Abstract
Free full text

Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells
Abstract
We reported previously that well-characterized enhancers but not promoters for typical tissue-specific genes, including the classic Alb1 gene, contain unmethylated CpG dinucleotides and evidence of pioneer factor interactions in embryonic stem (ES) cells. These properties, which are distinct from the bivalent histone modification domains that characterize the promoters of genes involved in developmental decisions, raise the possibility that genes expressed only in differentiated cells may need to be marked at the pluripotent stage. Here, we demonstrate that the forkhead family member FoxD3 is essential for the unmethylated mark observed at the Alb1 enhancer in ES cells, with FoxA1 replacing FoxD3 following differentiation into endoderm. Up-regulation of FoxD3 and loss of CpG methylation at the Alb1 enhancer accompanied the reprogramming of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (iPS) cells. Studies of two genes expressed in specific hematopoietic lineages revealed that the establishment of enhancer marks in ES cells and iPS cells can be regulated both positively and negatively. Furthermore, the absence of a pre-established mark consistently resulted in resistance to transcriptional activation in the repressive chromatin environment that characterizes differentiated cells. These results support the hypothesis that pluripotency and successful reprogramming may be critically dependent on the marking of enhancers for many or all tissue-specific genes.
Recent studies have revealed key properties of embryonic stem (ES) cells that may be important for their self-renewal and pluripotency. Foremost among these properties are the expression of a small number of transcriptional regulators—including Oct4, Sox2, and Nanog—that are central components of a pluripotency network (Boyer et al. 2005; Loh et al. 2006; J Wang et al. 2006; Jaenisch and Young 2008). These factors promote self-renewal and suppress differentiation, while contributing to the poised state of genes that will ultimately regulate differentiation toward specific lineages. Genes involved in early developmental decisions are often characterized by the presence of bivalent histone modification domains, consisting of repressive histone modifications combined with modifications associated with transcriptional activation (Bernstein et al. 2006; Boyer et al. 2006; Lee et al. 2006). Notably, the reprogramming of differentiated cells into induced pluripotent stem (iPS) cells is accompanied by the establishment of a histone modification profile mirroring that found in ES cells (Takahashi and Yamanaka 2006; Maherali et al. 2007; Takahashi et al. 2007; Wernig et al. 2007; Yu et al. 2007; Mikkelsen et al. 2008; Park et al. 2008).
ES cells are also characterized by hyperdynamic chromatin. Heterochromatin protein 1 (HP1), histone H1, and core histones are associated with chromatin less tightly in ES cells than in differentiated cells (Meshorer and Misteli 2006; Meshorer et al. 2006). Moreover, transcripts from a large number of tissue-specific genes can be detected in ES, although often at very low levels (Guenther et al. 2007; Efroni et al. 2008). Recently, the chromatin remodeling factor Chd1 was found to be required for the formation of this hyperdynamic chromatin state and for pluripotency (Gaspar-Maia et al. 2009). These properties are thought to be important for developmental plasticity, although the precise relevance of hyperdynamic chromatin in pluripotent cells remains unknown.
Although most studies of pluripotency have focused on genes encoding regulators of early developmental decisions, typical tissue-specific genes may also possess important epigenetic marks in ES cells. Szutorisz et al. (2005) reported that histone acetylation and H3 Lys 4 (H3K4) methylation at the B-lineage-specific Igll1/VpreB locus may be nucleated at a specific distant site in ES cells, with the modifications spreading through the locus during B-cell differentiation.
More recently, we found that well-characterized enhancers for representative tissue-specific genes possess windows of unmethylated CpGs in ES cells, long before the genes are transcribed (Xu et al. 2007). In contrast, the promoters of these genes appear to be fully methylated in pluripotent cells. For example, at the liver-specific Alb1 enhancer, an unmethylated CpG was observed in ES cells that coincided with a recognition site for FoxA1. FoxA1 binds the enhancer in endoderm and acts as a pioneer factor by enabling chromatin remodeling and transcriptional activation upon liver specification (Gualdi et al. 1996; Bossard and Zaret 1998; Cirillo et al. 2002). However, FoxA1 is not expressed in ES cells.
