeast epitope mapping had correctly identified four of these contact residues (P75, T76, G106 and L107). Epitope recognition is primarily facilitated by the three complementarity-determining regions (CDRs) of the E53 heavy chain. CDR3 is used predominantly as it projects out of the combining site to intimately contact both E protein loops. Only one light chain residue makes contact in the antibody–antigen interface (Supplementary Table II). Four potential hydrogen bonds form in the interface, three of which involve exposed backbone oxygen and nitrogen atoms of the E protein fusion loop. The binding interface has a low degree of shape complementarity (Sc=0.49) (Sundberg and Mariuzza, 2002) and occludes approximately 800 Å2 of surface area. The entire sequence of the E53 epitope is conserved between WNV and all four DENV serotypes with the exception of residue M77, which is a glutamine in DENV. This difference might explain the stronger binding and neutralization activity of this antibody against WNV (DH Fremont and MS Diamond, unpublished data).
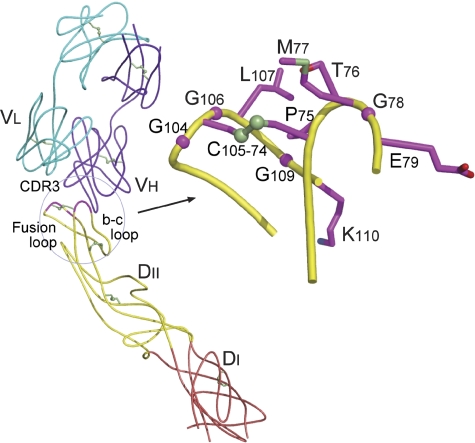
Crystal structure of E53 Fab in complex with the WNV envelope glycoprotein. On the left, ribbon diagram of the complex, with DII of E depicted in yellow, DI in red, the antibody heavy chain in purple and the light chain in cyan. The structurally defined epitope is displayed in magenta. On the right, close-up view of the E53 epitope, with the side chains of contacted residues coloured using magenta for carbon, red for oxygen, green for sulphur and blue for nitrogen atoms (<4.0 Å distance).
CryoEM studies of E53 antibody Fab fragment in complex with WNV and DENV
CryoEM experiments failed to form complexes of E53 Fab fragments with mature WNV, although it is possible that a low occupancy of potential E53 binding sites would not become evident in the icosahedral averaging procedure required for the image reconstruction. In contrast, when E53 Fab was incubated with immature WNV or DENV particles, it formed stable icosahedral complexes. The cryoEM reconstructions of immature WNV and DENV complexed with E53 Fab were determined to 15 and 23 Å resolution, respectively. These showed the anticipated trimeric E protein spikes (molecules A, B and C in Figure 1A) as observed in studies of the uncomplexed immature viruses (Zhang et al, 2003, 2007) (Figure 3; Supplementary Figure 1). The separation of the lipid membrane from the nucleocapsid core, a measure of the quality of the cryoEM reconstruction, was seen only in the DENV complex (Supplementary Figure 1). Fitting of the atomic structures of E and pr into the electron density of the complex required only minor movement of the individual E and pr molecules, compared with the known icosahedral arrangement in immature virions (Supplementary Tables III and IV). In both reconstructions, density representing the bound E53 Fab molecules was associated with the fusion loops on molecules A and B, but not with molecule C (Figure 3B and C). Indeed, in immature flavivirus particles, the fusion loops on molecules A and B (Figure 1A) are marginally accessible to water (accessible surface areas (Hubbard and Thornton, 1993) are 678 and 596 Å2, respectively), but the fusion loop on molecule C is completely covered by the pr peptide and neighbouring E proteins.
