Abstract
Free full text

Localization, Purification, and Functional Reconstitution of the P4-ATPase Atp8a2, a Phosphatidylserine Flippase in Photoreceptor Disc Membranes*
Abstract
P4-ATPases comprise a relatively new subfamily of P-type ATPases implicated in the energy-dependent translocation of aminophospholipids across cell membranes. In this study, we report on the localization and functional properties of Atp8a2, a member of the P4-ATPase subfamily that has not been studied previously. Reverse transcription-PCR revealed high expression of atp8a2 mRNA in the retina and testis. Within the retina, immunofluorescence microscopy and subcellular fractionation studies localized Atp8a2 to outer segment disc membranes of rod and cone photoreceptor cells. Atp8a2 purified from photoreceptor outer segments by immunoaffinity chromatography exhibited ATPase activity that was stimulated by phosphatidylserine and to a lesser degree phosphatidylethanolamine but not by phosphatidylcholine or other membrane lipids. Purified Atp8a2 was reconstituted into liposomes containing fluorescent-labeled phosphatidylserine to measure the ability of Atp8a2 to flip phosphatidylserine across the lipid bilayer. Fluorescence measurements showed that Atp8a2 flipped fluorescent-labeled phosphatidylserine from the inner leaflet of liposomes (equivalent to the exocytoplasmic leaflet of cell membranes) to the outer leaflet (equivalent to cytoplasmic leaflet) in an ATP-dependent manner. Our studies provide the first direct biochemical evidence that purified P4-ATPases can translocate aminophospholipids across membranes and further implicates Atp8a2 in the generation and maintenance of phosphatidylserine asymmetry in photoreceptor disc membranes.
Introduction
Lipids are asymmetrically distributed across cell membranes (1). Phosphatidylserine (PS)3 and phosphatidylethanolamine (PE) are confined to the cytoplasmic leaflet of the plasma membrane, whereas phosphatidylcholine (PC) and sphingolipids, including sphingomyelin and glycolipids, are preferentially if not exclusively localized on the extracellular leaflet (2). Membranes of intracellular organelles and vesicles also display transbilayer lipid asymmetry (1, 3).
Lipid asymmetry has been implicated in a number of important cellular functions. These include generating tight lipid packing to increase membrane impermeability; establishing the shape of intracellular organelles through membrane; bending; cell division; phagocytosis and cell death; fertilization; vesicle transport and fusion; regulating the functional activity of membrane-associated proteins, including enzymes, receptors, transporters, and channels; and sequestering protein complexes to membrane surfaces (4,–6).
A number of recent genetic studies have implicated members of the P4-ATPase family of membrane proteins in translocation of the aminophospholipids from the exocytoplasmic to the cytoplasmic leaflet of membranes (7). In Caenorhabditis elegans, knockdown of the P4-ATPase TAT-1 results in the exposure of PS on the surface of germ cells (8). Although no effect on reproduction was found, there was a significant loss of neural and muscle cells, which could be rescued by deletion of a PS-binding phagocyte receptor. In yeast, a deficiency in the P4-ATPases Drs2 and Dnf3 causes a loss of PE and PS asymmetry and a reduced rate of transport of fluorescent phospholipid derivatives across membranes (3, 9). Mutants of these P4- ATPases in yeast cause defects in vesicular trafficking (10, 11), suggesting a possible role for P4-ATPases in vesicle formation. In the spermatozoa of mice, Atp8b3 is necessary for PS asymmetry and fertilization (12).
The importance of P4-ATPases is underscored by the finding that mutations in several members of this subfamily of proteins are known to cause severe human disorders. Mutations in the gene encoding Atp8b1 cause progressive familial cholestasis, a disease associated with defects in bile secretion (13), as well as impaired hearing (14). Mutations in the gene encoding Atp10c have been linked to Angelman syndrome, a severe form of mental retardation (15).
In earlier biochemical studies, membranes of erythrocytes and bovine chromaffin granules were reported to contain aminophospholipid translocase activity (16,–19). PE and PS but not PC derivatives were translocated from the outer to the inner cytoplasmic leaflet of the membrane in an ATP-dependent manner. The ATPase implicated in aminophospholipid translocase activity in chromaffin granules has been identified as Atp8a1, a member of the P4-ATPase family (20). Recent studies have identified isoforms of Atp8a1 in murine erythrocytes, suggesting that this member of the P4 ATPase family may also be responsible for aminophospholipid translocase activity in erythrocyte membranes (21).
Outer segments are specialized light sensing compartments of rod and cone photoreceptor cells. They consist of a stack of highly organized disc membranes packed with the photopigment protein, rhodopsin, in rod outer segments (ROS) and cone opsin in cone outer segments. Rhodopsin and enzymes involved in the G-protein-mediated visual cascade in ROS have been extensively studied, leading to a comprehensive understanding of phototransduction (22). Mutations in the genes encoding many photoreceptor-specific proteins are known to cause inherited retinal degenerative diseases, which are a major cause of blindness in the population (23, 24). However, despite the progress made in understanding phototransduction and retinal degenerative diseases, we still know relatively little about proteins involved in photoreceptor outer segment morphogenesis and structure. Furthermore, although phospholipids were reported to be asymmetrically distributed across the disc membrane over 25 years ago, the protein(s) responsible for generating and maintaining this transbilayer asymmetry have not been identified (25, 26).
Recently, we have carried out a mass spectrometry-based proteomic study designed to identify low abundance membrane and soluble proteins of photoreceptor outer segments with the goal of defining their role in outer segment structure, function, and morphogenesis (27). The P4-ATPase Atp8a2 was identified as a membrane protein present in photoreceptor outer segment preparations. atp8a2 has been previously reported to be expressed in the testis as a 4.5-kb mRNA with high levels occurring during early spermatid development (28). The biochemical properties of this member of the P4-ATPase family, however, have not been investigated to date.
To begin to define the role of Atp8a2 in retinal photoreceptors, we have generated several monoclonal antibodies to Atp8a2 and used these immunoreagents to localize, purify, and characterize the functional properties of Atp8a2. Here, we show that Atp8a2 is expressed in the retina as well as testes and is present in outer segment disc membranes of rod and cone photoreceptors. Importantly, we have purified Atp8a2 from disc membranes by immunoaffinity chromatography for analysis of its aminophospholipid-dependent ATPase and flippase activities. The ATPase activity of Atp8a2 was activated by PS and to a lesser extent PE. Upon reconstitution into lipid vesicles, Atp8a2 was found to flip fluorescent-labeled PS to the cytoplasmic side of the membrane, confirming the aminophospholipid translocase activity of Atp8a2. To our knowledge, this is the first report in which a specific P4-ATPase has been directly shown to display flippase activity by functional reconstitution of the purified protein and the first membrane protein linked to phospholipid asymmetry in photoreceptor cells.