Unmethylated CpGs were also observed in ES cells at a tissue-specific enhancer for the macrophage/dendritic cell-specific Il12b gene, which encodes the p40 subunit of interleukin-12 (IL-12) and IL-23. This enhancer exhibits DNase I hypersensitivity only in terminally differentiated macrophages stimulated with microbial products, such as lipopolysaccharide (LPS) (Zhou et al. 2004). Macrophage activation is also accompanied by increased histone acetylation and H3K4 methylation at the enhancer, as well as by the recruitment of SWI/SNF remodeling complexes and specific transcription factors (Zhou et al. 2007). These observations suggested that chromatin at the Il12b enhancer is unperturbed until mature macrophages are activated. However, a pronounced window of unmethylated CpGs was observed in unstimulated macrophages, as well as in ES cells, hematopoietic progenitors, and nonhematopoietic tissues, suggesting that the enhancer is initially marked at the pluripotent stage (Xu et al. 2007).
A third tissue-specific enhancer found to contain an unmethylated window in ES cells is associated with the thymocyte-specific Ptcra gene, which encodes the pre-Tα protein. This enhancer was responsible for the thymocyte specificity of Ptcra transcription in both conventional and bacterial artificial chromosome (BAC) transgenic mice (Reizis and Leder 2001). Despite thymocyte-specific function and DNase I hypersensitivity, the Ptcra enhancer, like the Alb1 and Il12b enhancers, possesses a window of unmethylated CpG dinucleotides in ES cells and most other cell types (Xu et al. 2007).
Further examination of the Ptcra gene provided initial evidence that the ES cell marks at tissue-specific enhancers may be important for transcriptional activation in differentiated cells (Xu et al. 2007). When a plasmid containing the Ptcra enhancer and promoter upstream of a reporter gene was premethylated and stably transfected into ES cells, the unmethylated window at the enhancer was readily detected when individual clones were selected and examined by bisulfite sequencing. However, this same premethylated plasmid remained fully methylated and silent upon stable transfection into a thymocyte cell line that contains all factors required for efficient transcription of the endogenous Ptcra gene. These results suggested that enhancer marks are readily established in pluripotent cells, but that tissue-specific genes lacking pre-existing enhancer marks may be resistant to activation in differentiated cells.
In this study, we identified DNA motifs and transcription factors responsible for the establishment of enhancer marks at representative genes, and we examined the significance of the marks in both ES cells and iPS cells. The results provide strong support for a model in which the marking of tissue-specific enhancers is an important property of the pluripotent state, with susceptibility to establishment of these marks distinguishing ES and iPS cells from differentiated cells. The results further suggest that one reason ES cells may require a hyperdynamic chromatin environment is to allow the establishment of these critical enhancer marks.
Results
FoxD3 maintains an unmethylated CpG mark at the Alb1 enhancer in ES cells
The unmethylated CpG observed at the Alb1 enhancer in ES cells is located within one of two enhancer motifs known to bind FoxA1 in definitive gut endoderm (−10,695 in Fig. 1G; Cirillo et al. 2002; Xu et al. 2007). FoxA1 is not expressed in ES cells, but a previous chromatin immunoprecipitation (ChIP) experiment suggested that FoxD3 may bind the Alb1 enhancer in these cells (Xu et al. 2007). FoxD3 is abundantly expressed in ES cells, is necessary for their pluripotency, and is down-regulated upon differentiation into embryoid bodies (Fig. 1A; Clevidence et al. 1993; Sutton et al. 1996; Hanna et al. 2002).
