Abstract
Free full text

Notch1 Signaling Promotes the Maturation of CD4 and CD8 SP Thymocytes
Summary
Notch proteins regulate many developmental processes. Notch1 is highly expressed on thymocytes, but its role in regulating their development is not known. We show that activation of Notch1 signaling in CD4+CD8+ double positive thymocytes promotes the maturation of both CD4+ and CD8+ single positive thymocytes and that this occurs in the absence of interactions between the T cell receptor and MHC molecules expressed on thymic epithelial cells. We have also identified several genes that are transcriptionally regulated by Notch1 in T cells and show that they are upregulated during maturation into both single positive lineages. These observations suggest that Notch1 signaling plays a role in promoting maturation into both the CD4 and CD8 T cell lineages.
Introduction
The development of mature T cells from lymphoid progenitor cells involves a series of cell fate choices and differentiation steps that direct cells along one of several distinct developmental pathways. Of particular interest is the stage during which CD4+CD8+ double positive (DP) thymocytes differentiate into either CD4+ or CD8+ single positive (SP) thymocytes. This step serves the important function of allowing only those DP thymocytes that express T cell receptors (TCRs) capable of interacting with self-MHC ligands to fully mature and become part of the peripheral T cell pool. Signals transduced through the TCR/CD3 complex during interactions with MHC ligands expressed on thymic epithelial cells rescue DP thymocytes from programmed cell death and induce them to differentiate into mature SP thymocytes. These signals also influence whether DP thymocytes differentiate into the CD4 or CD8 SP lineages. The CD8 coreceptor and TCR bind cooperatively to MHC class I ligands and promote maturation into the CD8 SP lineage, whereas the CD4 coreceptor and TCR bind cooperatively to MHC class II ligands and promote maturation into the CD4 SP lineage. Although considerable progress has been made in defining membrane-proximal events and signal transduction pathways downstream of the TCR and coreceptors involved in this step of development, little is known about how these pathways and others function to regulate T cell maturation at the transcriptional level.
Recent interest has focused on the role of Notch proteins during T cell development (Osborne and Miele, 1999; Deftos and Bevan, 2000). Notch proteins are a family of large transmembrane receptors that play an important role in regulating cell fate decisions during many developmental processes. Signals transduced through Notch receptors during cell–cell interactions directly regulate the expression of genes that play important roles in cell fate choices and differentiation (Kimble and Simpson, 1997). Notch1, a mammalian homolog of Drosophila Notch, was first identified as a gene involved in chromosomal translocations with the TCR β gene in a subset of human T cell leukemias (Ellisen et al., 1991). These translocations result in the expression of truncated Notch1 polypeptides that lack most of the extra-cellular domain and constitutively activate the Notch signaling pathway. Expression of constitutively active Notch1 in bone marrow stem cells causes T cell leukemia, indicating a causative role for Notch1 in T cell oncogenesis (Pear et al., 1996). The involvement of Notch1 in T cell leukemia and its high expression on developing thymocytes suggested that Notch signaling may play a role in regulating normal T cell development (Ellisen et al., 1991; Hasserjian et al., 1996). This is further supported by the expression of Jagged2, a ligand for Notch1, on both thymocytes and thymic epithelial cells (Luo et al., 1997; Felli et al., 1999).
A role for Notch signaling in promoting the commitment of lymphoid progenitor cells to the T cell lineage has recently been proposed. Inducible deletion of Notch1 in bone marrow stem cells results in a severe block in the development of the most immature CD4−CD8− (DN) thymocytes, while having no apparent effect on the development of other hematopoietic cell lineages (Radtke et al., 1999). Conversely, expression of constitutively active Notch1 in bone marrow stem cells results in the development of a thymus-independent population of cells in the bone marrow that express markers of T cell lineage commitment (Pui et al., 1999). These results imply that Notch1 signaling plays a critical role in the commitment of lymphoid progenitor cells to the T cell lineage. Since Notch1 is expressed on thymocytes throughout their maturation (Hasserjian et al., 1996), it is likely that Notch signaling plays a role in subsequent steps of development. Indeed, expression of constitutively active Notch1 in thymocytes was reported to result in the development of excess CD8+ SP thymocytes at the expense of CD4+ SP thymocytes, leading to the hypothesis that Notch1 signaling regulates the CD4 versus CD8 cell fate choice (Robey et al., 1996). An analogous role for Notch1 in regulating the αβ versus γδ cell fate choice has also been suggested (Washburn et al., 1997).
Recent observations suggest that Notch1 may play a more general role in promoting the maturation of DP thymocytes into both the CD4+ and CD8+ SP lineages. The constitutively active intracellular domain of Notch1 was repeatedly isolated in a screen to identify cDNAs capable of conferring glucocorticoid resistance to a glucocorticoid-sensitive DP thymoma cell line (Deftos et al., 1998). Since glucocorticoid resistance is a marker of maturation into both the CD4 and CD8 SP lineages, this suggested a role for Notch in promoting maturation into both SP lineages. Indeed, expression of constitutively active Notch1 in DP thymoma cell lines also conferred additional phenotypes that correlate with maturation into both SP lineages. Moreover, the expression of Deltex, a gene that is transcriptionally regulated by Notch signaling in T cells, is upregulated during the maturation of both CD4+ and CD8+ SP thymocytes, suggesting that Notch signaling is activated during the maturation of both SP lineages.
To further characterize the role of Notch signaling in T cell development, we generated transgenic mice that express the constitutively active intracellular domain of Notch1 under the control of the Lck-proximal promoter. Analysis of thymocyte development in these mice supports the hypothesis that Notch1 signaling promotes the maturation of DP thymocytes into both the CD4 and CD8 lineages. The expression pattern of a number of genes regulated by Notch1 signaling lends further support to this interpretation.
Results
Notch Signaling Promotes the Maturation of Both CD4 and CD8 SP Thymocytes
We previously showed that expression of the constitutively active intracellular domain of Notch1 (NotchIC) in DP thymoma cell lines is sufficient to induce several phenotypic changes that correlate with maturation into both the CD4 and CD8 SP lineages (Deftos et al., 1998). This suggested that Notch signaling may play a role in regulating the DP to SP transition during normal T cell development. To test this hypothesis, we generated transgenic mice expressing NotchIC under the control of the Lck proximal promoter and analyzed the effect of this transgene on thymocyte development. RT-PCR analysis indicated these mice express NotchIC at high levels in the thymus and low levels in peripheral lymph nodes (Figures 1A and 1B), consistent with the activity of the Lck-proximal promoter, which is high in immature thymocytes and low in mature T cells (Allen et al., 1992). Western blot analysis confirmed expression of NotchIC in thymocytes and revealed an increase in the level of endogenous Notch1 expression (Figure 1C), confirming the previously described ability of NotchIC to upregulate endogenous Notch1 expression in T cells (Deftos et al., 1998). These results imply that NotchIC is expressed at high levels in immature thymocytes and at low levels in mature T cells in this line of transgenic mice.