EXPERIMENTAL PROCEDURES
Materials
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1, 2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS), brain polar lipids (porcine), l-α-phosphatidylinositol (bovine, liver), 1,2-dioleoyl- sn-glycero-3-phosphate, 1,2-dioleoyl-sn-glycero-3-phosphoglycerol, sphingomyelin (porcine, brain), cholesterol (ovine, wool), l-α-phosphatidylcholine (egg, chicken), C6 NBD-PS, C6 NBD-PE, C6 NBD-PC, and C12 NBD-PS were purchased from Avanti Polar Lipids (Alabaster, AL). ATP and n-octyl-β-d-glucopyranoside were purchased from Sigma, dithionite was from Fisher, CHAPS was from Anatrace (Maumee, OH), and synthetic 6C11 peptide (Ac-RDRLLKRLS) was purchased from Biomatik (Cambridge, Canada). The Rho 1D4 antibody was obtained from UBC-UILO.
DNA Constructs
Total bovine, human, and murine retinal RNA was isolated using the guanidinium thiocyanate phenol chloroform method (29). Because the only available sequences for bovine atp8a2 were incomplete, a 5′-rapid amplification of cDNA ends was performed (30). Random primed cDNA was prepared using the RT-PCR Master Mix Kit (GE Healthcare). Full-length human, bovine, and mouse atp8a2 were amplified by PCR using Pfu polymerase (Fermentas, Burlington, Canada). Restriction sites were introduced by PCR. Bovine and mouse atp8a2 were cloned into pcDNA3 and pCEP4 using the BamHI and NotI restriction sites, and human atp8a2 was cloned into pcDNA3 using KpnI and NotI. 1D4-tagged Atp8a2 contained a 9-amino acid C-terminal tag (TETSQVAPA). The sequence of bovine atp8a2 was deposited in GenBank™ (GQ303567). The sequences of human and murine atp8a2 had previously been deposited (NM_015803.2, NM_016529.4).
In Situ Hybridization
Detection of gene expression using DIG-labeled riboprobes was performed (31) with modifications. A fragment was isolated from an EcoRI/HindIII digest of murine atp8a2 and ligated into pcDNA3 (nucleotides 923–1641). Sense and antisense cRNA were made using the DIG RNA labeling kit (Roche Applied Science) with the SP6 and T7 polymerases, respectively. Probes were treated with DNase I (Fermentas) and purified by LiCl precipitation. Eyes from 6-month-old BALB/c mice were fixed for 40 min in 4% paraformaldehyde in PBS (10 mm phosphate, pH 7.4, 140 mm NaCl, 3 mm KCl), frozen in Tissue-Tek OCT, and cut into 12-μm sections. Probes were diluted to 400 ng/ml and incubated with sections for 16 h at 60 °C. Sections were treated with anti-DIG-alkaline phosphatase (Roche Applied Science) and developed in 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium for 15 h.
Gene Expression by RT-PCR
RNA was isolated from tissues from 6-month-old C57/B6 mice to prepare random primed cDNA. atp8a2 gene expression was measured using the following forward and reverse primers: 5′-ACGAGGGACGTGCTCATGAAGC-3′ and 5′-CCTCAAGTGTCACCAGCAGGCT-3′. Glyceraldehyde-3-phosphate dehydrogenase was used for comparison employing the following forward and reverse primers: 5′-ATCAAATGGGGTGAGGCCGGTG-3′ and 5′-CGGCATCGAAGGTGGAAGAGTG-3′. The primers were annealed at 55 °C, and the PCR was run for 25 cycles using Taq polymerase (New England Biolabs).
Generation of Monoclonal Antibodies against Atp8a2
Fragments corresponding to the C-terminal 103 amino acids and the P-domain (amino acids 371–873) of bovine Atp8a2 were cloned in frame with glutathione S-transferase (GST) in the pGEX-4T-1 vector (GE Healthcare). Mice were immunized with GST fusion proteins purified on glutathione-Sepharose 4B (GE Healthcare). Hybridoma cell lines were generated as previously described (32) and screened for reactivity against Atp8a2 using Western blots of bovine ROS. The epitope for the Atp6C11 monoclonal antibody was identified by measuring its immunoreactivity to synthetic overlapping 9-amino acid peptides spanning the C terminus of Atp8a2 as described previously (33).
Expression of Atp8a2 in HEK293T
HEK293T cells (American Type Culture Collection, Manassas, VA) in 10 cm dishes were transfected at 30% confluence with 20 μg of Atp8a2–1D4 in pcDNA3 by the calcium phosphate method (34) and harvested 48 h later. For control experiments, cells were transfected with empty pcDNA3. HEK293T membranes were isolated as described (35).
Immunofluorescence Microscopy
Cryosections of retina tissue from bovine eyes fixed in 4% paraformaldehdye, 100 mm phosphate buffer (PB), pH 7.4, for 1 h were blocked and permeabilized with 10% normal goat serum and 0.2% Triton X-100 in PB for 30 min. The sections were then labeled overnight at room temperature with Atp2F6 hybridoma culture fluid diluted 1:3 in PB containing 2.5% normal goat serum and 0.1% Triton X-100. Sections were washed with PB and labeled for 1 h with Cy3-conjugated goat anti-mouse Ig secondary antibody (diluted 1:1000) and counterstained with 4′,6-diamidino-2-phenylindole nuclear stain. For controls, Atp2F6 was preabsorbed with 20 μg of GST fusion protein for 30 min before the antibody was added to the sections. For double labeling studies, sections were labeled with Atp2F6 and a mixture of cone opsin antibodies (JH 492, JH 455; diluted 1:4000), a kind gift of Jeremy Nathans (36). The sections were labeled with secondary 1:1000 diluted anti-mouse and anti-rabbit antibodies conjugated to Alexa-488 and Alexa-594, respectively. Samples were visualized under a Zeiss LSM 700 confocal microscope.
Isolation of Retina and ROS Membranes
Retina and ROS membranes from frozen bovine retinas were prepared as previously described (27, 37, 38). Highly enriched ROS disc membranes were prepared by an immunogold density perturbation procedure (27).
Purification of Atp8a2
Purified Atp6C11 monoclonal antibody was coupled to CNBr-activated Sepharose at a concentration of 1.5–2 mg of protein/ml of packed beads as described previously (39). Atp8a2 was purified by solubilizing 3.5 mg of ROS in 1 ml of buffer A (50 mm Hepes, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT, 20 mm CHAPS, and 0.5 mg/ml DOPC) containing complete protease inhibitor (Roche Applied Science) for 30 min at 4 °C. Insoluble material was removed by centrifugation (100,000 × g for 10 min), and the soluble fraction was incubated with Atp6C11-Sepharose at 4 °C for 2 h. The matrix was washed six times with 500 μl of buffer B (50 mm Hepes, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT, 10 mm CHAPS, 0.5 mg/ml DOPC), and the protein was eluted twice in buffer B containing 0.2 mg/ml of 6C11 peptide at room temperature in 50 μl for 1 h.