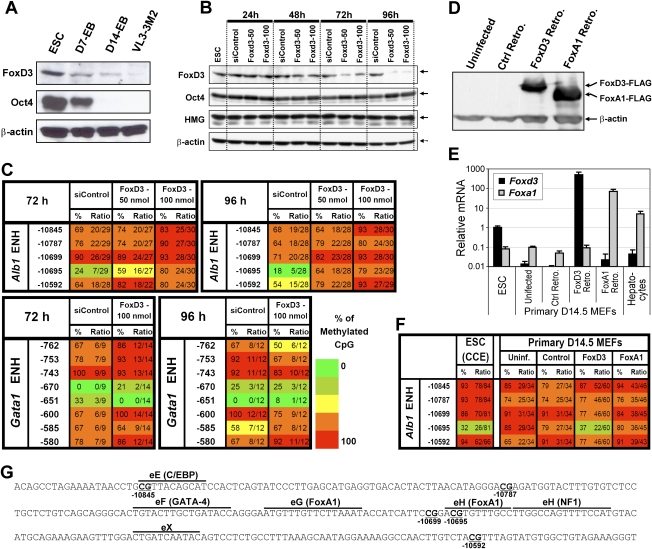
FoxD3 regulates the Alb1 enhancer mark in ES cells. (A) FoxD3 and Oct4 protein levels were monitored by Western blot using whole-cell extracts prepared from CCE ES cells, day 7 and day 14 embryoid bodies (EBs), and VL3-3M2 thymocytes (as a negative control). β-Actin was analyzed as a loading control. (B) FoxD3 was depleted from ES cells transfected with 50 or 100 nmol of a Foxd3 siRNA pool (Dharmacon). In parallel experiments, ES cells were transfected with a nontargeting control siRNA pool (siControl; Dharmacon). At four time points after transfection, knockdown efficiency was monitored by Western blot using FoxD3 and Oct4 antibodies. HMG1 and β-actin were analyzed as controls. (C) Methylation state of the Alb1 enhancer was monitored by bisulfite sequencing. A region of a Gata1 enhancer that also contains unmethylated CpGs in ES cells was monitored as a control. Results are shown for DNA isolated at two different time points after siRNA transfection. The locations of the CpGs relative to the Alb1 or Gata1 start site are shown at the left. The percentage of plasmid clones that exhibited CpG methylation at each position in the bisulfite sequencing analysis is indicated, along with the number of methylated clones and the total number of clones analyzed (presented as a ratio). Methylation levels are represented in a gradation of colors: 0%–20% (dark green), 21%–40% (light green), 41%–60% (yellow), 61%–80% (orange), and 81%–100% (red). (D) FoxD3 and FoxA1 were overexpressed in day 14.5 MEFs by retroviral transduction. Both proteins were expressed with a Flag epitope at their C terminus. MEFs were also transduced with a control retrovirus containing an EGFP cDNA. The expression of FoxD3 and FoxA1 proteins was monitored by Western blot using an anti-Flag antibody. (E) Foxd3 and Foxa1 mRNA levels were monitored by real-time RT–PCR in uninfected ES cells, day 14.5 MEFs, and hepatocytes, as well as day 14.5 MEFs transduced with a control retrovirus or retroviruses containing Foxd3 or Foxa1 expression cassettes. (F) Bisulfite sequencing was used to examine DNA methylation at the Alb1 enhancer in untransduced MEFs and in MEFs transduced with a control retrovirus or retroviruses expressing FoxD3 or FoxA1. Results obtained with CCE ES cells are shown for comparison. (G) The sequence of the Alb1 enhancer (−10,566 to −10,895) is shown. Known transcription factor-binding sites are marked above the sequence, and CpGs are underlined.
To determine whether FoxD3 is responsible for the unmethylated CpG, FoxD3 expression was reduced in ES cells by transfection of an siRNA pool that specifically targets the Foxd3 transcript. The Foxd3 siRNA reduced the concentrations of FoxD3, but not the concentrations of Oct4, Sox2, or Nanog (Fig. 1B; data not shown). Interestingly, bisulfite sequencing analysis revealed that methylation of the Alb1 enhancer CpG increased substantially following FoxD3 knockdown, especially at late time points when FoxD3 concentrations were most strongly reduced (Fig. 1C). In contrast, FoxD3 knockdown had no effect on an unmethylated CpG mark at a tissue-specific enhancer for the Gata1 gene (Fig. 1C, bottom). Similar results were found when two individual Foxd3 siRNAs were tested independently (Supplemental Fig. 1A). Thus, FoxD3 is required for maintenance of the unmethylated CpG at the Alb1 enhancer in ES cells.