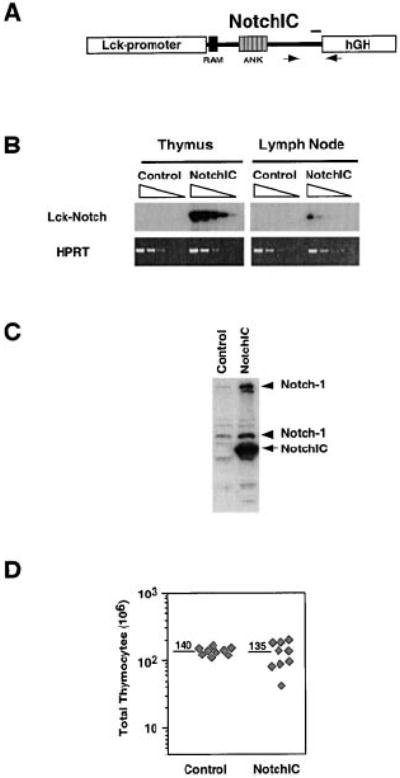
(A) Schematic representation of the NotchIC transgene. NotchIC corresponds to amino acids 1751–2444 of murine Notch1. Arrows correspond to the locations of primers used for RT-PCR. The bar corresponds to the region of Notch1 recognized by the antiserum used for Western blot analysis.
(B) RT-PCR analysis of NotchIC transgene expression in thymocytes and peripheral lymphocytes. cDNA prepared from thymocytes and lymph node cells from NotchIC and littermate control mice was serially diluted 5-fold and amplified by PCR using transgene specific primers. The PCR products were analyzed by Southern blot using a Notch1 cDNA probe.
(C) Western blot analysis of transgene expression and endogenous Notch1 expression in thymocytes. Thymocyte lysates from NotchIC or littermate control mice were analyzed by Western blot using an antiserum specific for amino acids 2426–2445 of Notch1. The arrow indicates the NotchIC transgene, and the arrowheads indicate the full-length and proteolytically processed forms of endogenous Notch1.
(D) Total thymocyte numbers from NotchIC and littermate control mice. Total thymocyte cell numbers were determined for NotchIC and control mice from three representative litters between 5 and 12 weeks of age. Diamonds represent cell numbers from individual mice, and the numbers above the bars represent the average of these values.
The NotchIC mice had normal thymocyte cellularity, although there was higher variability in thymocyte cell numbers compared to littermate controls (Figure 1D). FACS analysis of CD4 and CD8 expression revealed a large decrease in the percentage of DP thymocytes and a corresponding increase in the percentage of CD8+ SP thymocytes (Figure 2A). The magnitude of this effect was variable, with the percentage of DP thymocytes ranging from 75% to 24% (compared to 80% ± 5% for control mice) and the percentage of CD8+ SP thymocytes ranging from 8% to 44% (compared to 3% ± 1% for control mice) (Figure 2A and data not shown). In mice that had a “weak” phenotype (a small change in the percentage of DP and CD8+ SP thymocytes), we sometimes observed a small reduction in the percentage of CD4+ SP thymocytes (Figure 2A, center panel). However, in mice that had a “strong” phenotype (a larger change in the percentage of DP and CD8+ SP thymocytes), there was an increase in the percentage of CD4+ SP thymocytes (Figure 2A, right panel).
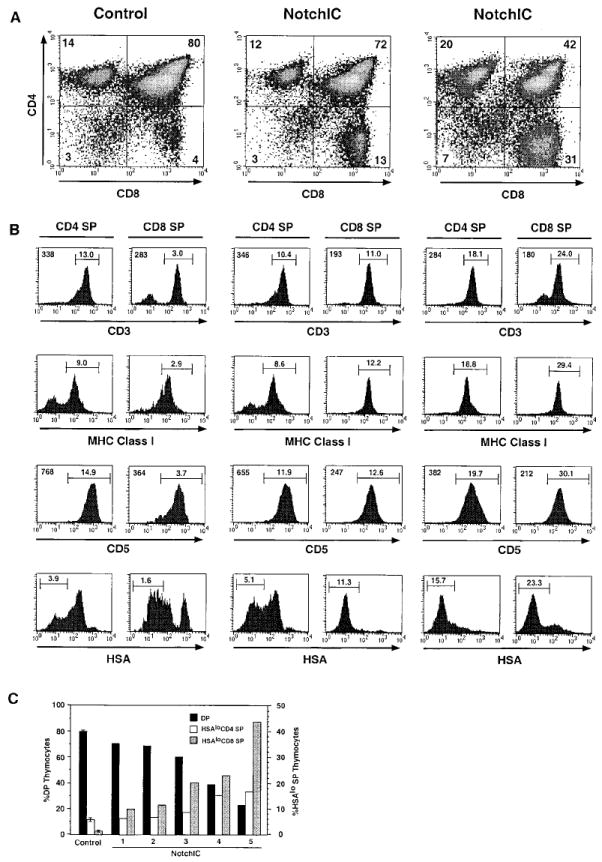
(A) FACS analysis of thymocytes from NotchIC and control mice. Thymocytes from two NotchIC mice and one littermate control were analyzed for CD4 and CD8 expression. Numbers indicate the percent of total thymocytes that fall within the indicated quadrants.
(B) FACS analysis of maturation markers on CD4+ and CD8+ SP thymocytes from NotchIC and littermate control mice. CD4+ SP and CD8+ SP thymocytes from the mice shown in (A) were analyzed for expression of CD3, MHC class I, CD5, and HSA. Numbers above the bars represent the percent of total thymocytes that fall within the indicated gate. Numbers in the upper left corners of the histograms represent the mean florescence intensity of staining for the cells in this same gate.
(C) Percent of DP, HSAlo CD4+ SP, and HSAlo CD8+ SP thymocytes in representative NotchIC mice. NotchIC mice and control mice from several litters were analyzed by FACS for CD4, CD8, and HSA expression. CD4+ SP and CD8+ SP thymocytes were electronically gated and analyzed for HSA expression as in (B). The percent of CD4+CD8+ (DP), HSAlo CD4+ SP, and HSAlo CD8+ SP thymocytes for individual NotchIC mice are graphed separately. The average value for five littermate control mice is shown for reference. The axis on the left gives the percent of total thymocytes that are DP, and the axis on the right gives the percent of total thymocytes that are HSAlo CD4+ SP or HSAlo CD8+ SP.
To determine whether the change in the percentage of CD4+ and CD8+ SP thymocytes is due to an effect on the number of phenotypically mature SP thymocytes, we examined the expression of additional cell surface markers that are developmentally regulated during the DP to SP transition. DP thymocytes express low levels of CD3 and MHC class I, intermediate levels of CD5, and high levels of HSA. Following maturation into either the CD4 or CD8 lineage, they upregulate expression of CD3, MHC class I, and CD5 and downregulate expression of HSA. FACS analysis of CD8+ SP thymocytes from NotchIC mice revealed that they expressed high levels CD3, MHC class I, and CD5, although the levels of CD3 and CD5 were lower than the levels expressed on CD8+ SP thymocytes from control mice (Figure 2B). Interestingly, CD8+ SP thymocytes from NotchIC mice expressed lower levels of HSA than the majority of CD8+ SP thymocytes from control mice (Figure 2B). Downregulation of HSA expression normally occurs as a late event during SP thymocyte maturation, and low levels of HSA expression correlate with increasing maturation. Together, these results suggest that the NotchIC transgene promotes the development of an increased number of CD8+ SP thymocytes that are phenotypically similar to mature CD8+ SP thymocytes.