Reconstitution of Atp8a2 into Lipid Vesicles
Atp8a2 was purified from ROS as described above with the following modifications. Atp6C11-Sepharose was washed in buffer B containing 0.75% (w/v) n-octyl-β-d-glucopyranoside and 0.5 mg/ml l-α-phosphatidylcholine instead of CHAPS and DOPC. Purified protein was mixed 1:1 with buffer C (50 mm Hepes, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT, 1% n-octyl-β-d-glucopyranoside, 10% sucrose, 5 mg/ml l-α-phosphatidylcholine) and stirred for 1 h at room temperature. Subsequently, the sample was dialyzed against 1 liter of buffer D (10 mm Hepes, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT, 10% sucrose) for ~20 h with two changes to remove the detergent.
ATPase Activity Assay
Typically 25 ng of Atp8a2 was diluted in 25 μl of buffer E (50 mm Hepes, pH 7.5, 150 mm NaCl, 12.5 mm MgCl2, 1 mm DTT, and 5 mm ATP). The concentration of Atp8a2 was determined by comparison with known amounts of bovine serum albumin or with known amounts of Atp8a2 by quantitative Western blots. For ATPase activity assays in detergent, buffer E contained 10 mm CHAPS and 2.5 mg/ml DOPC and the stimulating lipid at the indicated concentration. For ATPase assays of reconstituted Atp8a2, the buffer contained 10% sucrose and a final lipid concentration of 1 mg/ml. Samples were incubated at 37 °C for 15–30 min and stopped by the addition of 25 μl of 12% SDS. The concentration of phosphate was determined as described (40). Briefly, color was developed for 5 min following the addition of 75 μl of Solution D (6% ascorbic acid, 1% ammonium molybdate in 1 n HCl). The reactions were stopped by the addition of 120 μl Solution E (2% sodium citrate, 2% sodium meta-arsenite, 2% acetic acid) and read at 850 nm in a microplate reader. The amount of phosphate released was determined by comparison with known phosphate concentrations. All data points were performed in triplicate, and each experiment was repeated at least three times independently with similar results. Data were analyzed with GraFit 6.0 (Erithacus Software).
Flippase Assay
The dithionite NBD-lipid assay was used as described (41,–43) with modifications. Atp8a2 was purified from 3.5 mg of ROS and reconstituted into 250 μl of liposomes at a lipid concentration of 2.5 mg/ml containing 2.5% (w/w) NBD-lipid. Transport was initiated by mixing 20 μl of reconstituted Atp8a2 with nucleotide in buffer D to 50 μl (final nucleotide concentration 0.5 mm) and incubated at 23 °C for 2.5 min. In some cases, NEM or sodium orthovanadate were also added at a concentration of 5 and 1 mm, respectively. The sample was then diluted to 1 ml in buffer D, transferred to a cuvette (path length 1 cm), and read in a fluorescence spectrophotometer (Varian, Palo Alto, CA) using excitation and emission wavelengths of 478 and 540 nm, respectively, and a slit width of 5 nm. Sodium dithionite prepared in 1 m Tris, pH 10, was added at a final concentration of 2 mm once a base-line fluorescence was achieved. After a stable base line was reached, Triton X-100 was then added to a final concentration of 1%. The percentage of NBD-lipid that was accessible to dithionite treatment was defined as %NBDout and was used to calculate the extent of NBD-lipid transport using the following equation,

where control, trial, and ATP represent liposomes treated with AMP-PNP, various nucleotides or inhibitors, and ATP, respectively. All data points were performed in triplicate, and each experiment was repeated independently at least three times.
SDS-PAGE and Western Blots
Proteins were separated by SDS gel electrophoresis on 9% polyacrylamide gels and either stained with Coomassie Blue or transferred to Immobilon FL membranes (Millipore, Bedford, MA) in buffer containing 25 mm Tris, 192 mm glycine, 10% methanol, pH 8.3. Membranes were blocked with 1% milk in PBS for 30 min, incubated with culture supernatant diluted in PBS for 40 min, washed stringently with PBST (PBS containing 0.05% Tween 20), incubated for 40 min with secondary antibody (goat anti-mouse conjugated with IR dye 680 (LI-COR, Lincoln, NE) diluted 1:20,000 in PBST containing 0.5% milk), and washed with PBST prior to data collection on a LI-COR Odyssey infrared imaging system. The concentrations of various antibody supernatants used in this study for Western blot are given (supplemental Table 1).
RESULTS
atp8a2 mRNA Expression in the Retinal Photoreceptor Cells and Testis
The expression of atp8a2 mRNA in various tissues of 6-month-old C57/B6 mice was examined by RT-PCR. A 500-bp atp8a2 fragment was detected in retina and testis but not heart, kidney, spleen, liver, brain, or lung (Fig. 1A). Detection in the testis by RT-PCR is in general agreement with earlier studies showing high atp8a2 expression in the testis by Northern blot analysis (28).
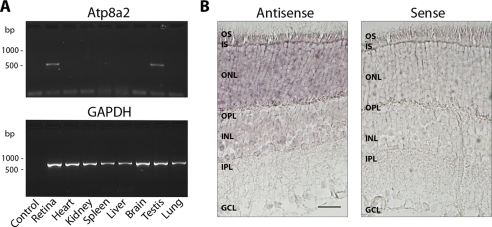
Gene expression of atp8a2 by RT-PCR and in situ hybridization. A, the relative gene expression of atp8a2 was measured using gene-specific primers on cDNA prepared from RNA isolated from different mouse tissues. The relative gene expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Expression of atp8a2 is only detected in the retina and the testis. B, labeling of antisense probe and control sense probe to atp8a2 in mouse retina. atp8a2 mRNA is detected in the outer nuclear layer and inner segment in mouse retina. OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Bar, 30 μm.
In situ hybridization using DIG-labeled antisense probes to atp8a2 was carried out to identify cells in the retina that express atp8a2. atp8a2 mRNA expression was detected in photoreceptor cells with the most intense staining observed in the inner segment (IS) and outer nuclear layer (ONL) (Fig. 1B). Weak staining was also detected in the inner nuclear and ganglion cell layers. In control samples, no staining was observed when the sense probe was used.
Monoclonal Antibodies to Atp8a2
Monoclonal antibodies were generated against GST fusion proteins for use as probes to study the biochemical properties and subcellular localization of Atp8a2. The Atp6C11 antibody was obtained from a mouse immunized with a GST fusion protein comprising the C-terminal 103 amino acids of bovine Atp8a2, and the Atp2F6 antibody was produced from a mouse immunized with a GST fusion protein containing amino acids 371–873 of the P-domain of Atp8a2.