Ectopically expressed FoxD3 promotes methylation loss at the Alb1 enhancer in mouse embryonic fibroblasts (MEFs)
We next asked whether FoxD3 was sufficient for establishment of the unmethylated CpG mark at the endogenous Alb1 enhancer when overexpressed in differentiated cells. This experiment was performed with primary day 14.5 MEFs in which the endogenous Alb1 enhancer was fully methylated (see Fig. 1F), consistent with previous evidence that the enhancer is fully methylated in most primary mouse tissues that do not express Alb1 (Xu et al. 2007). FoxD3 and FoxA1, both tagged at their C terminus with a Flag epitope, were overexpressed in the MEFs by transduction of recombinant retroviruses, resulting in expression at comparable concentrations (Fig. 1D). Foxd3 mRNA was substantially more abundant in MEFs transduced with the Foxd3 retrovirus than in untransduced ES cells (Fig. 1E).
Bisulfite sequencing performed 4 d after retroviral transduction revealed that ectopically expressed FoxD3 promoted the efficient loss of methylation at the Alb1 enhancer CpG at nucleotide −10,695 (Fig. 1F; see Supplemental Fig. 1 for five independent experiments). Surprisingly, ectopically expressed FoxA1 failed to promote the loss of methylation, suggesting that this property of FoxD3 is not shared by all members of the Fox family.
Establishment of the Alb1 enhancer mark during reprogramming of MEFs into iPS cells
Recent studies have demonstrated that differentiated cells can be reprogrammed to iPS cells by expression of a cocktail of defined transcription factors (Takahashi and Yamanaka 2006; Maherali et al. 2007; Takahashi et al. 2007; Wernig et al. 2007; Yu et al. 2007; Park et al. 2008). The availability of iPS cells and the parental MEFs from which they were generated provides an opportunity to examine the extent to which the Alb1 enhancer mark correlates with pluripotency.
Bisulfite sequencing of DNA from two iPS lines (1A2 resorted and 2D4) (Maherali et al. 2007) and from the parental MEFs revealed that the Alb1 enhancer CpG at −10,695 was converted from a fully methylated state in MEFs to an unmethylated state in the iPS cells (Fig. 2A). Interestingly, partial loss of methylation was also observed at two flanking CpGs in the iPS cells, whereas these CpGs were fully methylated in ES cells. Consistent with the loss of methylation at the Alb1 enhancer CpG, Foxd3 transcripts were much more abundant in the iPS cell lines than the MEFs (Fig. 2B).
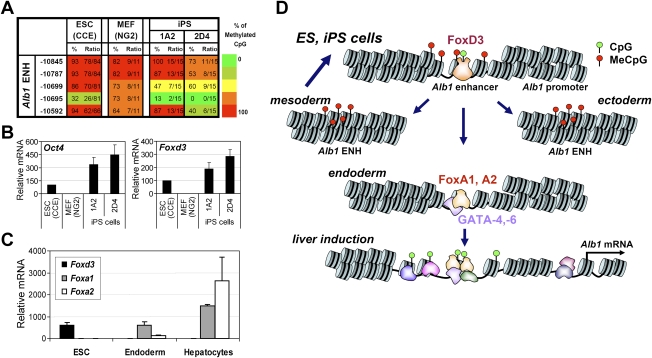
Loss of methylation at an Alb1 enhancer CpG accompanies reprogramming. (A) Methylation at the Alb1 enhancer was monitored by bisulfite sequencing in primary NG2 MEFs and in two iPS lines derived from the NG2 MEFs, 1A2 resorted and 2D4. (Left) Methylation levels observed in CCE ES cells are shown for comparison. (B) Oct4 and Foxd3 mRNA levels were monitored by real-time RT–PCR in CCE ES cells, NG2 MEFs, and the 1A2 resorted and 2D4 iPS lines. (C) Foxd3, Foxa1, and Foxa2 mRNA levels were monitored by real-time RT–PCR in CCE ES cells, definitive endoderm (FoxA2+ FoxA3+) obtained by in vitro differentiation, and mature hepatocytes. Transcript levels were normalized against Gapd transcript levels. (D) A model of Alb1 gene activation during liver development is shown (see the text).
To gain further insight into the relationship between the Alb1 enhancer mark and the expression of FoxD3, FoxA1, and FoxA2, transcript levels for the three Fox family genes were analyzed in ES cells, definitive endoderm (FoxA2+ FoxA3+) obtained by in vitro differentiation (Gadue et al. 2006, 2009; Gouon-Evans et al. 2006), and mature hepatocytes. Foxd3 transcripts were abundant in ES cells, but were greatly reduced in endoderm and hepatocytes (Fig. 2C). In contrast, Foxa1 and Foxa2 transcripts were absent in ES cells, but were strongly up-regulated in endoderm and hepatocytes.