CD4+ SP thymocytes from NotchIC mice also expressed high levels of CD3, MHC class I, and CD5, although the levels of CD3 and CD5 were lower than the levels expressed on CD4+ SP thymocytes from control mice (Figure 2B). Analysis of HSA expression revealed that the reduction in CD4+ SP thymocyte numbers observed in some NotchIC mice occurs primarily within a subset of cells that express intermediate levels of HSA, whereas there was either no change or an increase in the percentage of HSAlo CD4+ SP thymocytes (Figure 2B). Moreover, in NotchIC mice with a “strong” phenotype, the majority of CD4+ SP thymocytes expressed HSA at lower levels than the majority of CD4+ SP thymocytes from control mice (Figure 2B). Analysis of a large number of NotchIC mice revealed a correlation between the number of DP, HSAlo CD4+ SP, and HSAlo CD8+ SP thymocytes (Figure 2C). At one end of the spectrum were mice with a relatively small decrease in the percentage of DP thymocytes, a corresponding increase in the percentage of HSAlo CD8+ SP thymocytes, and no change or a small increase in the percentage HSAlo CD4+ SP thymocytes. At the other end of the spectrum were mice with a larger decrease in the percentage of DP thymocytes and a substantial increase in the percentage of both HSAlo CD4+ SP and HSAlo CD8+ SP thymocytes. In all cases, there were more HSAlo CD8+ SP thymocytes than HSAlo CD4+ SP thymocytes. Together, these results indicate that expression of NotchIC in immature thymocytes promotes the development of phenotypically mature CD4+ SP and CD8+ SP thymocytes, with the promotion of CD8+ SP development occurring more efficiently than the promotion of CD4+ SP development.
The increase in the number of phenotypically mature CD4+ and CD8+ SP thymocytes in NotchIC mice raised the question of whether these cells have completed the maturation process and are exported to the periphery. FACS analysis of T cells from the spleen and peripheral lymph nodes revealed a reduction in the number of both CD4+ and CD8+ T cells, with no significant alteration in the CD4/CD8 ratio, suggesting that the excess SP thymocytes that develop in these mice may die before reaching the periphery (Figure 3A and data not shown). Consistent with this, we observed a significant increase in the number of Annexin V+ thymocytes in NotchIC mice, which were predominantly DP and CD8 SP thymocytes (Figure 3B). This analysis likely underestimates the degree of apoptosis, since apoptotic cells are rapidly engulfed by phagocytic cells in the thymus (Surh and Sprent, 1994). While the NotchIC mice have an increase in the percentage of Annexin V− CD4 and CD8 SP thymocytes (data not shown), the reduction in mature T cells in the periphery suggests that many of these do not survive to populate peripheral lymphoid tissues.
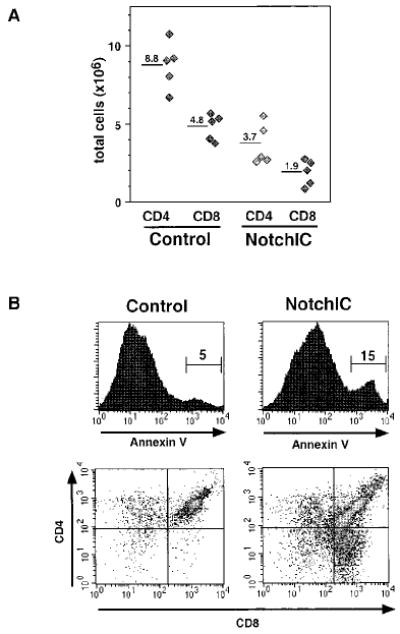
(A) Total numbers of CD4+ and CD8+ T cells in the peripheral lymph nodes of NotchIC and littermate control mice. Lymph nodes were collected from NotchIC and control mice from two representative litters. Total CD4+ and CD8+ T cell numbers were determined by multiplying total cell numbers by the percent CD4+ SP and CD8+ SP cells determined by FACS. Diamonds represent cell numbers from individual mice, and the numbers above the bars represent the average of these values.
(B) FACS analysis of Annexin V+ thymocytes in NotchIC and littermate control mice. Thymocytes were analyzed for CD4 and CD8 expression and Annexin V binding. The upper panels show Annexin V staining for total thymocytes. The numbers represent the percent of total thymocytes that fall within the indicated region. Cells in this region were gated and analyzed for CD4 and CD8 expression in the lower panels.
Notch1 Signaling Promotes CD4+ and CD8+ SP Thymocyte Maturation in the Absence of MHC Expression on Thymic Epithelial Cells
The development of mature CD4+ and CD8+ SP thymocytes is dependent upon interactions between the TCR on DP thymocytes and MHC molecules on thymic epithelial cells (Goldrath and Bevan, 1999). The ability of constitutively active Notch1 to promote the development of both CD4+ and CD8+ SP thymocytes raised the question of whether this is similarly dependent upon TCR–MHC interactions. To determine this, we transferred bone marrow cells from NotchIC or littermate control mice into irradiated MHC-deficient hosts and analyzed T cell development in these chimeric mice. As expected, MHC-deficient hosts reconstituted with bone marrow from control mice did not develop a significant number of mature CD4+ or CD8+ SP thymocytes (Figures 4A and 4B). In contrast, a significant but highly variable number of CD4+ and CD8+ SP thymocytes developed in MHC-deficient hosts reconstituted with bone marrow from NotchIC mice (Figure 4A). These SP thymocytes expressed low levels of HSA and high levels of MHC class I, consistent with their having undergone several phenotypic changes that occur during normal maturation (Figure 4B). Analysis of a number of NotchIC→MHC−/− chimeric mice revealed a direct correlation between the number of HSAlo CD4+ and HSAlo CD8+ SP thymocytes that developed, with the number of CD8+ SPs being greater than the number of CD4+ SPs (Figure 4C). As is the case for SP thymocytes in the parental NotchIC mice (Figure 2B), the SP thymocytes that develop in MHC-deficient hosts express lower levels of CD3 and CD5 than SP thymocytes from control mice (Figure 4D and data not shown). Moreover, many of these cells are Annexin V+, and they do not accumulate in peripheral lymphoid tissues (data not shown). Together, these results imply that ectopic activation of Notch1 signaling in DP thymocytes can bypass the normal requirement for TCR and coreceptor interactions with MHC ligands on thymic epithelial cells for development into SP thymocytes.
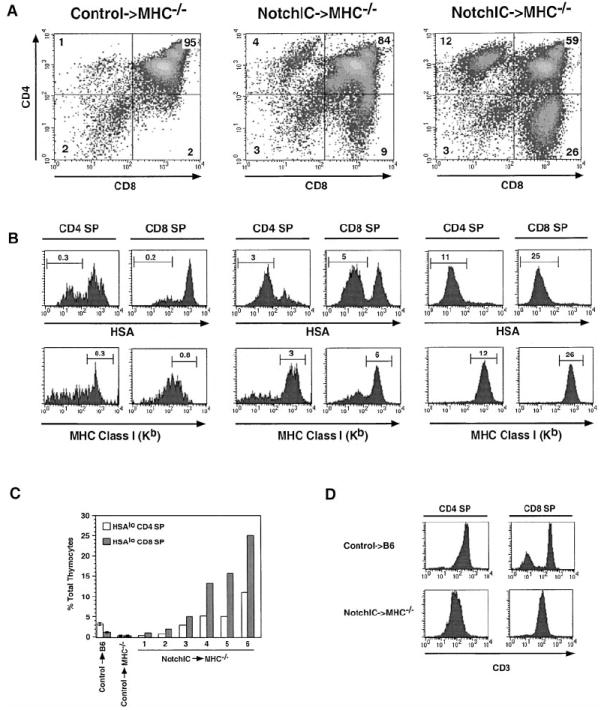
(A) FACS analysis of CD4 and CD8 expression on thymocytes from one littermate control→MHC−/− and two NotchIC→MHC−/− bone marrow chimeric mice. Numbers indicate the percent of total thymocytes that fall within the indicated quadrants.