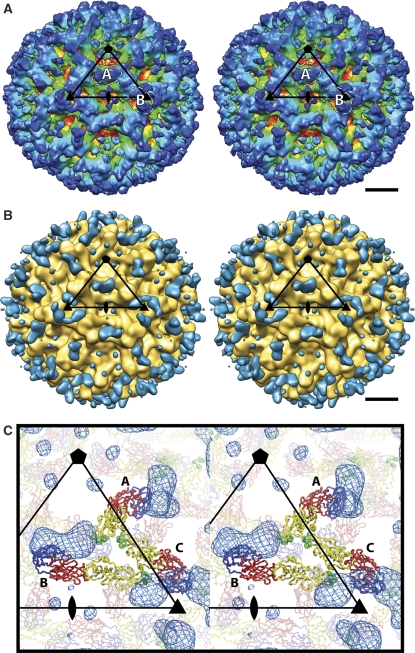
CryoEM reconstruction of immature WNV in complex with Fab fragments of the anti-fusion-loop antibody E53. (A) Stereoview showing the immature WNV–E53 Fab complex at 15 Å resolution. The density is coloured according to radius (red: 190 Å; yellow: 220 Å; green: 235 Å; light blue: 260 Å; blue: 305 Å). The black triangle shows an icosahedral asymmetric unit. The dashed circles mark the positions where the E53 Fab is bound to the fusion loop of molecules A and B, respectively. The scale bar represents 100 Å. Similar results were obtained for immature DENV in complex with E53 Fab (Supplementary Figure 1). (B) Stereoview showing the difference density between the immature WNV–E53 Fab complex and immature WNV (blue) superpositioned onto the surface of immature WNV (gold) at 24 Å resolution. The black triangle marks an icosahedral asymmetric unit. The scale bar represents 100 Å. (C) The difference density (blue mesh) is shown relative to the immature E protein structure coloured as in Figure 1.
Fitting of the E53 crystal structure into the cryoEM maps
The atomic coordinates of the E53 Fab and E glycoprotein were fitted separately into the cryoEM densities (Figure 4). The resultant complex of molecule A and the variable domain of E53 in the cryoEM map was the same, within experimental error, as observed in the crystal structure of the E53 Fab–WNV E complex. Furthermore, the epitope of E53 on the E protein as determined crystallographically and by cryoEM is consistent with the yeast display epitope mapping data (Oliphant et al, 2006).
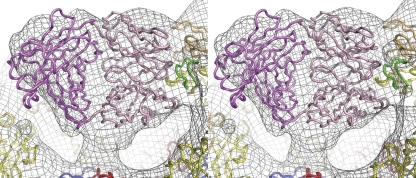
Stereoview showing E53 Fab, pr and E as fitted into the cryoEM reconstruction of the immature WNV–E53 Fab complex (grey mesh). Shown is the fit into the density associated with molecule A. Domain DII of the E protein and the fusion loop are coloured in yellow and green, respectively. The E53 Fab variable and constant domains are coloured in light and dark magenta, respectively. The pr molecules are coloured in orange.
When the E53 Fab–E glycoprotein crystal structure was superimposed onto the B molecule fitted into the EM map, clashes occurred between the three-fold symmetry-related Fab molecules. Fitting of the E53 Fab variable domain into the density adjacent to the fusion loop of the B molecule showed that the E53 variable domain had rotated by about 35° compared with that observed in the crystal structure, to avoid steric conflicts between the symmetry-related Fabs. Presumably, the loops containing the epitope residues have twisted to follow the limited motion of the Fab molecule, necessitating a small (1.6–5.3 Å), almost unidirectional movement of these residues. Comparison of cryoEM density heights showed ~75% occupancy of the Fab molecules bound to the A molecules and 50% occupancy to the B molecules (see Materials and methods). Similar results were obtained when the atomic coordinates of E53 Fab and DENV E were fitted into the cryoEM density of the immature DENV–E53 Fab complex (Supplementary Table III).
In the mature virus, only about 45, 30 and 35% of the area of the E53 epitope is accessible to water in molecules A', B' and C', respectively. Furthermore, E53 Fabs bound to the mature virus would be expected to cause severe clashes with neighbouring E molecules (Figure 5). An earlier study (Lok et al, 2008) showed that Fab fragments can bind to epitopes that are partially occluded in mature DENV. In doing so, the Fab fragments lock the virus into a conformation that is presumably present transiently while the virus is in continuous dynamic motion about a mean ‘mature' conformation. Thus, if E53 Fabs occasionally bind to mature virus as might perhaps be possible, parts of the virus may become locked into the immature conformation. In summary, E53 Fabs do not bind efficiently to mature infectious virus, consistent with earlier functional observations (Nelson et al, 2008), raising the question how E53 and similar fusion-loop-specific antibodies can neutralize flavivirus infection.