These results support the hypothesis that FoxD3 gains access to the Alb1 enhancer in ES cells and is responsible for the efficient loss of methylation at a single CpG (Fig. 2D). FoxD3 may serve as a “placeholder” for FoxA1 and FoxA2, which are up-regulated during endoderm differentiation and bind the Alb1 enhancer after FoxD3 is down-regulated. Then, as documented in previous studies, FoxA1 initiates a cascade of events that culminates in extensive chromatin accessibility, the binding of numerous transcription factors, and transcriptional activation in hepatocytes (Fig. 2D; Gualdi et al. 1996; Bossard and Zaret 1998; Cirillo et al. 2002).
iPS cells, but not MEFs, are permissive for establishment of the Ptcra enhancer mark
We next investigated the utility of protein–DNA interactions at a tissue-specific enhancer in pluripotent cells. One possibility is that, in the absence of a mark established in pluripotent cells, loci may become assembled into a repressive chromatin structure in differentiated cells that is resistant to activation. Enhancer marks may be readily established in pluripotent cells due to the specialized properties of transcription factors expressed in ES cells, including FoxD3, combined with the hyperdynamic state of chromatin in pluripotent cells.
To examine the relationship between chromatin structure and the establishment of enhancer marks at tissue-specific genes, we focused on the Ptcra and Il12b enhancers. As described above, we found previously that premethylation of a Ptcra enhancer–promoter–reporter–insulator plasmid conferred resistance to the establishment of an unmethylated CpG window at the enhancer in a thymocyte line that efficiently expresses the endogenous Ptcra gene. In contrast, ES cells were readily susceptible to establishment of the unmethylated window, providing initial evidence that enhancer marks are readily established during pluripotency and may be necessary for transcriptional activation in differentiated cells. However, one major deficiency of this experiment was that it involved a comparison between an ES cell line and an unrelated transformed thymocyte line. Therefore, the variable behavior of the premethylated plasmid in the two lines may have reflected the difference between an untransformed and a transformed line, rather than a difference between pluripotent cells and differentiated cells.
The availability of iPS cells and the primary parental MEFs from which they were generated provides an opportunity to more carefully examine the relationship between pluripotency and susceptibility to the establishment of enhancer marks. Toward this end, the Ptcra enhancer–promoter–reporter–insulator plasmids were premethylated with the SssI CpG methylase and were stably transfected into iPS cells and the parental MEFs. Several stably transfected MEF and iPS cell clones were selected, and DNA methylation at the integrated enhancer was analyzed by bisulfite sequencing (Fig. 3A).
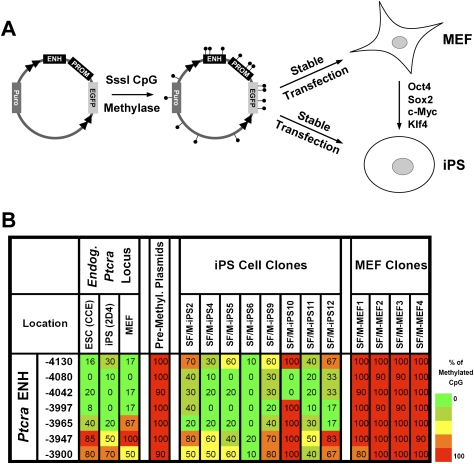
Reprogramming of MEFs into iPS cells is accompanied by increased susceptibility to the establishment of an unmethylated window at the Ptcra enhancer. (A) The Ptcra enhancer–promoter–reporter–insulator construct was premethylated and stably transfected into the 2D4 iPS cell line or the parental MEFs. (B) Several individual iPS and MEF clones were analyzed by bisulfite sequencing. Primers were designed to amplify only the stably integrated transgenes but not the endogenous alleles. Methylation profiles of the endogenous Ptcra enhancer in CCE ES cells, iPS 2D4, and MEF cells are also shown, along with a profile of the methylated plasmid prior to transfection.