(B) FACS analysis of HSA and MHC class I expression on CD4+ and CD8+ SP thymocytes from control→MHC−/− and NotchIC→MHC−/− bone marrow chimeric mice. CD4+ SP and CD8+ SP thymocytes from the mice shown in (A) were analyzed by FACS for expression of HSA and MHC class I. Numbers above the bars represent the percent of total thymocytes that fall within the indicated gate.
(C) Percent of HSAlo CD4+ SP and HSAlo CD8+ SP thymocytes in NotchIC→MHC−/− chimeric mice. CD4+ SP and CD8+ SP thymocytes from six NotchIC→MHC−/− chimeric mice, two littermate control→MHC−/−, and two littermate control→B6 chimeric mice were analyzed by FACS for HSA expression as in (B). The percent of HSAlo CD4+ SP and HSAlo CD8+ SP thymocytes for the NotchIC→MHC−/− chimeras mice are graphed individually. The average values for the littermate control→MHC−/− and littermate control→B6 chimeric mice are shown for reference.
(D) FACS analysis of CD3 expression on CD4+ and CD8+ SP thymocytes from NotchIC→MHC−/− and littermate control→B6 bone marrow chimeric mice.
Notch1 Signaling Induces the Expression of CD25 and CD44 on DP Thymocytes
We observed that a large proportion of DP thymocytes from NotchIC mice express high levels of CD25 (Figure 5A). Additional analysis revealed that CD25+ DP thymocytes from NotchIC mice express higher levels of CD3 and CD5 than CD25− DP thymocytes from these mice or littermate control mice (Figure 5B). A large fraction of these CD25+ DP thymocytes also expressed high levels of CD44 (Figure 5B). There was no consistent increase in CD25 expression in DN or SP thymocytes (data not shown). The increased expression of CD25 and CD44 on DP thymocytes raised the question of whether this is a direct effect of NotchIC expression or is due to the expansion of a small subset of CD25+ DP thymocytes found in normal mice. To address this, we expressed NotchIC in the AKR1 DP thymoma cell line (Hyman et al., 1980) and analyzed its effect on CD25 and CD44 expression. Interestingly, a large proportion of cells expressing NotchIC expressed high levels of CD25 and CD44 (Figure 5C). Together, these data indicate that activation of Notch1 signaling in DP thymocytes and thymoma cell lines results in the upregulation of CD25 and CD44 expression.
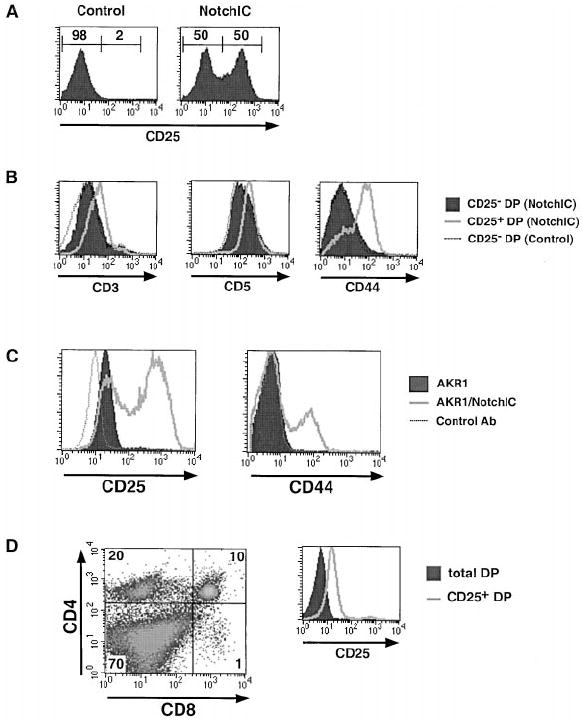
(A) FACS analysis of CD25 expression on DP thymocytes from NotchIC and littermate control mice. Numbers above the bars represent the percent of DP thymocytes in indicated region.
(B) FACS analysis of CD3, CD5, and CD44 expression on CD25+ DP thymocytes from NotchIC mice.
(C) FACS analysis of CD25 expression on AKR1 cells expressing NotchIC. The AKR1 DP thymoma cell line and a polyclonal population of AKR1 cells expressing NotchIC were analyzed for CD25 and CD44 expression.
(D) FACS analysis of CD25+ DP thymocytes from B6 mice. CD25+ thymocytes were enriched by panning on antibody-coated plates and analyzed for expression of CD4, CD8, and CD25. The expression of CD25 on panned and unpanned DP thymocytes is shown in the histogram.
Although we could detect a small number of CD25+ DP thymocytes in normal mice, it was not clear whether these represented a true subset of DPs. To determine this, we enriched for CD25+ thymocytes by panning on antibody-coated plates and analyzed these enriched cells by FACS (Figure 5D). As expected, CD25+ thymocytes from normal mice included a large number of DN thymocytes, a significant number of CD4+ SP thymocytes, and very few CD8+ SP thymocytes (Itoh et al., 1999). A small number of CD25+ DP thymocytes was also observed. However, in contrast to the CD25+ DP thymocytes from NotchIC mice, CD25+ DP thymocytes from normal mice did not express detectable levels of CD44 and did not express higher levels of CD3 and CD5 than CD25− DP thymocytes, indicating that CD25+ DP thymocytes in normal mice are distinct from CD25+ DP thymocytes from NotchIC mice. Moreover, CD25+ DP thymoctyes from normal mice do not express elevated levels of the Notch1 target gene Deltex (data not shown). This suggests that CD25 expression on a subset of DP thymocytes in normal mice is not due to Notch signaling.
Identification of Notch Target Genes and Characterization of Their Pattern of Expression
We previously identified Deltex as a gene that is transcriptionally regulated by Notch1 in a DP thymoma cell line. Analysis of the expression of Deltex in thymocyte subsets revealed low expression in DP thymocytes and high expression in CD4+ and CD8+ SP thymocytes, suggesting that Notch signaling is activated during the DP to SP transition (Deftos et al., 1998). We used the cDNA–RDA technique to identify additional genes regulated by Notch signaling in T cells and characterized their pattern of expression in developing thymocytes. This led to the identification of Meltrin β, three members of the Ifi-200 gene family (Ifi-202, Ifi-204, and Ifi-D3), and Pre-Tα. Meltrin β is an ADAM family metalloprotease (Inoue et al., 1998), Ifi-200 proteins are a family of proteins implicated in transcriptional regulation and cell cycle control (Johnstone and Trapani, 1999), and Pre-Tα is a component of the Pre-TCR complex that plays a critical role during early thymocyte development (Saint-Ruf et al., 1994). To confirm that these genes are regulated by Notch1 signaling in T cells, we examined their expression in three different T cell lines following infection with retrovirus that directs expression of NotchIC (Figure 6A). All of these genes are upregulated by NotchIC in the AKR1010 cell line. Deltex, Meltrin β, Ifi-202, and Ifi-204 were also upregulated by NotchIC in the 2B4.11 T cell hybridoma, whereas Ifi-D3 was expressed constitutively at relatively high levels and slightly upregulated by NotchIC, and Pre-Tα was not expressed. Deltex, Meltrin β, Pre-Tα, Ifi-202, and Ifi-204 were upregulated by NotchIC expression in the AKR1 DP thymoma cell line, whereas Ifi-D3 was not expressed. In addition, HES1, a gene that is regulated by Notch signaling in other cell types (Hsieh et al., 1997; Jarriault et al., 1998), was upregulated by NotchIC in all three cell lines (Figure 6A). To confirm that these genes are regulated by Notch signaling in thymocytes, we characterized their expression in DP thymocytes from NotchIC and control mice (Figure 6B). This revealed that Deltex, Meltrin β, Ifi-204, and HES1 are expressed at low levels in DP thymocytes from normal mice and were strongly upregulated in DP thymocytes from NotchIC mice. Pre-Tα was expressed at relatively high levels in DP thymocytes from normal mice and was upregulated approximately 5-fold in DP thymocytes from NotchIC mice. Ifi-D3 was expressed at relatively high levels in DP thymocytes and was not upregulated by NotchIC, whereas Ifi-202 was not detected in thymocytes from either control or NotchIC mice (Figure 6B and data not shown). Collectively, these results indicate that Deltex, Meltrin β, HES1, Ifi-204, and Pre-Tα are transcriptionally regulated by Notch1 signaling in thymocytes.