The specificity of these antibodies was confirmed on Western blots of expressed bovine 1D4-tagged Atp8a2 (Atp8a2-1D4) and ROS membranes. As shown in Fig. 2, both the Atp6C11 and Atp2F6 antibodies labeled a prominent 130-kDa protein in both transfected HEK293T cells and bovine ROS. The size of the protein was consistent with the molecular mass of ~132 kDa determined from the amino acid sequence of bovine Atp8a2. The identity of the 130-kDa protein as Atp8a2 was further confirmed on Western blots of atp8a2–1D4-transfected cells labeled with the Rho 1D4 antibody (Fig. 2). In controls, no labeling was observed in membranes from mock-transfected HEK293T cells. The Atp6C11 antibody against bovine Atp8a2 cross-reacted with both human and mouse Atp8a2, whereas the Atp2F6 antibody only labeled the bovine protein (Fig. 2). Mouse Atp8a2 expressed in HEK293T cells appeared less stable than the human and bovine proteins and accordingly showed weaker staining by both the Rho 1D4 and Atp6C11 antibodies.
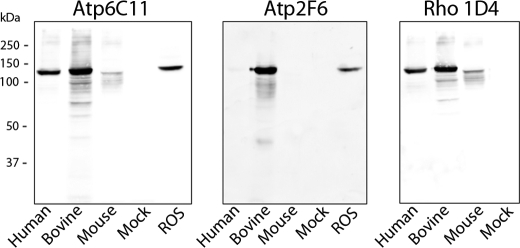
Characterization of Atp8a2 antibodies by Western blotting. Western blots of membranes (10 μg of protein) from HEK293T cells transfected with human, bovine, or mouse Atp8a2–1D4 vector or empty vector (Mock) together with bovine ROSs (30 μg of protein) were labeled with the Atp6C11 and Atp2F6 monoclonal antibodies to Atp8a2 and the Rho 1D4 monoclonal antibody to the 1D4 epitope. The same 130-kDa bovine protein was detected in both the HEK293T and ROS membrane samples. The Atp6C11 labeled the human, bovine, and mouse Atp8a2, whereas the Atp2F6 antibody was specific for the bovine protein. No labeling was observed in membranes from mock-transfected cells.
Synthetic peptides and GST fusion proteins were used to further localize the epitope for these monoclonal antibodies. The epitope for the Atp6C11 antibody was mapped to a 9- amino acid sequence RDRLLKRLS (amino acids 1,118–1,126 of bovine Atp8a2) on the basis of immunoreactivity to synthetic peptides, whereas the epitope for the Atp2F6 antibody was localized to a region between amino acids 440 and 468 using GST fusion proteins.
Localization of Atp8a2 in the Retina by Immunofluorescence Microscopy
Cyrosections of bovine retina were labeled with the Atp2F6 antibody to determine the cellular and subcellular distribution of Atp8a2 within the retina (Fig. 3A). Immunolabeling was observed in the outer segment layer of photoreceptors consisting of mainly ROS but not in other layers of the retina. Double labeling studies using the Atp2F6 monoclonal antibody and cone opsin-specific polyclonal antibodies revealed that cone as well as rod outer segments were stained with the Atp2F6 antibody (Fig. 3B). In control samples, immunolabeling was abolished by preabsorbing the Atp2F6 antibody with a GST fusion protein containing the Atp2F6 epitope (Fig. 3A). The Atp6C11 antibody could not be used for immunolocalization of Atp8a2 because the epitope was sensitive to paraformaldehyde fixation. Furthermore, additional studies indicated that this antibody cross-reacted with an unidentified soluble protein in retinal extracts by Western blot (supplemental Fig. 1).
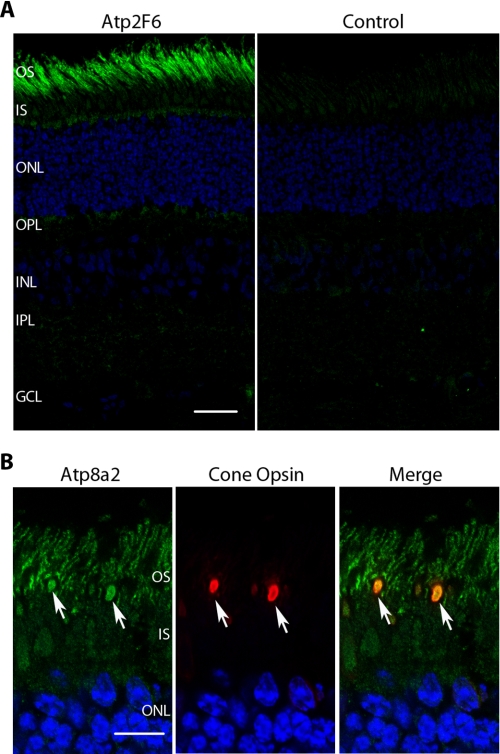
The localization of Atp8a2 in the bovine retina by immunofluorescence microscopy. A, Atp8a2 was immunolabeled with Atp2F6 antibody (green) and nuclei with 4′,6-diamidino-2-phenylindole (blue). In the control sample, Atp2F6 antibody was blocked by excess GST fusion protein prior to immunolabeling. Atp8a2 is localized to the outer segment layer consisting primarily of ROS. OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Bar, 30 μm. B, double labeling of Atp8a2 (green) and cone opsin (red) and merged image showing co-localization of Atp8a2 and cone opsin in cone outer segments (arrows). Bar, 10 μm.
Atp8a2 Is Present in ROS Disc Membranes
The distribution of Atp8a2 within ROS was further evaluated by subcellular fractionation. Disc membranes, which comprise 95% of total ROS membranes, were separated from the ROS plasma membrane by an immunogold density perturbation method (27, 37). Western blots of crude retinal membranes, isolated ROS membranes, and disc membranes were probed for Atp8a2 for comparison with ABCA4, a marker for the disc membrane, and the cyclic nucleotide-gated channel subunit α-subunit CNGA1, a marker for the plasma membrane of ROS (33, 44). Fig. 4 shows that Atp8a2 was present at similar levels in ROS and disc membranes and at lower amounts in crude retinal membranes. This profile was similar to that of ABCA4 and distinctly different from CNGA1, which is absent in disc membranes. These results indicate that Atp8a2 is present in disc membranes of ROS.