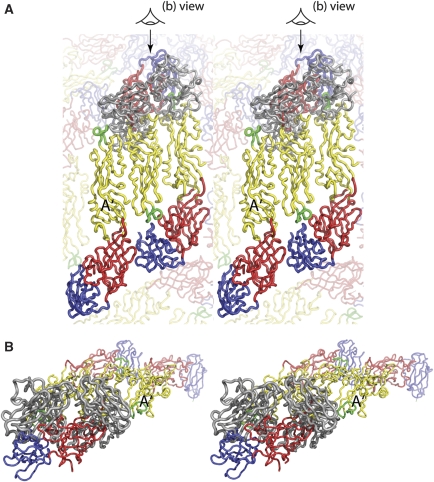
Stereoviews showing that E53 Fab cannot bind to the mature virus. (A) The crystal structure of the E53 Fab–E complex was superpositioned onto DII of molecule A' to show that severe clashes would occur between the Fab and the E molecules. The Fab is shown in grey. Each protein is shown as a Cα backbone. Domains DI, DII and DIII of the E protein are coloured in red, yellow and blue, respectively. The fusion loop is coloured in green. (B) Side view of structure. The direction of view is indicated in (A) by a cartoon eye.
Discussion
CryoEM images show that preparations of WNV or DENV contain a mixture of smooth mature, spiky immature and ‘partially mature' particles, most likely due to incomplete processing by furin during virus maturation (Figure 6). E53 has almost no neutralizing effect on fully mature WNV produced in cells overexpressing furin (Nelson et al, 2008), presumably because it binds poorly, if at all, to mature virus. Moreover, completely immature particles are poorly or not infectious regardless of E53 binding, because they enter target cells less efficiently and/or are unable to undergo the pH-dependent structural transition required for endosomal fusion (Guirakhoo et al, 1991; Stadler et al, 1997; Elshuber et al, 2003). In contrast, E53 reduces infectivity of WNV preparations that contain virions with both mature and immature patches on the same particle surface (Nelson et al, 2008). The structural data support the functional experiments that suggested that the neutralizing activity of E53 is associated with its binding to partially immature heterogeneous virions.
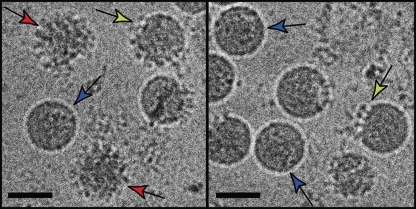
CryoEM images of two different WNV samples. Some immature (red arrows) and partially immature particles (green arrows) are frequently found in preparations of mature virus. Fully mature particles are marked with blue arrows. The same type of partially immature virus is usually also present in mature DENV samples. The scale bars represent 500 Å.
During cell entry, partially mature particles should encounter the low pH environment of the endosome, inducing a conformational rearrangement of the E protein in the immature portion of the virion, similar to the structural changes described for virus maturation during viral egress (Yu et al, 2008). Even though furin is present in the endosome and could cleave prM, dissociation of the pr peptide would be unlikely because of the mildly acidic environment (Li et al, 2008; Yu et al, 2008). Conversely, the mature part of the virus lacks bound pr and thus, in the low pH environment of the endosome, can change its conformation to form fusogenic trimeric spikes. Thus, fusion of the low-pH form of the partially mature particle with the endosomal membrane might occur despite the presence of pr/prM on parts of the virion. Presumably, as long as E molecules on the immature part of the virus have adopted a mature conformation at low pH, a cooperative formation of a fusion active state initiated from the mature portion of the particle should be possible. However, E53-decorated E molecules on the immature spikes of the partially mature virus would be inhibited from adopting the mature conformation and thus, interfere with the cooperative transition required to form fusion active trimers.
Highly cross-reactive fusion-loop antibodies, such as E53, bind to a conserved region of the flavivirus E protein and are generally weakly neutralizing. Because of their cross-reactivity, fusion-loop antibodies generated during a primary DENV infection should bind to partially or mostly immature viruses produced during a secondary infection. Therefore, these fusion-loop antibodies may promote ADE of infection by augmenting attachment and/or entry of partially immature virions. Indeed, it has been shown recently that fusion-loop-specific MAbs promote ADE in animals (Goncalvez et al, 2007).