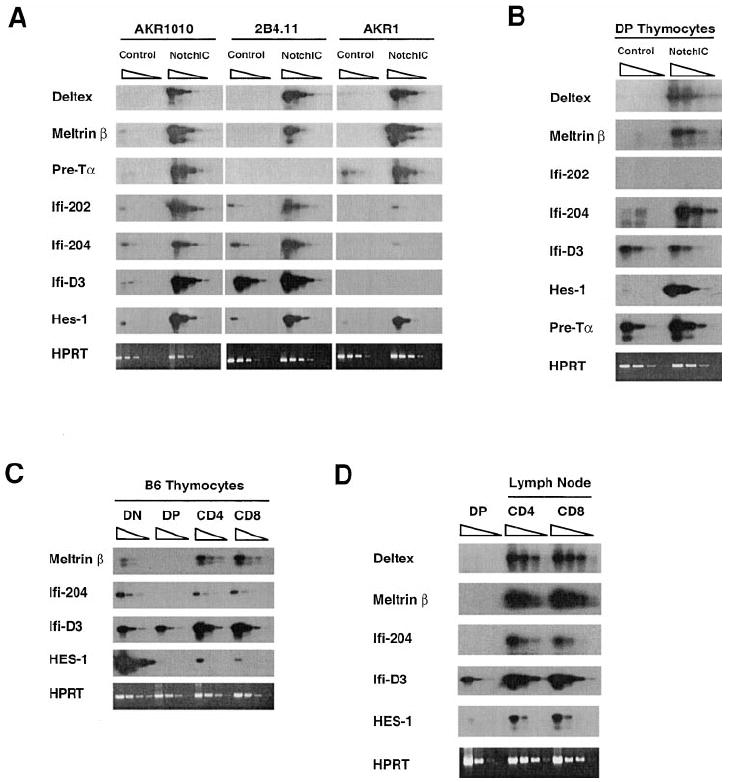
(A) RT-PCR analysis of the expression of Notch target genes in the AKR1 and AKR1010 thymomas and the 2B4.11 T cell hybridoma. Equivalent amounts of cDNA prepared from control and NotchIC expressing cells were serially diluted 5-fold and amplified using primers specific to the indicated genes. PCR products were analyzed by Southern blotting using specific probes.
(B) RT-PCR analysis of the expression of Notch target genes in DP thymocytes from NotchIC and littermate control mice. DP thymocytes were sorted from NotchIC and littermate control mice.
(C) RT-PCR analysis of the expression of Notch1 target genes in thymocyte subsets. Thymocyte subsets were sorted from B6 mice.
(D) RT-PCR analysis of the expression of Notch1 target genes in peripheral T cells. CD4+ and CD8+ T cells were sorted from peripheral lymph nodes of B6 mice.
We previously showed that Deltex is expressed at relatively high levels in DN thymocytes and CD4+ and CD8+ SP thymocytes and at low levels in DP thymocytes (Deftos et al., 1998). Analysis of the expression of Meltrin β, Ifi-204, Ifi-D3, and HES1 in thymocyte subsets from normal mice revealed a similar pattern of expression (Figure 6C). As reported by others (Saint-Ruf et al., 1994), Pre-Tα expression was detected in DN and DP thymocytes, but not SP thymocytes (data not shown). Collectively, these results indicate that most of the Notch target genes we identified are expressed at relatively low levels in DP thymocytes and are upregulated in both CD4+ and CD8+ SP thymocytes. Although Pre-Tα and Ifi-D3 are expressed at relatively high levels in DP thymocytes, we note that NotchIC had no effect or a weak effect on the expression of these genes in DP thymocytes, suggesting that their expression in DP thymocytes may be regulated by other factors, or that low levels of Notch signaling may be sufficient for their maximal expression in these cells. Collectively, these data support the hypothesis that Notch1 signaling is low in DP thymocytes and becomes activated during the maturation of both CD4+ and CD8+ SP thymocytes.
We also examined the expression of these Notch target genes in CD4+ and CD8+ T cells from lymph nodes of normal mice. Interestingly, Deltex, Meltrin β, Ifi-204, Ifi-D3, and HES1 are expressed at high levels in both CD4+ and CD8+ T cells relative to DP thymocytes (Figure 6D), raising the possibility that Notch signaling may remain on in mature T cells. Alternatively, Notch1 signaling may activate the expression of these genes during development, and other factors may maintain their expression in mature T cells.
Discussion
We have presented two lines of evidence suggesting that Notch1 signaling plays a role in promoting the DP to SP transition. First, we showed that expression of NotchIC in DP thymocytes promotes the maturation of both CD4+ SP and CD8+ SP thymocytes in the absence of interactions between the TCR and MHC ligands on thymic epithelial cells. Second, we identified several genes that are transcriptionally regulated by Notch1 in T cells and showed that expression of these genes is upregulated during the maturation of both SP lineages. Together, these results suggest that Notch1 signaling is activated during the DP to SP transition and promotes maturation into both the CD4 and CD8 SP lineages.
Analysis of a different line of transgenic mice expressing NotchIC under control of the Lck-proximal promoter led Robey and coworkers to a different conclusion (Robey et al., 1996). They reported that Lck–NotchIC transgenic mice generate an excess of CD8+ SP thymocytes at the expense of CD4+ SP thymocytes and that this effect is dependent on the expression of either MHC class I or MHC class II on thymic epithelial cells. Based on these results, they suggest that Notch signaling regulates the CD4 versus CD8 cell fate choice in a manner analogous to the role of Notch in regulating binary cell fate choices during development in Drosophila and C. elegans. We believe that our data contradict this model. Consistent with the work of Robey et al., we observe that expression of NotchIC leads to the generation of an excess of phenotypically mature CD8+ SP thymocytes. However, in contrast to their results, we observe that the NotchIC transgene promotes, rather than inhibits, the development of phenotypically mature CD4+ SP thymocytes. Moreover, in our line of transgenic mice, phenotypically mature CD4+ SP and CD8+ SP thymocytes can develop in the absence of MHC expression on thymic epithelial cells. The reason for the differences between these two lines of transgenic mice is not clear but may be due to differences in the level of transgene expression or to differences in the region of Notch1 expressed as a transgene. For example, our transgene contains the complete C-terminal transcriptional activation domain (TAD) of NotchIC, which enhances the ability of NotchIC to activate CBF1-dependent transcription 5-to 10-fold, whereas the transgene used by Robey and colleagues contains a truncation of the TAD that compromises its function (Kurooka et al., 1998). We also note that the NotchIC mice reported by Robey and colleagues showed a relatively small reduction in the number of CD4+ SP thymocytes, whereas in systems in which Notch plays a well-established role in binary cell fate choices, the block in differentiation into the alternative cell lineage caused by constitutively activated Notch is relatively complete (Greenwald, 1998). Consistent with this, recent experiments demonstrate that ectopic activation of Notch1 signaling in bone marrow stem cells results in a virtually complete block in differentiation into the B cell lineage, while promoting differentiation into the T cell lineage (Pui et al., 1999).