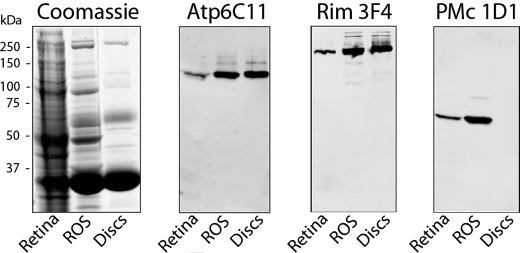
Atp8a2 localizes to the disc membranes of outer segments. Approximately 30 μg of crude retina membranes (Retina), isolated ROS, and purified disc membranes (Discs) were resolved by SDS gel electrophoresis. The gels were stained with Coomassie Blue, and Western blots were labeled with the Atp6C11 antibody to Atp8a2, PMc 1D1 antibody to CNGA1 as a plasma membrane-specific marker, and the Rim 3F4 to ABCA4 as a disc-specific marker. The profile of Atp6C11 resembled Rim3F4, indicating that Atp8a2 is localized to disc membranes.
Purification of Atp8a2 by Immunoaffinity Chromatography
Atp8a2 was purified from detergent-solubilized ROS in order to study its functional properties. Our purification strategy involved 1) solubilization of Atp8a2 from ROS with CHAPS in the presence of PC as a stabilizing lipid; 2) binding of Atp8a2 to an Atp6C11-Sepharose immunoaffinity matrix; 3) removal of unbound protein by extensive washing; and 4) elution of functional Atp8a2 with buffer containing the competing 9-amino acid synthetic 6C11 peptide. The eluted protein identified as Atp8a2 by Western blotting was largely free of other detectable proteins, as shown by Coomassie Blue staining (Fig. 5). The remaining Atp8a2 that was not eluted with peptide could be recovered under denaturing conditions by elution with 2% SDS. The purification and enzymatic activity of purified Atp8a2 obtained under nondenaturing conditions is given in Table 1. This immunoaffinity purification method resulted in a 51% yield with purification in the specific PS-activated ATPase activity of nearly 2,000-fold.
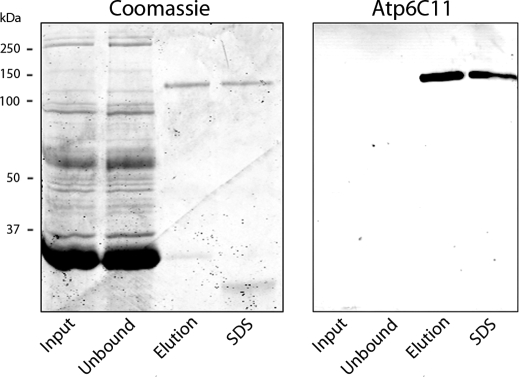
Purification of Atp8a2 from bovine ROS membranes by immunoaffinity chromatography. ROS membranes solubilized in CHAPS detergent (precolumn) were applied to an Atp6C11-Sepharose column. The flow-through (unbound) and bound Atp6C11 eluted first with the competing 6C11 peptide (peptide) and subsequently with SDS were resolved by SDS gel electrophoresis. Gels were stained with Coomassie Blue, and Western blots were labeled with the Atp6C11 antibody to Atp8a2. The following amount of protein was applied to the indicated lanes: precolumn (30 μg), unbound (30 μg), peptide elution (100 ng), and SDS elution (100 ng).
TABLE 1
Purification of Atp8a2 from bovine rod outer segments by immunoaffinity chromatography
The activity of Atp8a2 was measured in the presence of 5 mm ATP and endogenous lipid concentrations following solubilization for the lysate and unbound fractions, respectively. For the elution fraction, 10% exogenous DOPS was added to maximally stimulate the ATPase activity.
| Fraction | Volume | Protein | Total PS-stimulated activity | Specific activity | Yield | Purification |
|---|---|---|---|---|---|---|
| μl | mg | nmol ATP/min | nmol ATP/min/mg | % | -fold | |
| Lysate | 490 | 1.59 | 0.232 | 28.67 | 100 | 1.0 |
| Unbound | 490 | 1.59 | 0.152 | 18.74 | ||
| Elution | 65 | 4.3 × 10−4 | 1.010 | 54,087 | 51 | 1,887 |
The abundance and ATPase activity of Atp8a2 were determined in ROS preparations. Atp8a2 comprised less than 0.1% of the total ROS membrane protein, in general agreement with earlier proteomic studies (27). Despite its low abundance, however, the ATPase activity of Atp8a2 accounted for more than 35% of total ATPase activity in bovine ROS preparations. This was determined by comparing the ATPase activity of solubilized ROS membranes before and after the complete removal of Atp8a2 on the immunoaffinity column under basal conditions for other ATPases (Table 1). Additional studies indicated that phospholipids, such as PC, and reducing agents, such as DTT, were required during purification in order to stabilize Atp8a2 and prevent disulfide bond formation.
Kinetics of ATP Hydrolysis by Atp8a2
Because the ATPase activities of various members of the P4-ATPase family of proteins have been shown to be stimulated by phospholipids, we studied the effect of different phospholipids on the ATPase activity of detergent-solubilized, immunoaffinity-purified Atp8a2. Fig. 6A shows that the ATPase activity of Atp8a2 is strongly stimulated by the addition of 10 mol % brain polar lipids and 10 mol % synthetic DOPS, resulting in a specific activity of 45–55 μmol of ATP/min/mg of protein. Atp8a2 was only weakly activated by the 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine and not activated by other lipids, including DOPC, DOPI, 1,2-dioleoyl-sn-glycero-3-phosphate, cholesterol, and sphingomyelin.
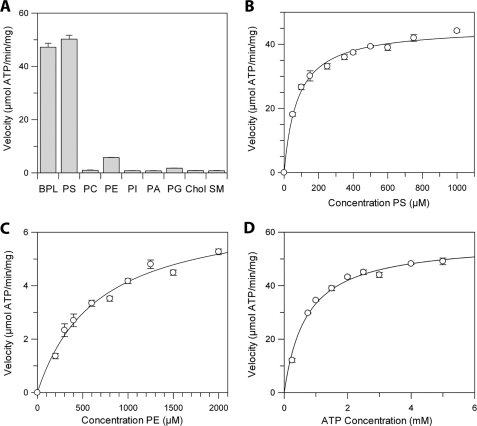
Effect of different lipids and ATP on the ATPase activity of Atp8a2. A, the velocity of ATP hydrolysis of detergent-solubilized and purified Atp8a2 in the presence of various lipids (10 mol %) and phosphatidylcholine (90 mol %) at 5 mm ATP. BPL, brain polar lipids; PI, phosphatidylinositol; PA, phosphatidic acid; PG, phosphatidylglycerol; Chol, cholesterol; SM, sphingomyelin. B, the effect of varying PS concentration on the velocity of ATP hydrolysis in the presence of PC and 5 mm ATP. C, the effect of varying PE concentration on the velocity of ATP hydrolysis of Atp8a2 in the presence of PC and 5 mm ATP. D, the effect of varying ATP concentration on the velocity of ATP hydrolysis of Atp8a2 in the presence of 10% PS.