Although E53 is capable of inhibiting viral attachment to some cells (Nybakken et al, 2005), our data strongly suggest that E53 also has the capacity to block the transition from an immature to a mature structure by steric hindrance. Many animal viruses undergo structural reorganization during both viral egress and entry (Steven et al, 2005). Novel information about these conformational transitions during the viral life cycle might be acquired by finding antibodies that block these processes and might, therefore, allow identification of new antiviral targets for drug development. Indeed, we have recently identified a new intermediate in the fusion process by using an anti-WNV antibody that blocks pH-dependent structural transitions (B Kaufmann et al, in preparation).
The structural and biochemical observations regarding the interaction of WNV or DENV with E53 MAb show that partially immature virions, which are the result of incomplete processing and/or subunit rearrangements, are infectious. Therefore, partially immature particles expand the pool of infectious virus and likely contribute to the pathogenesis of flavivirus infection. Exposing mature and immature surface features on the same particle may be important for evading recognition by some antibodies or other immune system recognition molecules (e.g. lectins). Moreover, because of the variable number of epitopes on mixed particles, it may be difficult for antibodies such as E53 to reach a stoichiometric threshold for neutralization, setting the stage for possible antibody enhancement of infection. Thus, antibodies, such as E53, that recognize epitopes on immature and partially immature virions may in some cases control infection yet in others contribute to enhanced infection and disease pathogenesis. This balance should be considered during development and evaluation of flavivirus vaccines.
Materials and methods
Protein production and purification
E53 Fab was generated and purified as described earlier (Oliphant et al, 2006).
WNV E (residues 1–415) was expressed in DE3 CodonPlus (RIL) bacteria as an insoluble aggregate. After isolation of the inclusion bodies, E was solubilized and then refolded oxidatively by dilution. E was then purified over a Superdex 75 (16/60) gel filtration column, pre-equilibrated in 20 mM HEPES (pH 7.5) containing 150 mM NaCl and 0.01% sodium azide. Antibody/antigen complexes were formed at room temperature by mixing E53 Fab with E and then purified by gel filtration chromatography. The purified complex was buffer exchanged into 25 mM HEPES (pH 7.4) and 0.01% sodium azide. Crystals were obtained using the hanging drop vapour diffusion technique. Aliquots of protein solution were mixed with an equal volume of reservoir solution (30% PEG 300, 0.1 M MES, pH 6.5) before equilibration against reservoir solution at 20°C. Crystals were rapidly cooled in liquid nitrogen for data collection.
Crystallographic structure determination
Data were collected at the Advanced Light Source beamline 4.2.2 (Lawrence Berkeley Laboratories, Berkeley, CA) by the oscillation method at a wavelength of 1.24 Å at 100 K and recorded with a CCD detector. Data were processed, scaled and merged with d*trek (Pflugrath, 1999). The crystals belong to space group P21212 with unit cell dimensions of a=146.41 Å, b=160.14 Å and c=43.88 Å, containing one E53 Fab–E complex per asymmetric unit. Initial phases were obtained by molecular replacement (Collaborative Computational Project, 1994) using the coordinates of WNV E (PDB accession code 2HG0) and an IgG1 Fab (PDB accession code 2IGF). An atomic model was iteratively built in O (Jones et al, 1991) and refined with CNS (Brunger et al, 1998) and Phenix (Adams et al, 2002) and contains residues 7–144, 163–297 of E, 1–208 of the E53 light chain, 1–127 and 136–211 of the E53 heavy chain, and no solvent molecules. The electron density for the Fab variable domains and DII of the E protein including the fusion and AB-loop epitope were of good quality, while significant regions of the Fab constant domains and E protein DI and DII regions distal from the combining site were of very poor quality. Electron density corresponding to DIII was highly ambiguous and no coordinates for this region have been incorporated. The final 3.0-Å resolution model was refined using six translation-libration-screw domains (two per chain) with the program Phenix (Adams et al, 2002) to an Rwork=24.7% and Rfree=34.4% for all F>0 (Supplementary Table I). Procheck analysis of the coordinates indicates that 98.9% of residues are located in favoured and additionally allowed regions of a Ramachandran plot.