The phenotype of the NotchIC transgenic mice suggested that Notch1 signaling may be involved in promoting the DP to SP transition during normal thymocyte development. To test this hypothesis, we identified Deltex, Meltrin β, Ifi-204, and HES1 as genes that are transcriptionally regulated by Notch1 signaling in thymocytes and analyzed their expression in developing thymocytes. These genes were found to be expressed at low levels in DP thymocytes and high levels in CD4 and CD8 SP thymocytes, suggesting that Notch signaling is low or absent in DP thymocytes and upregulated during the DP to SP transition. Moreover, these Notch target genes are expressed at high levels in DN thymocytes, consistent with recent data that indicating that Notch1 signaling is involved in the development of these cells (Pui et al., 1999; Radtke et al., 1999). The hypothesis that Notch signaling is low or absent in DP thymocytes is further supported by the observation that ectopic activation of Notch signaling in DP thymocytes induces the appearance of a population of CD25+ DP thymocytes that is not present in normal mice. Although we have detected expression of CD25 in a small subset of DP thymocytes in normal mice, these cells differ from the CD25+ DP thymocytes observed in the NotchIC mice in that they do not express higher levels of CD44, CD3, and CD5 relative to CD25− DP thymocytes. Moreover, we have not detected elevated Deltex expression in CD25+ DP thymocytes, further arguing that these cells do not have active Notch signaling.
We found that DP thymocytes expressing constitutively active Notch1 develop into both CD4 and CD8 SP thymocytes in the absence of MHC expression on thymic epithelial cells. This implies that ectopic Notch signaling can overcome the normal requirement for TCR–MHC interactions for SP thymocyte maturation. However, we cannot exclude the possibility that TCR interaction with MHC expressed on bone marrow–derived cells is necessary for the development of these cells. Regardless, this observation strongly supports a role for Notch signaling in promoting the DP to SP transition. Since TCR-dependent signaling events play a well-established role in regulating the DP to SP transition, this suggests that signals downstream of the TCR may interact with Notch to control this step of development. We favor a model for the DP to SP transition in which positively selecting TCR signals allow DP thymocytes to receive a productive Notch signal, with the Notch signal then acting to induce changes in gene expression that promote maturation. In line with accumulating evidence, we also favor the idea that the strength of the initial TCR signal influences the CD4 versus CD8 cell fate choice (Itano et al., 1996; Matechak et al., 1996; Hernandez-Hoyos et al., 2000). According to this model, thymocytes that receive a weak TCR signal are induced by Notch to mature into the CD8 SP lineage, whereas thymocytes that receive a strong TCR signal are induced by Notch to mature into the CD4 SP lineage. In NotchIC→MHC−/− chimeric mice, the DP thymocytes may receive a Notch signal in the absence of a TCR signal, and the CD4 versus CD8 cell fate choice may be made stochastically. It should also be noted that SP thymocytes that develop in the NotchIC→MHC−/− chimeric mice do not survive to populate the periphery. This may be due to the fact that these cells do not develop properly in the absence of a TCR signal or that they lack TCR signals required for survival in the periphery (Goldrath and Bevan, 1999). Alternatively, the complete maturation and survival of mature T cells in the periphery may require a prolonged Notch signal that is not provided by the Lck–NotchIC transgene.
Notch signaling regulates development by inducing changes in gene expression, and it is likely that Notch1 regulates the expression of genes that play important roles during T cell development. We identified Deltex, Meltrin β, Pre-Tα, HES1, and members of the Ifi-200 gene family as Notch1 target genes in T cells. Deltex was originally identified in Drosophila as a gene that genetically interacts with the Notch signaling pathway and may function as a positive regulator of Notch signaling (Matsuno et al., 1995). Meltrin β is a member of the ADAM (a disintegrin and metalloprotease) family of metalloproteases and is expressed in a wide variety of tissues (Inoue et al., 1998; Kurisaki et al., 1998). Ifi-200 proteins are a family of nuclear proteins that includes at least four members (Ifi-202, Ifi-203, Ifi-204, and Ifi-D3) in mice and have been implicated in cell cycle regulation and gene transcription (Johnstone and Trapani, 1999). HES1 is a bHLH transcription factor that is best known for its role in inhibiting cell differentiation during lateral specification (Artavanis-Tsakonas et al., 1999). HES1-deficient mice have a block in thymocyte development at the DN stage (Tomita et al., 1999), consistent with the high level of expression of HES1 in these cells. A role for HES1 in negatively regulating CD4 expression during CD8+ SP thymocyte maturation has been proposed (Kim and Siu, 1998); however, the equivalent expression of HES1 in CD4+ SP and CD8+ SP thymocytes and in mature T cells argues against this model. Pre-Tα is a component of the Pre-TCR and plays a well-established role during the development of the αβ lineage of T cells at the DN to DP transition (von Boehmer et al., 1999). We previously showed that expression of NotchIC in the AKR1010 cell line induces the expression of CD3 and the TCR β chain on the cell surface (Deftos et al., 1998). The identification of Pre-Tα as a gene regulated by Notch1 signaling raises the possibility that the effect of NotchIC on CD3 and TCR β surface expression in this cell line may be due to expression of the Pre-TCR rather than the αβ TCR as we previously hypothesized. Moreover, the induction of Pre-Tα expression by NotchIC raises the possibility that the reported increase in αβ versus γδ T cell development in NotchIC transgenic mice may be due in part to elevated expression of Pre-Tα in DN thymocytes (Washburn et al., 1997; Bruno et al., 1999).
Experimental Procedures
NotchIC Transgenic Mice and Bone Marrow Chimeric Mice
A cDNA fragment corresponding to amino acid numbers 1751–2444 of mNotch1 (del Amo et al., 1993) was removed from the pMI/ NotchIC plasmid (Deftos et al., 1998) and cloned into the BamHI site of the p1017 vector (Chaffin et al., 1990). This construct includes the majority of the intracellular domain of Notch1, including the RAM, ankyrin repeat, and C-terminal transcriptional activation domain (TAD), but lacks the C-terminal PEST domain. The 7.3 kb SpeI fragment from p1017–NotchIC was microinjected into fertilized (B6 × DBA2)F2 embryos to generate founders. Transgenic founders were identified by PCR of tail DNA using primers specific for human growth hormone. Of the eight founders generated, four died prior to breeding with evidence of leukemia, consistent with the known oncogenic properties of Notch1 in T cells (Ellisen et al., 1991; Girard et al., 1996; Pear et al., 1996). Transgene expression was detected in progeny of two of the remaining founders, and transgenic lines were established from both of these by breeding to B6 mice. Both lines of mice had a qualitatively similar phenotype, and the one with the stronger phenotype was characterized in detail. Although the majority of mice from this line were healthy up to 4 weeks of age, some of them subsequently developed leukemia. FACS analysis of the leukemic cells indicated that the leukemic cells expressed variable levels of both CD4 and CD8, intermediate levels of CD3, high levels of HSA and CD25, and low levels of CD69, suggesting they were derived from immature thymocytes. The level of expression of HSA and CD25 on these leukemic cells was significantly higher than the levels expressed on the CD25+ DP thymocytes observed in healthy NotchIC mice. Cells with a similar phenotype to the leukemic cells were detected in the thymi of some apparently healthy mice, and these animals were excluded from the analysis presented here. FACS analysis using antibodies against a variety of TCRα and TCRβ families indicated that the CD4 and CD8 SP thymocytes in the NotchIC mice express a large repertoire of TCRs, arguing that they do not represent mono-or oligoclonal expansion of transformed cells.