The effect of increasing PS and PE concentrations on the ATPase activity of Atp8a2 was examined. Michaelis-Menten kinetics was observed for both PS and PE. The Km for PS and PE was 78 ± 7 and 660 ± 90 μm, respectively (Table 2). The Vmax for PS was ~6.5 times higher than for PE. The resulting specificity constant (Kcat/Km) was 60-fold greater for PS compared with PE (Table 2).
TABLE 2
The kinetic parameters of the ATPase activity of Atp8a2
The ATPase activity was measured in 5 mm ATP under varying concentrations of PS and PE to determine the Km/Vmax for each lipid. Similarly, varying concentrations of ATP were used in the presence of 10% PS to determine the Km/Vmax for ATP.
| Substrate | Kf | Vmax | Kcat | Kcat/Km |
|---|---|---|---|---|
| μm | μmol ATP/min/mg | s−1 | s−1 μm−1 | |
| PS | 78 ± 7 | 45.4 ± 0.8 | 100 ± 2 | 1.3 ± 0.1 |
| PE | 660 ± 90 | 6.9 ± 0.4 | 15.2 ± 0.9 | 0.023 ± 0.007 |
| ATP | 704 ± 7 | 57 ± 2 | 125 ± 4 | 0.18 ± 0.04 |
The effect of ATP concentration on the ATPase activity was also studied at a saturating concentration of PS (Fig. 6D). Michaelis-Menten kinetics revealed a Km for ATP of 704 ± 7 μm and a Vmax of 57 ± 2 μmol of ATP/min/mg of protein. For comparison with other P4-ATPases purified from natural sources, Atp8a1 from bovine chromaffin granules was reported to have a maximal specific activity of 8 μmol of ATP/min/mg and a Km of 350 μm (18), whereas the protein isolated from human erythrocytes had a specific activity of 790 nmol of ATP/min/mg and a Km of between 200 and 260 μm (45). The kinetic parameters of Atp8a2 are summarized (Table 2).
The effect of ATPase inhibitors on the activity of Atp8a2 was also investigated (supplemental Fig. 2A). The sulfhydryl-modifying reagent NEM at 1 mm and vanadate at 0.1 mm inhibited the ATPase activity of Atp8a2 by over 90%. In contrast, ouabain and azide, inhibitors of Na/K-ATPase and mitochondrial ATPases, respectively, had no effect. Atp8a2 was specific for ATP because GTP was not hydrolyzed by Atp8a2 (supplemental Fig. 2A).
Finally, we have studied the effect of pH on the activity of Atp8a2. Maximum activity was observed between pH 7 and 9 when PS was used as a substrate. In contrast, PE showed a narrow pH optimum at pH 8 with activity dropping to 40% at pH 9 (supplemental Fig. 2B). This loss in activity above pH 8 may be the result of deprotonation of the primary amino group of PE.
Kinetics of Reconstituted Atp8a2
To investigate the activity of Atp8a2 incorporated into a lipid bilayer, Atp8a2 purified from ROS was reconstituted into PC vesicles containing various amounts of PS by a dialysis procedure. These vesicles were unilamellar, as observed by cryoelectron microscopy (data not shown). As in the case of detergent-solubilized enzyme, the ATPase activity of reconstituted Atp8a2 increased, with PS concentration reaching a maximal activity of ~35 μmol of ATP/min/mg of protein for 5 mol % DOPS, a value that is ~25% less than Atp8a2 in CHAPS detergent (supplemental Fig 3A).
We reasoned that the lower ATPase activity of reconstituted Atp8a2 could arise from an unfavorable orientation of a fraction of Atp8a2 in lipid vesicles, such as to exclude ATP. The orientation of Atp8a2 was determined by comparing the effect of limited trypsin digestion of Atp8a2 in hypotonically lysed ROS with that in reconstituted lipid vesicles. As shown in supplemental Fig. 4A, the Atp6C11 epitope was rapidly degraded by trypsin in ROS discs, consistent with the C terminus of Atp8a2 being accessible on the cytoplasmic side of disc membranes. In contrast, the Atp2F6 epitope was largely resistant to trypsin digestion. In the case of Atp8a2 reconstituted into lipid vesicles, trypsin had a more limited effect on the Atp6C11 epitope (supplemental Fig. 4B). Quantification of the Atp6C11 immunolabeling following trypsin treatment suggested that 30% of the reconstituted Atp8a2 was inaccessible to trypsin and correspondingly ATP. Hence, the lower specific activity observed for reconstituted Atp8a2 relative to the detergent-solubilized protein can be accounted for by the observation that only ~70% of reconstituted Atp8a2 is accessible to ATP in the lipid vesicles.
Phospholipid Flippase Activity of Atp8a2
NBD-labeled phospholipids have been previously used to measure lipid flippase activity in various biological systems by fluorescence measurements (9, 41, 42). To determine the feasibility of using this assay for measuring PS flippase activity of Atp8a2, we first determined if NBD-labeled PS could stimulate the ATPase activity of Atp8a2. As shown in supplemental Fig. 3B, both C6 NBD-PS and C12 NBD-PS activated the ATPase activity of Atp8a2 although to a lesser extent than unlabeled PS at the same lipid concentration. C12 NBD-PS was more effective than C6 NBD-PS as a substrate for Atp8a2.
Next, the ability of Atp8a2 to flip NBD-labeled PS was studied in reconstituted liposomes by fluorescence spectroscopy. Reconstituted vesicles containing NBD-labeled PS were incubated with or without ATP. Dithionite was subsequently added to bleach the fluorescence signal resulting from NBD-labeled lipid on the exposed leaflet of the vesicles, whereas the NBD-lipid on the inside is not affected (43). Fig. 7A shows an example of the fluorescence traces for Atp8a2-reconstituted vesicles containing NBD-labeled PS. In the absence of ATP or in the presence of ATP and the ATPase inhibitor NEM, dithionite resulted a 65% decrease in fluorescence, corresponding to the bleaching of NBD-labeled PS on the outer leaflet of the unilamellar liposomes. Atp8a2-containing vesicles pretreated with ATP in the absence of inhibitor showed a larger reduction in fluorescence upon the addition of dithionite. The difference between the ATP-treated and -untreated is a measure of the extent of flipping of NBD-labeled PS from the inner (lumen) to the outer (cytoplasmic) leaflet of the vesicles. The subsequent addition of Triton X-100 resulted in the complete loss of fluorescence due to the accessibility of all NBD-labeled PS to dithionite. Both C6 NBD PS and C12 NBD-PS were transported by Atp8a2 in an ATP-dependent manner. In control studies, lipid vesicles devoid of Atp8a2 showed no ATP-dependent difference in fluorescence (Fig. 7B). No detectable ATP-dependent flipping of NBD-labeled PC or PE was observed (Fig. 7C). The inability to observe NBD-labeled PE flipping may be due to the lack in sensitivity of this assay or the inability of NBD-labeled PE to serve as a substrate for Atp8a2. Finally, the transport of NBD-labeled PS was inhibited by unlabeled PS (DOPS), supporting the physiological significance of the observed Atp8a2 flippase activity (Fig. 7D). The finding that equimolar DOPS inhibits NBD-PS flippase greater than 75% is consistent with DOPS being a better substrate than NBD-PS, as found for the PS-activated ATPase activity of Atp8a2 (supplemental Fig 3B).