The results revealed that the iPS cells readily supported establishment of the unmethylated window at the Ptcra enhancer (Fig. 3B). The precise CpGs converted to an unmethylated state varied from clone to clone, as observed previously in ES cells (Xu et al. 2007), but general sensitivity to loss of enhancer methylation was consistently observed. Methylation was not lost at two other regions of the integrated plasmid (data not shown). In striking contrast, the same premethylated plasmid remained fully methylated and silent in the MEFs (Fig. 3B). Thus, the generation of iPS cells from MEFs is accompanied by a dramatic increase in susceptibility to establishment of an unmethylated window at the premethylated Ptcra enhancer.
The results in Figure 3B (left) further reveal that, in primary MEFs, the endogenous Ptcra enhancer is unmethylated, despite the strong resistance of the premethylated enhancer to the loss of methylation. This observation suggests that the unmethylated state of the endogenous Ptcra enhancer in MEFs was established at an earlier stage of embryogenesis and was maintained through differentiation, even though the Ptcra gene is not expressed in MEFs. Together, these findings support the hypothesis that tissue-specific enhancers become marked in pluripotent cells for the purpose of preventing assembly of the enhancer into repressive chromatin that is resistant to transcriptional activation in differentiated cells (see below). Although the early marking of the Ptcra enhancer may have no functional significance in MEFs, it may be critical for Ptcra transcriptional competence in thymocytes.
iPS cells and ES cells are permissive for establishment of the Il12b enhancer mark, whereas MEFs, macrophages, and myeloid progenitors are resistant
We next asked whether the Ptcra results are relevant to other tissue-specific genes by performing analogous experiments with the macrophage/dendritic cell-specific Il12b enhancer (see above). A premethylated plasmid containing the Il12b enhancer upstream of the Il12b promoter and GFP reporter gene (Fig. 4A), and flanked by chicken β-globin insulator sequences, was stably transfected into iPS cells and the parental MEFs. Bisulfite sequencing revealed that an unmethylated window was readily established at the transfected Il12b enhancer in iPS cells (Fig. 4B). In contrast, the Il12b enhancer, like the Ptcra enhancer, remained resistant to establishment of an unmethylated window in MEFs.
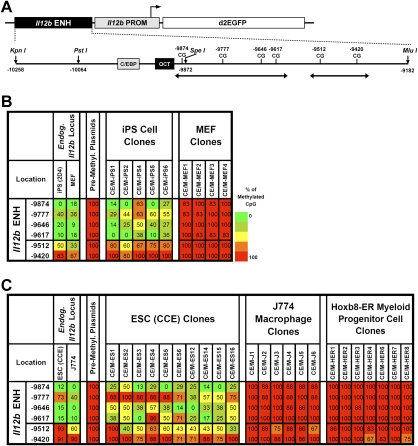
Selective establishment of an unmethylated window at the Il12b enhancer. (A) A diagram of the Il12b enhancer–promoter–EGFP plasmid is shown. A 1.1-kb Il12b enhancer fragment and 0.4-kb promoter fragment were inserted upstream of a destabilized EGFP reporter (pd2EGFP). The cloned Il12b enhancer contains six CpGs (−9874 to −9420). Putative transcription factor-binding sites are shown as shaded boxes. Regions analyzed by bisulfite sequencing are indicated by double-headed arrows. (B) Methylation profiles for five iPS cell clones and four MEF clones transfected with the premethylated Il12b plasmid are shown. Methylation profiles of the endogenous Il12b enhancer in MEFs and iPS 2D4 cells, and of the premethylated Il12b plasmid prior to transfection, are also shown. (C) Methylation profiles of 10 ES cell clones, six J774 macrophage clones, and seven Hoxb8-ER myeloid progenitor cell clones transfected with premethylated Il12b plasmid are shown. Methylation profiles of the endogenous Il12b enhancer in CCE ES cells and J774 cells, and of the premethylated Il12b plasmid before transfection, are also shown.
Susceptibility to establishment of the Il12b enhancer mark was also observed in stably transfected ES cells (Fig. 4C). However, the J774 macrophage line, which actively expresses the endogenous Il12b gene following LPS stimulation, was resistant to establishment of the Il12b enhancer mark at a stably transfected premethylated plasmid (Fig. 4C). Consistent with the resistance to loss of methylation, the premethylated plasmid remained transcriptionally silent following LPS stimulation of the J774 clones (data not shown).