To generate bone marrow chimeric mice, bone marrow was harvested from NotchIC and littermate control mice and 10 × 106 cells were injected into the tail vein of irradiated (1000 rad) MHC-deficient mice (Grusby et al., 1993). Thymocytes and lymph node cells from these mice were analyzed by FACS 4–6 weeks later.
Cell Lines and Tissue Culture
Transduction of the AKR1010 and 2B4.11 cell lines with retrovirus prepared from the pMI/NotchIC vector was previously described (Deftos et al., 1998). The AKR1 cell line is a DP thymoma cell line derived from AKR mice (Hyman et al., 1980). All cells were cultured in DMEM containing 10% FCS, 2 mM glutamine, 25 mM HEPES, 50 μM β-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Flow Cytometric Analysis and Cell Sorting
The following conjugated mAbs were purchased from Pharmingen: CyC-anti-CD4 (H129.19), APC-anti-CD8α (53-6.7), PE-anti-CD3 (145-2C11), biotin-anti-HSA (M1/69), biotin-anti-Kb (AF6-88.5), biotin-anti-CD5 (53-7.3), biotin-anti-CD44 (IM7), and bio-anti-CD25 (7D4). FITC-avidin (Vector) and PE-streptavidin (Caltag) were used to reveal staining by biotin-conjugated antibodies. FITC–Annexin V (Pharmingen) was used according to the manufacturer’s instructions. Cells were stained and analyzed by flow cytometry using standard procedures. Data were collected on 5 × 104 events using a FACSCalibur flow cytometer (Becton Dickinson) and analyzed using Cell-Quest software (Becton Dickinson).
For FACS analysis of CD25+ thymocytes, 1.5 × 107 thymocytes were incubated with excess anti-CD25 antibody (PC61) for 30 min at 4°C, washed, and incubated on tissue culture plates coated with goat anti-rat Ig (Sigma). After 1 hr at 4°C, unbound cells were removed by extensive washing with PBS, and the remaining bound cells removed by direct pipeting. We recovered and stained 2 × 106 cells with anti-CD4, anti-CD8, and anti-CD25 (7D4). Unpanned thymocytes from the same mouse were stained simultaneously for comparison.
For FACS sorting of thymocyte subsets, thymocytes from B6 mice, NotchIC mice, or littermate control mice were stained with mAbs against CD4, CD8, and CD3 and sorted using a FACStar Plus flow cytometer (Becton Dickinson). CD3−CD4−CD8− (DN), CD3loCD4+CD8+ (DP), and CD3hi CD4+CD8− (CD4 SP) and CD3hiCD4−CD8+ (CD8 SP) thymocytes and lymph node cells were sorted. An aliquot of the sorted cells were reanalyzed by FACS, and in all cases greater than 95% of the cells fell in the sorting gate.
Representational Difference Analysis
RDA was performed as described (Hubank and Schatz, 1994) using cDNA prepared from the parental AKR1010 cell line as the “driver” and cDNA prepared from AKR1010 expressing NotchIC as the “tester.” Multiple distinct products were observed by agarose gel electrophoresis following two rounds of subtractive hybridization. Sequence analysis identified these as DpnII fragments derived from Deltex, Ifi-202, Ifi-204, and Ifi-D3 (Choubey et al., 1989; Tannenbaum et al., 1993; Pampeno and Meruelo, 1996). An additional round of RDA was performed using cloned mNotch1, Deltex, Ifi-202, Ifi-204, and Ifi-D3 to supplement the driver. This yielded cDNA fragments corresponding to Pre-Tα (Saint-Ruf et al., 1994) and Meltrin β (Inoue et al., 1998).
RT-PCR Analysis of Notch Target Gene Expression
RNA was prepared from cell lines or FACS-sorted thymocyte subsets using RNA STAT-60 (Tel-Test). cDNA was prepared using Superscript II reverse transcriptase (GIBCO-BRL) and normalized for equivalent template amounts by serial dilution and amplification using primers specific to HPRT (5′-GATACAGGCCAGACTTTG TTG-3′ and 5′-GGTAGGCTGGCCTATAGGCT-3′). Normalized cDNA was serially diluted 5-fold and amplified for 25–35 cycles (94°C, 1 min; 55°C, 1 min; 72°C, 1 min) using the following primers: Deltex (5′-CACTGGCCCTGTCCACCCAGCCTTGGCAGG-3′ and 5′-ATGC GAATTCGGGAAGGCGGGCAACTCAGG-3′), Meltrin β (5′-AGTCC CAGGGATGCCAAGTGTGG-3′ and 5′-ATCACGGGACCCACACTC TTAGG-3′), Ifi-202 (5′-GGGAAACCAATATTACACTC-3′ and 5′-AAT TTCCACCATTGAATTGG-3′), Ifi-204 (5′-AGGCAACCAAAGTTAGT GTG-3′ and 5′-GTTCTCCCGACTGAGTCTGG-3), Ifi-D3 (5′-ACTTC CTCTGTGTTAGAGGCTGC-3′ and 5′-AAAGCTGTCATTTAGAG GTG-3′), Pre-Tα (5′-TGGCTGCAACTGGGTCATGCTTC-3′ and 5′-GGC TCAGAGGGGTGGGTAAGATC-3′), HES1 (5′-GCCAGTGTCAACAC GACACCGG-3′ and 5′-TCACCTCGTTCATGCACTCG-3′). PCR products were separated by agarose gel electrophoresis, transferred to nitrocellulose, and probed by Southern blot using probes prepared by PCR amplification from cloned cDNAs.
Acknowledgments
We thank Jacqueline Kirchner for critically reading this manuscript, You-Wen He and Anne Norment for insightful discussions during the course of this work, and Xiao-Cun Pan for care of the mice. This work was supported by grants from the National Institutes of Health, AI29802 and CA09537, and the Howard Hughes Medical Institute.