WNV and DENV propagation and purification
Mature and immature WNV was produced and purified from Vero cells as described earlier (Kaufmann et al, 2006; Zhang et al, 2007). For immature DENV, C6/36 Aedes albopictus mosquito cells were grown in MEM supplemented with 10% fetal bovine serum (FBS) following standard cell culture procedures. Confluent cells were infected with DENV-2 at a multiplicity of infection of 0.2 in the presence of 5% FBS. Medium was replaced 24 h after infection with MEM containing 20 mM NH4Cl. Cell culture supernatant was harvested 3 days after infection and virus was purified as described for WNV.
Complex formation, cryoEM and 3D image reconstructions
Purified WNV particles were incubated with E53 Fab in the presence of 100 mM NaCl at 4°C overnight, using a ratio of about five Fab fragments per E protein. Purified immature DENV particles were incubated with Fab at 37°C for 30 min and then at 4°C for 2 h, using a ratio of about two Fab fragments per E protein. Micrographs of the frozen-hydrated sample were recorded on Kodak (Rochester, NY) SO-163 films with a CM300 FEG transmission electron microscope (Philips, Eindhoven, The Netherlands). Images were taken at a nominal magnification of × 47 000 and a total electron dose of 12–15 e−/Å2. The cryoEM micrographs were digitized on a Nikon 9000 scanner (Tokyo, Japan) with a 6.35-μm step size, and subsequently sets of four pixels were averaged to sample the specimen at 2.69 Å intervals. The program RobEM (Baker, 2004) was used to select a total of 4143 particles from 84 micrographs for the immature WNV–E53 Fab complex and a total of 2741 particles from 23 micrographs for the complex of immature DENV with E53 Fab. The defocus level was determined by fitting the theoretical microscope contrast transfer functions (CTFs) to the incoherent sum of the Fourier transforms of all particle images from each micrograph. The 3D reconstruction was computed using CTF phase-corrected images. The reconstruction was initiated by using a cryoEM density map of immature WNV as a model. The particle orientations were determined with SPIDER (Frank et al, 1996), and the 3D electron density map was calculated with a modified version of XMIPP (Sorzano et al, 2004) assuming icosahedral symmetry. Only 3927 and 2741 particles of the WNV and DENV complex, respectively, were selected to calculate the final 3D electron density maps. Selection was based on correlation with the model projections and stability of the particle centre position used. The resolution of the resultant map was estimated by comparing structure factors for the virus shell computed from two independent half-data sets. The estimated resolution was based on determining the spacing frequency at which the correlation between the two independent data sets became less than 0.5.
One measure of the map quality is the resolution of the lipid leaflets. The above procedure did not give a good representation of the lipid bilayer in the immature WNV–E53 Fab reconstruction (Supplementary Figure 1). Thus, as an alternative reconstruction technique, the Polar Fourier Transform (PFT) (Baker and Cheng, 1996) reciprocal space procedure was used for both WNV and DENV. This gave considerably better representations of the membrane region of these viruses, but the quality of the density representing the glycoprotein was reduced. This might suggest that the PFT method is the better procedure indicating that the interpretation of the Fab density in the XMIPP reconstruction could be inaccurate. However, the excellent agreement of the cryoEM density of the E53 Fab–virus complex with the crystal structure of the soluble E ectodomain in complex with E53 Fab showed that the reconstruction based on the modified XMIPP procedure was accurate. Thus, the lack of a clear separation of the lipid bilayers in the membrane of the reconstructions is not the result of disorder in the lipid due to the E53 binding, but a limitation of the reconstruction techniques.
Fitting of X-ray coordinates into the cryoEM electron density
The fitting of the E and E53 Fab atomic coordinates to the immature virusFab complexes followed earlier procedures (Kaufmann et al, 2006) using the program EMfit (Rossmann et al, 2001). First, the E glycoprotein (PDB codes 3C6D (Li et al, 2008) and 2OF6 (Zhang et al, 2007) of DENV and WNV, respectively) and pr (PDB code 3C5X (Li et al, 2008) for DENV and a model for the pr of WNV generated by SWISS-MODEL (Arnold et al, 2006)) were fitted independently into the cryoEM density (Supplementary Table III). Each E molecule was divided into two rigid bodies, DI+DIII (residues 1–51+133–196+281–400 for WNV and residues 1–49+132–192+280–395 for DENV) and DII+pr. The position and orientation of each rigid body was determined one at a time by making a complete 3D angular search, followed by rotational and translational refinement. The density corresponding to the best fit was set to zero before fitting the next rigid body. Domain DI+DIII was fitted by using a maximum distance restraint of 10 Å between the termini of strands connecting DI and DII. The results showed that the general arrangement of the E ectodomain and pr in the virus–E53 Fab complexes did not significantly change in comparison to the structure of the unliganded immature virus.