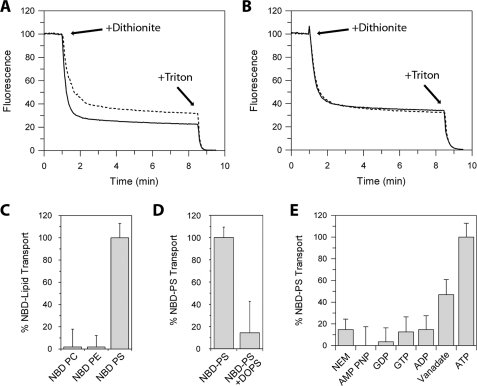
Transport or flippase activity of Atp8a2. Loss in NBD fluorescence upon the addition of 2 mm dithionite (+Dithionite) to Atp8a2 proteoliposomes containing C12 NBD-PS pretreated with ATP (solid line) or ATP + NEM (dotted line) (A) and empty liposomes containing C12 NBD-PS pretreated with ATP (solid line) or ATP + NEM (dotted line) (B). The difference in the fluorescence between samples containing ATP and samples containing ATP + NEM (or no ATP) reflects the transport activity. Triton X-100 (1%) was added at the end of the trace to make all C12 NBD-PS accessible to bleaching by dithionite. C, transport activity of C6 NBD-PC, -PE, and -PS. D, transport activity of C12 NBD-PS in the absence or the presence of an equimolar amount of DOPS (2.5%). E, effect of pretreating Atp8a2 proteoliposomes with various nucleotides (AMP-PNP, GDP, GTP, ADP, and ATP) and inhibitors (NEM and vanadate) on the transport of C12 NBD-PS. NBD-labeled PS is transported by Atp8a2 in an ATP-dependent manner from the inside of the vesicle (corresponding to the exocytoplasmic side) to the outside of vesicles (corresponding to the cytoplasmic side of cell membranes). The transport of NBD-labeled PS is inhibited by PS, the physiological substrate of Atp8a2.
The effect of various nucleotides and ATPase inhibitors on the Atp8a2 flippase activity was also studied. Little or no Atp8a2 flippase activity was observed when AMP-PNP, GDP, GTP, or ADP was used in place of ATP (Fig. 7E). The addition of NEM and to a lesser extent vanadate reduced the ATP-dependent flippase activity of Atp8a2.
DISCUSSION
P4-ATPases constitute a relatively new subfamily of P-type ATPases. Fourteen members of this subfamily have been identified in mammals and five in yeast based on sequence similarities (7, 11, 28). However, despite studies linking a number of P4-ATPases to important cellular processes and inherited human diseases (4, 7), relatively little is known about the biochemical properties of most members of this subfamily of P-type ATPases.
In this study, we have examined the expression, subcellular localization, and biochemical activities of Atp8a2, a member of the P4-ATPase subfamily that shares a 67% identity in amino acid sequence with Atp8a1, the first cloned member of this subfamily. Unlike most other members of the P4-ATPase subfamily, which are widely expressed in different tissues (7), atp8a2 shows a restricted tissue distribution in adult mice with high expression in the retina as well as the testis. Within the retina, atp8a2 mRNA expression was primarily observed in photoreceptor cells, and the protein was localized to disc membranes of rod and cone outer segments, as revealed by immunocytochemical and subcellular fractionation techniques. The presence of Atp8a2 in disc membranes of photoreceptor outer segments is consistent with our recent proteomic screen in which Atp8a2 was identified as a low abundant protein in outer segment disc membrane preparations (27).
To gain insight into the functional properties of Atp8a2, we have developed a simple and efficient single-step immunoaffinity purification procedure to isolate Atp8a2 from detergent-solubilized ROS membranes. This preparation displayed a single intense 130-kDa protein corresponding to Atp8a2 and was essentially free of other proteins, as analyzed by Coomassie Blue staining of SDS gels. Several other P4-ATPases have recently been reported to form a heteromeric complex with members of the Cdc50/Lem3p family of membrane proteins (4, 7, 46, 47). Cdc50 proteins contain two transmembrane segments that are separated by a heavily N-glycosylated exocytoplasmic domain. Recent studies suggest that these proteins play important roles in the stability and trafficking of P4-ATPases out of the endoplasmic reticulum as well as in the proper functioning of these transporters (4, 7, 47). Cdc50 proteins have been considered to be the counterpart of the β-subunit of the Na/K-ATPase oligomeric complex. Cdc50a was detected in our recent proteomic analysis of outer segments (27). It is possible that the Cdc50a associates with Atp8a2 and is present in our immunoaffinity-purified preparations but not readily detected by conventional gel staining techniques. Alternatively, Atp8a2 may not require an accessory subunit for expression and activity. The generation of suitable anti-Cdc50 antibodies should clarify whether Atp8a2 exists as a heteromeric complex with Cdc50a or another member of the Cdc50 family of proteins.
The catalytic and lipid transport properties of purified and reconstituted Atp8a2 were examined. The addition of phospholipid together with DTT was required during detergent solubilization and purification in order to stabilize the protein in its functional state. PC was typically used because this phospholipid does not function as a substrate for Atp8a2. Kinetic studies showed that PS activated the ATPase activity of purified Atp8a2 in detergent and after reconstitution into liposomes. An increase in ATPase activity was also observed for PE, but this aminophospholipid was 60 times less effective than PS as a substrate. In agreement with other P-type ATPases, both NEM and vanadate inhibited the enzymatic activity of Atp8a2. Hence, the substrate specificity and inhibition properties of Atp8a2 resemble those of Atp8a1, although the specific activity of Atp8a2 measured in this study is generally greater than that reported for Atp8a1 (18).
Although aminophospholipid translocase activity has been inferred for a number of P4-ATPases from both genetic and PS/PE-dependent ATPase activation studies, the phospholipid flippase activity of purified and reconstituted P4-ATPases has not been reported previously. In this study, we have used NBD-labeled PS to measure the flippase activity of purified Atp8a2 reconstituted into lipid vesicles. Our studies using a fluorescence-based assay (42) indicate that Atp8a2 is able to translocate PS from the inner leaflet (equivalent to the exocytoplasmic side of biological membranes) to the outer leaflet (cytoplasmic side) of vesicles in an ATP-dependent manner. This process requires energy derived from ATP hydrolysis because ADP, AMP-PNP, and GTP were ineffective as nucleotide substrates. Together, these studies provide direct evidence for the role of Atp8a2 as an aminophospholipid translocase with a strong preference for PS as the transported phospholipid substrate.