To determine the stage of development at which cells lose their susceptibility to the establishment of an enhancer mark, we examined primary myeloid progenitor cells that are maintained at an early developmental stage by expression of a Hoxb8-ER fusion protein (GG Wang et al. 2006). Interestingly, the premethylated Il12b enhancer plasmid was found to be strongly resistant to establishment of the unmethylated window in these cells, suggesting that susceptibility is lost at a very early stage of development (Fig. 4C, right).
Establishment of an Il12b enhancer mark on a premethylated BAC transgene in ES cells
To determine whether enhancer marks can also be established in ES cells when an enhancer is in a more native context, we examined a 191-kb BAC spanning the Il12b locus (Fig. 5A). An EGFP cDNA was introduced into the second exon of the Il12b gene within the Il12b BAC by homologous recombination in Escherichia coli. Short DNA sequence tags were also introduced by homologous recombination adjacent to both the Il12b promoter and enhancer; these tags allowed us to distinguish the BAC integrants from the endogenous Il12b alleles in bisulfite sequencing experiments.
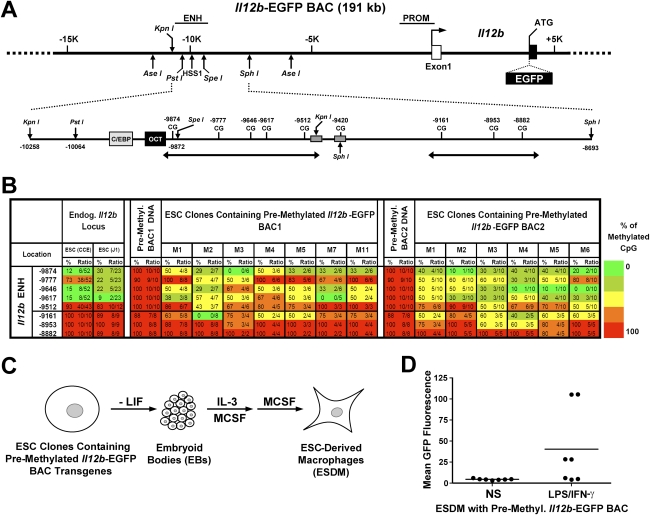
Establishment of the Il12b enhancer mark in a premethylated BAC. (A) A diagram of the 191-kb Il12b-EGFP BAC is shown, along with the Il12b promoter and inducible −10-kb enhancer, 5′ untranslated exon (open box), coding exon 2 (filled box), restriction enzyme recognition sites, and EGFP cDNA insertion site. (B) Methylation profiles are shown of ES cell clones stably transfected with two different premethylated Il12b-EGFP BACs. BAC1 and BAC2 differ in the locations of short DNA sequence tags inserted near the enhancer by homologous recombination in E. coli. The tags are used to distinguish the BAC transgenes from the endogenous Il12b locus in the bisulfite sequencing experiments. As controls, the methylation profiles of the premethylated BAC DNAs prior to transfection are shown. For comparison, the methylation profiles of the endogenous Il12b enhancer in CCE and J1 ES cells are shown at the left. (C) The strategy used to differentiate ES cell clones containing premethylated Il12b-EGFP BACs into macrophages is diagrammed. (D) GFP expression in ES cell-derived macrophages (ESDM) generated from several clonal ES cell lines containing a premethylated Il12b-EGFP BAC was measured by flow cytometry before stimulation (NS) and after stimulation with LPS + IFN-γ.
To determine whether the Il12b enhancer marks can be established at the modified Il12b-EGFP BAC in ES cells, we premethylated the BAC and stably transfected it into ES cells. Despite the large size of the BAC, the in vitro methylation was extremely efficient (Fig. 5B, see columns 3 and 11 for results obtained with two different versions of the Il12b-EGFP BAC containing different 6-base-pair [bp] sequence tags near the enhancer). After stable transfection of the premethylated BAC and the selection of clonal lines, bisulfite sequencing revealed the establishment of an unmethylated window at the Il12b enhancer (Fig. 5B). This unmethylated window was similar to that observed at the endogenous Il12b enhancer in ES cells, although it was broader in some clones. Thus, in the context of chromatin assembled on a 191-kb BAC, enhancer marks can be established in ES cells.