Similarly, the atomic coordinates of E53 Fab, extracted from the crystal structure of the E53 Fab–WNV E protein complex, were fitted into the cryoEM density of the complex. For independent fitting, the Fab was divided into two rigid bodies, the variable domain (residues 1–121 chain H and 1–107 chain L) and the constant domain (residues 122–221 chain H and 108–213 chain L). Although the variable domain fitted well into the density associated with molecule A (Figure 1A), the constant domain required some adjustment, accounting for a small change of the elbow angle from 144° to 153° compared with the crystal structure (Figure 4). Furthermore, a good fit of the E53 variable domain into the cryoEM density was obtained when DII and the Fab variable domain of the crystal structure were fitted as one rigid body, verifying the conservation of the mode of binding between the E protein and the variable domain of E53.
However, fitting of the E53 Fab variable domain into the difference density adjacent to the B molecule fusion loop showed that the E53 variable domain had rotated by about 35° relative to the E protein in comparison with the crystal structure. When the crystal structure of the E53 Fab–E complex was superpositioned onto the B molecule in the EM map there were clashes between the three-fold symmetry-related Fab molecules. Thus, the immature WNV structure, the complex of E53 Fab with the B molecule, is slightly different than that observed in the crystal structure and with molecule A. Moreover, to avoid steric hindrance, the elbow angle of the Fab associated with the B molecule changed from 144° to 171°.
A rough measure of the Fab occupancies was obtained by comparing the average height of the densities (‘sumf' (Rossmann et al, 2001)) between the DII+pr and the Fab variable domain. The height of density associated with the Fab molecules was lower compared with the density of DII of E (73 and 57% of the variable domain density for molecules A and B, respectively). This is most likely due to incomplete occupancy of the binding sites by the Fab molecules. The Fab bound to the B molecule has a lower occupancy compared with molecule A, probably as a result of steric hindrance. The height of the density for the more distant Fab constant domains is even further diminished (61 and 83% of the variable domain density in the A and B molecules, respectively). This suggests that either the elbow angle in the Fab molecule is somewhat flexible.
The elbow angle between Fab variable and constant domains was calculated using the program Elbow Angle Calculation (http://proteinmodel.org/AS2TS/RBOW/index.html).
Data deposition
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession code 3I50 (rcsb053971). The cryoEM density maps were deposited in the EM databank under accession number EMD-5103 (immature WNV–E53 Fab) and EMD-5102 (immature DENV–E53 Fab). The fitted complex structures were deposited in the Protein Data Bank under accession codes 3IXX (for WNV–E53 Fab) and 3IXY (for DENV–E53 Fab).
Figure preparation
Figures were created using the programs PYMOL (DeLano, 2002) and CHIMERA (Pettersen et al, 2004).