References
- Allen JM, Forbush KA, Perlmutter RM. Functional dissection of the lck proximal promoter. Mol Cell Biol. 1992;12:2758–2768. [Europe PMC free article] [Abstract] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. [Abstract] [Google Scholar]
- Bruno L, Scheffold A, Radbruch A, Owen MJ. Threshold of pre-T-cell-receptor surface expression is associated with αβ T-cell lineage commitment. Curr Biol. 1999;9:559–568. [Abstract] [Google Scholar]
- Chaffin KE, Beals CR, Wilkie TM, Forbush KA, Simon MI, Perlmutter RM. Dissection of thymocyte signaling pathways by in vivo expression of pertussis toxin ADP-ribosyltransferase. EMBO J. 1990;9:3821–3829. [Europe PMC free article] [Abstract] [Google Scholar]
- Choubey D, Snoddy J, Chaturvedi V, Toniato E, Opdenakker G, Thakur A, Samanta H, Engel DA, Lengyel P. Interferons as gene activators. Indications for repeated gene duplication during the evolution of a cluster of interferon-activatable genes on murine chromosome 1. J Biol Chem. 1989;264:17182–17189. [Abstract] [Google Scholar]
- Deftos ML, Bevan MJ. Notch signaling in T cell development. Curr Opin Immunol. 2000;12:166–172. [Abstract] [Google Scholar]
- Deftos ML, He YW, Ojala EW, Bevan MJ. Correlating Notch signaling with thymocyte maturation. Immunity. 1998;9:777–786. [Europe PMC free article] [Abstract] [Google Scholar]
- del Amo FF, Gendron-Maguire M, Swiatek PJ, Jenkins NA, Copeland NG, Gridley T. Cloning, analysis, and chromosomal localization of Notch-1, a mouse homolog of Drosophila Notch. Genomics. 1993;15:259–264. [Abstract] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. [Abstract] [Google Scholar]
- Felli MP, Maroder M, Mitsiadis TA, Campese AF, Bellavia D, Vacca A, Mann RS, Frati L, Lendahl U, Gulino A, Screpanti I. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int Immunol. 1999;11:1017–1025. [Abstract] [Google Scholar]
- Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. [Abstract] [Google Scholar]
- Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. [Abstract] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. [Abstract] [Google Scholar]
- Grusby MJ, Auchincloss H, Jr, Lee R, Johnson RS, Spencer JP, Zijlstra M, Jaenisch R, Papaioannou VE, Glimcher LH. Mice lacking major histocompatibility complex class I and class II molecules. Proc Natl Acad Sci USA. 1993;90:3913–3917. [Europe PMC free article] [Abstract] [Google Scholar]
- Hasserjian RP, Aster JC, Davi F, Weinberg DS, Sklar J. Modulated expression of notch1 during thymocyte development. Blood. 1996;88:970–976. [Abstract] [Google Scholar]
- Hernandez-Hoyos G, Sohn SJ, Rothenberg EV, Alberolalla J. Lck activity controls CD4/CD8 T cell lineage commitment. Immunity. 2000;12:313–322. [Abstract] [Google Scholar]
- Hsieh JJ, Nofziger DE, Weinmaster G, Hayward SD. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71:1938–1945. [Europe PMC free article] [Abstract] [Google Scholar]
- Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. [Europe PMC free article] [Abstract] [Google Scholar]
- Hyman R, Cunningham K, Stallings V. Evidence for a genetic basis for the Class A Thy-1-defect. Immunogenetics. 1980;10:261–271. [Google Scholar]
- Inoue D, Reid M, Lum L, Kratzschmar J, Weskamp G, Myung YM, Baron R, Blobel CP. Cloning and initial characterization of mouse Meltrin beta and analysis of the expression of four metalloprotease-disintegrins in bone cells. J Biol Chem. 1998;273:4180–4187. [Abstract] [Google Scholar]
- Itano A, Salmon P, Kioussis D, Tolaini M, Corbella P, Robey E. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J Exp Med. 1996;183:731–741. [Europe PMC free article] [Abstract] [Google Scholar]
- Itoh M, Takahashi T, Sakaguchi N, Kuniyasu Y, Shimizu J, Otsuka F, Sakaguchi S. Thymus and autoimmunity: production of CD25+CD4+ naturally anergic and suppressive T cells as a key function of the thymus in maintaining immunologic self-tolerance. J Immunol. 1999;162:5317–5326. [Abstract] [Google Scholar]
- Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Israël A. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnstone RW, Trapani JA. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol Cell Biol. 1999;19:5833–5838. [Europe PMC free article] [Abstract] [Google Scholar]
- Kim HK, Siu G. The notch pathway intermediate HES-1 silences CD4 gene expression. Mol Cell Biol. 1998;18:7166–7175. [Europe PMC free article] [Abstract] [Google Scholar]
- Kimble J, Simpson P. The LIN-12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol. 1997;13:333–361. [Abstract] [Google Scholar]
- Kurisaki T, Masuda A, Osumi N, Nabeshima Y, Fujisawa-Sehara A. Spatially-and temporally-restricted expression of meltrin alpha (ADAM12) and beta (ADAM19) in mouse embryo. Mech Dev. 1998;73:211–215. [Abstract] [Google Scholar]
- Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998;26:5448–5455. [Europe PMC free article] [Abstract] [Google Scholar]
- Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17:6057–6067. [Europe PMC free article] [Abstract] [Google Scholar]
- Matechak EO, Killeen N, Hedrick SM, Fowlkes BJ. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 1996;4:337–347. [Abstract] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. [Abstract] [Google Scholar]
- Osborne B, Miele L. Notch and the immune system. Immunity. 1999;11:653–663. [Abstract] [Google Scholar]
- Pampeno CL, Meruelo D. A novel cDNA transcript expressed in fractionated X-irradiation-induced murine thymomas. Cell Growth Differ. 1996;7:1113–1123. [Abstract] [Google Scholar]
- Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. [Europe PMC free article] [Abstract] [Google Scholar]
- Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. [Abstract] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, Mac-Donald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. [Abstract] [Google Scholar]
- Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. [Abstract] [Google Scholar]
- Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling HJ, von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science. 1994;266:1208–1212. [Abstract] [Google Scholar]
- Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. [Abstract] [Google Scholar]
- Tannenbaum CS, Major J, Ohmori Y, Hamilton TA. A lipopolysaccharide-inducible macrophage gene (D3) is a new member of an interferon-inducible gene cluster and is selectively expressed in mononuclear phagocytes. J Leukoc Biol. 1993;53:563–568. [Abstract] [Google Scholar]
- Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. [Europe PMC free article] [Abstract] [Google Scholar]
- von Boehmer H, Aifantis I, Feinberg J, Lechner O, Saint-Ruf C, Walter U, Buer J, Azogui O. Pleiotropic changes controlled by the pre-T-cell receptor. Curr Opin Immunol. 1999;11:135–142. [Abstract] [Google Scholar]
- Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 1997;88:833–843. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/s1074-7613(00)00009-1
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1074761300000091/pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/s1074-7613(00)00009-1
Article citations
In Vitro Human Haematopoietic Stem Cell Expansion and Differentiation.
Cells, 12(6):896, 14 Mar 2023
Cited by: 12 articles | PMID: 36980237 | PMCID: PMC10046976
Review Free full text in Europe PMC
Notch Signaling in Acute Inflammation and Sepsis.
Int J Mol Sci, 24(4):3458, 09 Feb 2023
Cited by: 7 articles | PMID: 36834869 | PMCID: PMC9967996
Review Free full text in Europe PMC
DLL4 and VCAM1 enhance the emergence of T cell-competent hematopoietic progenitors from human pluripotent stem cells.
Sci Adv, 8(34):eabn5522, 24 Aug 2022
Cited by: 15 articles | PMID: 36001668 | PMCID: PMC9401626
Notch Signalling in Breast Development and Cancer.
Front Cell Dev Biol, 9:692173, 06 Jul 2021
Cited by: 33 articles | PMID: 34295896 | PMCID: PMC8290365
Review Free full text in Europe PMC
iWhale: a computational pipeline based on Docker and SCons for detection and annotation of somatic variants in cancer WES data.
Brief Bioinform, 22(3):bbaa065, 01 May 2021
Cited by: 7 articles | PMID: 32436933 | PMCID: PMC8557746
Go to all (220) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Correlating notch signaling with thymocyte maturation.
Immunity, 9(6):777-786, 01 Dec 1998
Cited by: 242 articles | PMID: 9881968 | PMCID: PMC2789684
A reassessment of the effect of activated Notch1 on CD4 and CD8 T cell development.
J Immunol, 169(4):1817-1821, 01 Aug 2002
Cited by: 72 articles | PMID: 12165504
Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength.
Immunity, 14(3):253-264, 01 Mar 2001
Cited by: 122 articles | PMID: 11290335
New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision.
Curr Opin Immunol, 14(2):207-215, 01 Apr 2002
Cited by: 106 articles | PMID: 11869894
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (3)
Grant ID: CA09537
Grant ID: T32 CA009537
Grant ID: T32 CA009537-13
NIAID NIH HHS (3)
Grant ID: R01 AI029802
Grant ID: R01 AI029802-11
Grant ID: AI29802