Previous studies have shown that ROS disc membranes display lipid asymmetry, with PS and PE preferentially localized on the cytoplasmic leaflet (25, 26). Our studies showing that Atp8a2 is present in disc membranes support the view that this P4-ATPase functions in the generation and maintenance of transbilayer asymmetry with respect to PS and possibly PE. The specific role of PS/PE asymmetry in disc membranes is not known. However, we speculate that the negatively charged surface of disc membranes contributed by PS may play roles in generating and stabilizing the structure of disc membranes either by inducing non-random lipid distributions within the bilayer or facilitating interactions with cytoskeletal or soluble proteins, thereby inducing the rim region of disc membranes (48). PS asymmetry may also be important in modulating the interaction of enzymes of the visual cascade with disc membranes (49). Finally, it is possible that PS asymmetry may play a role in vesicle transport/fusion and phagocytosis of outer segment discs by RPE cells as part of the outer segment renewal process.
The importance of Atp8a2 in photoreceptor outer segments is supported by analysis of ATPase activities of outer segments. Although Atp8a2 makes up less than 0.1% of the total membrane protein of outer segments, it is one of the most active ATPases in outer segment preparations. Analysis of outer segment ATPase activity before and after removal of Atp8a2 suggests that Atp8a2 is responsible for over 50% of the basal ATPase activity of outer segments. The Na/K-ATPase that is a major contributor to total ATPase activity in most cells is not present in outer segment preparations (27, 50). ABCA4, an ATP transporter implicated in the flipping of N-retinylidene-PE across disc membranes (51,–53), is present in outer segments in greater abundance than Atp8a2 but displays a specific activity that is several orders of magnitude lower than Atp8a2 (54). Hence, Atp8a2 most likely consumes a considerable amount of the ATP of outer segments to maintain aminophospholipid asymmetry. Atp8a2 is needed to maintain this lipid asymmetry over the life of the outer segment, typically 10 days.
It will be important to generate a mouse deficient in Atp8a2 to further evaluate its role in photoreceptor structure and function. Likewise, it is of interest to determine if mutations in atp8a2 are responsible for human retinal degenerative diseases. Although our data strongly suggest an important role for Atp8a2 as an aminophospholipid translocase in photoreceptors, Atp8a2 may also play an important role in reproductive and developmental biology because Atp8a2 is expressed at high levels in the testes and during early sperm development.
In summary, our studies show that Atp8a2 is present in the disc membranes of rod and cone photoreceptors. ATPase and flippase measurements provide direct evidence for the function of Atp8a2 as an aminophospholipid translocase with a high specificity for PS. These studies support the role of this member of the P4-ATPase subfamily in the generation and maintenance of phospholipid asymmetry in photoreceptor disc membranes.
Acknowledgments
We thank Laurie Molday and Theresa Hii for technical assistance and Dr. Frank Dyka and Dr. Seiffolah Azadi for helpful discussions related to the cloning of atp8a2 and in situ hybridization.
*This work was supported, in whole or in part, by National Institutes of Health Grant EY 02422. This work was also supported by Canadian Institutes of Health Research Grant MT 5822.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
3The abbreviations used are:
- PS
- phosphatidylserine
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phosphoserine
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- GST
- glutathionine S-transferase
- AMP-PNP
- adenylyl-imidodiphosphate
- ROS
- rod outer segment
- NEM
- N-ethylmaleimide
- DTT
- dithiothreitol
- PB
- phosphate buffer
- PBS
- phosphate-buffered saline
- DIG
- digoxigenin
- RT
- reverse transcription
- NBD
- 7-nitrobenz-2-oxa-1,3-diazol-4-yl
- C6 NBD-PS
- 1-oleoyl-2-{6-[7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoserine
- C6 NBD-PE
- 1-oleoyl-2-{6-[7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoethanolamine
- C6 NBD-PC
- 1-oleoyl-2-{6-[7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine
- C12 NBD-PS
- 1-oleoyl-2-{12-[7-nitro-2–1,3- benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoserine.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m109.047415
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/content/284/47/32670.full.pdf
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/284/47/32670
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/content/full/284/47/32670
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/284/47/32670.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Membrane structure-responsive lipid scrambling by TMEM63B to control plasma membrane lipid distribution.
Nat Struct Mol Biol, 18 Oct 2024
Cited by: 0 articles | PMID: 39424995
Role of Enzymes Capable of Transporting Phosphatidylserine in Brain Development and Brain Diseases.
ACS Omega, 9(32):34243-34249, 01 Aug 2024
Cited by: 0 articles | PMID: 39157110 | PMCID: PMC11325426
Review Free full text in Europe PMC
The lipid flippase ATP8A1 regulates the recruitment of ARF effectors to the trans-Golgi Network.
Arch Biochem Biophys, 758:110049, 13 Jun 2024
Cited by: 0 articles | PMID: 38879142
Functional and in silico analysis of ATP8A2 and other P4-ATPase variants associated with human genetic diseases.
Dis Model Mech, 17(6):dmm050546, 24 Apr 2024
Cited by: 3 articles | PMID: 38436085 | PMCID: PMC11073571
Direct evidence of lipid transport by the Drs2-Cdc50 flippase upon truncation of its terminal regions.
Protein Sci, 33(3):e4855, 08 Dec 2023
Cited by: 1 article | PMID: 38063271 | PMCID: PMC10895448
Go to all (120) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - GQ303567
RefSeq - NCBI Reference Sequence Database (2)
- (1 citation) RefSeq - NM_016529.4
- (1 citation) RefSeq - NM_015803.2
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2.
J Biol Chem, 286(19):17205-17216, 18 Mar 2011
Cited by: 86 articles | PMID: 21454556 | PMCID: PMC3089563
Phospholipid flippase ATP8A2 is required for normal visual and auditory function and photoreceptor and spiral ganglion cell survival.
J Cell Sci, 127(pt 5):1138-1149, 10 Jan 2014
Cited by: 39 articles | PMID: 24413176 | PMCID: PMC3937779
Phosphatidylserine flipping by the P4-ATPase ATP8A2 is electrogenic.
Proc Natl Acad Sci U S A, 116(33):16332-16337, 01 Aug 2019
Cited by: 13 articles | PMID: 31371510 | PMCID: PMC6697784
Substrates of P4-ATPases: beyond aminophospholipids (phosphatidylserine and phosphatidylethanolamine).
FASEB J, 33(3):3087-3096, 03 Dec 2018
Cited by: 31 articles | PMID: 30509129
Review
Funding
Funders who supported this work.
CIHR (1)
Grant ID: MT 5822
NEI NIH HHS (2)
Grant ID: R01 EY002422
Grant ID: EY 02422