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We are grateful to Sheryl Kelly and Carol J Greski for help in the preparation of the paper. The work was supported by the NIH grants RO1-AI76331 (MGR), R01-AI073755 (MSD), U01-AI061373 (MSD), and PDVI grants TR-17 (RJK and MGR) and DR-5 (DHF). We are grateful to the Keck Foundation grant for the purchase of the CM300 field emission gun electron microscope used in this study. MVC processed and analysed cryoEM data, fitted X-ray structures in cryoEM maps and wrote the paper. BK prepared Fab complexes of mature and immature WNV. SL prepared Fab complex with immature DENV. GEN, BRC, CAN, JTW and DHF produced the WNV E–Fab complex crystals and performed the crystallographic analysis. VAK provided computational support for cryoEM reconstruction. HAH and PRC collected cryoEM data. RJK was involved in study design. MSD produced E53 Fab for cryoEM. MGR was involved in study design, data analysis and paper writing. MVC, BK, SL, RJK, MSD, MGR and DHF discussed and analysed the results and edited the paper.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [Abstract] [Google Scholar]
- Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX (2001) Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol 75: 4268–4275 [Europe PMC free article] [Abstract] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [Abstract] [Google Scholar]
- Baker TS (2004) RobEM imaging and visualization program (http://cryoem.ucsd.edu/program Docs/runRobem.txt )
- Baker TS, Cheng RH (1996) A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol 116: 120–130 [Abstract] [Google Scholar]
- Brandt W (1982) Infection enhancement of dengue type 2 virus in the U-937 human monocyte cell line by antibodies to flavivirus cross-reactive determinants. Infect Immun 36: 1036–1041 [Europe PMC free article] [Abstract] [Google Scholar]
- Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA (2004) Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J 23: 728–738 [Europe PMC free article] [Abstract] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [Abstract] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [Abstract] [Google Scholar]
- Crill WD, Chang GJ (2004) Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J Virol 78: 13975–13986 [Europe PMC free article] [Abstract] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific (http://wwwpymolorg) [Google Scholar]
- Elshuber S, Allison SL, Heinz FX, Mandl CW (2003) Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol 84: 183–191 [Abstract] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [Abstract] [Google Scholar]
- Goncalvez A, Engle R, St Claire M, Purcell R, Lai C (2007) Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc Natl Acad Sci USA 104: 9422–9427 [Europe PMC free article] [Abstract] [Google Scholar]
- Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C (1991) Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol 72: 1323–1329 [Abstract] [Google Scholar]
- Hubbard SJ, Thornton JM (1993) NACCESS. Computer Program, Department of Biochemistry and Molecular Biology, University College London (http://www.bioinf.manchester.ac.uk/naccess/) [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [Abstract] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG (2006) West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc Natl Acad Sci USA 103: 12400–12404 [Europe PMC free article] [Abstract] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH (2002) Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–725 [Europe PMC free article] [Abstract] [Google Scholar]
- Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG (2008) The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319: 1830–1834 [Abstract] [Google Scholar]
- Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG (2008) Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol 15: 312–317 [Abstract] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC (2003) A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA 100: 6986–6991 [Europe PMC free article] [Abstract] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC (2004) Structure of the dengue virus envelope protein after membrane fusion. Nature 427: 313–319 [Abstract] [Google Scholar]
- Nelson S, Jost CA, Xu Q, Ess J, Martin JE, Oliphant T, Whitehead SS, Durbin AP, Graham BS, Diamond MS, Pierson TC (2008) Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog 4: e1000060. [Europe PMC free article] [Abstract] [Google Scholar]
- Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH (2006) Crystal structure of the West Nile virus envelope glycoprotein. J Virol 80: 11467–11474 [Europe PMC free article] [Abstract] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH (2005) Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437: 764–769 [Europe PMC free article] [Abstract] [Google Scholar]
- Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS (2007) Induction of epitope-specific neutralizing antibodies against West Nile virus. J Virol 81: 11828–11839 [Europe PMC free article] [Abstract] [Google Scholar]
- Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS (2006) Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol 80: 12149–12159 [Europe PMC free article] [Abstract] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [Abstract] [Google Scholar]
- Pflugrath JW (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr 55: 1718–1725 [Abstract] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375: 291–298 [Abstract] [Google Scholar]
- Roberson JA, Crill WD, Chang GJ (2007) Differentiation of West Nile and St Louis encephalitis virus infections by use of noninfectious virus-like particles with reduced cross-reactivity. J Clin Microbiol 45: 3167–3174 [Europe PMC free article] [Abstract] [Google Scholar]
- Rossmann MG, Bernal R, Pletnev SV (2001) Combining electron microscopic with X-ray crystallographic structures. J Struct Biol 136: 190–200 [Abstract] [Google Scholar]
- Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A (2004) XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol 148: 194–204 [Abstract] [Google Scholar]
- Stadler K, Allison SL, Schalich J, Heinz FX (1997) Proteolytic activation of tick-borne encephalitis virus by furin. J Virol 71: 8475–8481 [Europe PMC free article] [Abstract] [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF (2005) Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol 15: 227–236 [Europe PMC free article] [Abstract] [Google Scholar]
- Stiasny K, Kiermayr S, Holzmann H, Heinz FX (2006) Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J Virol 80: 9557–9568 [Europe PMC free article] [Abstract] [Google Scholar]
- Sundberg EJ, Mariuzza RA (2002) Molecular recognition in antibody-antigen complexes. Adv Protein Chem 61: 119–160 [Abstract] [Google Scholar]
- Wengler G, Wengler G (1989) Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J Virol 63: 2521–2526 [Europe PMC free article] [Abstract] [Google Scholar]
- Widman DG, Frolov I, Mason PW (2008) Third-generation flavivirus vaccines based on single-cycle, encapsidation-defective viruses. Adv Virus Res 72: 77–126 [Abstract] [Google Scholar]
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J (2008) Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319: 1834–1837 [Abstract] [Google Scholar]
- Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG (2003) Structures of immature flavivirus particles. EMBO J 22: 2604–2613 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG (2007) Structure of immature West Nile virus. J Virol 81: 6141–6145 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG (2004) Conformational changes of the flavivirus E glycoprotein. Structure 12: 1607–1618 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/emboj.2009.245
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2771083?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/134196874
Article citations
Predicting immune response targets in orthoflaviviruses through sequence homology and computational analysis.
J Mol Model, 30(8):295, 31 Jul 2024
Cited by: 0 articles | PMID: 39083139
Epitope(s) involving amino acids of the fusion loop of Japanese encephalitis virus envelope protein is(are) important to elicit protective immunity.
J Virol, 98(4):e0177323, 26 Mar 2024
Cited by: 0 articles | PMID: 38530012 | PMCID: PMC11019926
Prior flavivirus immunity skews the yellow fever vaccine response to cross-reactive antibodies with potential to enhance dengue virus infection.
Nat Commun, 15(1):1696, 24 Feb 2024
Cited by: 0 articles | PMID: 38402207 | PMCID: PMC10894228
Single B cell transcriptomics identifies multiple isotypes of broadly neutralizing antibodies against flaviviruses.
PLoS Pathog, 19(10):e1011722, 09 Oct 2023
Cited by: 5 articles | PMID: 37812640 | PMCID: PMC10586629
A Prototype-Pathogen Approach for the Development of Flavivirus Countermeasures.
J Infect Dis, 228(suppl 6):S398-S413, 01 Oct 2023
Cited by: 2 articles | PMID: 37849402 | PMCID: PMC10582523
Review Free full text in Europe PMC
Go to all (143) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Electron Microscopy Data Bank (2)
- (1 citation) Electron Microscopy Data Bank - EMD-5102
- (1 citation) Electron Microscopy Data Bank - EMD-5103
Protein structures in PDBe (Showing 8 of 8)
-
(1 citation)
PDBe - 2IGFView structure
-
(1 citation)
PDBe - 3C6DView structure
-
(1 citation)
PDBe - 2HG0View structure
-
(1 citation)
PDBe - 3I50View structure
-
(1 citation)
PDBe - 2OF6View structure
-
(1 citation)
PDBe - 3C5XView structure
-
(1 citation)
PDBe - 3IXXView structure
-
(1 citation)
PDBe - 3IXYView structure
Show less
Electron Microscopy Data Bank (EMDB) at PDBe (2)
Protocols & materials
Related Immune Epitope Information - Immune Epitope Database and Analysis Resource
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A fusion-loop antibody enhances the infectious properties of immature flavivirus particles.
J Virol, 85(22):11800-11808, 31 Aug 2011
Cited by: 42 articles | PMID: 21880758 | PMCID: PMC3209313
Structural basis of West Nile virus neutralization by a therapeutic antibody.
Nature, 437(7059):764-769, 01 Sep 2005
Cited by: 259 articles | PMID: 16193056 | PMCID: PMC7095628
Mechanism for maturation-related reorganization of flavivirus glycoproteins.
J Struct Biol, 185(1):27-31, 16 Nov 2013
Cited by: 16 articles | PMID: 24252771 | PMCID: PMC3901303
Flaviviruses and their antigenic structure.
J Clin Virol, 55(4):289-295, 21 Sep 2012
Cited by: 80 articles | PMID: 22999801
Review
Funding
Funders who supported this work.
NIAID NIH HHS (6)
Grant ID: R01-AI073755
Grant ID: U01 AI061373
Grant ID: R01 AI076331
Grant ID: R01-AI76331
Grant ID: U01-AI061373
Grant ID: R01 AI073755