To monitor transcriptional competence of the Il12b gene within the BAC, we differentiated seven of the stably transfected ES cell clones into macrophages (Fig. 5C; Keller et al. 1993). The mature post-mitotic macrophages were then stimulated with LPS + interferon-γ (IFNγ), which results in efficient activation of a large number of endogenous proinflammatory genes, with expression levels comparable with those found in primary bone marrow-derived macrophages (SD Pope, unpubl.). Transcription from the premethylated Il12b-EGFP BAC transgenes was monitored by measuring GFP fluorescence. Upon LPS and IFNγ treatment for 6 h, substantial fluorescence was observed with four of the seven clones (Fig. 5D), comparable with results obtained following transfection of unmethylated BACs (data not shown). These results demonstrate that the premethylated BACs introduced into ES cells are competent for transcriptional activation.
Mi-2β helps maintain resistance to the establishment of enhancer marks in differentiated cells
We hypothesized that the premethylated plasmids remain stably methylated and silent in all of the differentiated cells we examined because methylated DNA rapidly assembles into strongly repressive chromatin in differentiated cells. Consistent with this hypothesis, four VL3-3M2 thymocyte clones stably transfected with the premethylated Ptcra reporter plasmid were found to possess low levels of histone H3K9 acetylation (Ac-H3K9) and H3K4 trimethylation (3Me-H3K4) in ChIP experiments, whereas much higher levels were found in clones stably transfected with an unmethylated plasmid (Fig. 6A). ChIP experiments with RNA polymerase II antibodies also revealed weak signals at the integrated premethylated plasmids (Fig. 6A).
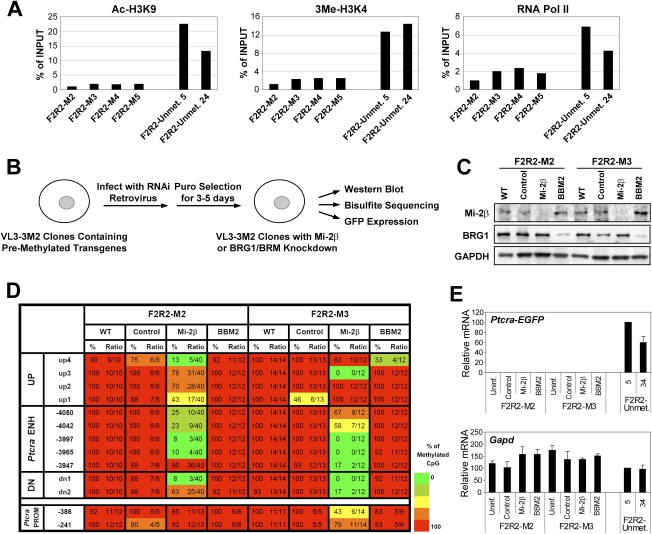
Mi-2β contributes to the repressive chromatin environment in differentiated cells. (A) Ac-H3K9 and 3Me-H3K4 levels, as well as RNA polymerase II levels, were examined by ChIP at the Ptcra enhancer within stably integrated Ptcra enhancer–promoter–reporter–insulator plasmids (construct F2R2) in VL3-3M2 thymocytes. Four independent clones generated by stable transfection with the premethylated plasmid (M2–M5) and two clones generated by stable transfection with the unmethylated plasmid (Unmet. 5 and Unmet. 34) were examined. Precipitated DNA samples were amplified using primers specific to the Ptcra enhancer region within the integrated plasmid. The ChIP signals are shown as a percentage of the input DNA signal and are representative of three independent experiments. (B) The experimental strategy used to examine the effect of Mi-2β or Brg1/Brm knockdown on DNA methylation at the integrated Ptcra enhancer is diagrammed. (C) Western blots were performed to examine the efficiency of Mi-2β and Brg1 knockdown after retroviral transduction of constructs expressing specific shRNAs. Extracts were examined from cells that were left untransduced (WT) or were transduced with a control retrovirus (Control, no shRNA cassette) or retroviruses expressing shRNAs specific for Mi-2β or Brg1/Brm (BBM2) transcripts. Retroviral transduction was performed with two independent VL3-3M2 clones containing the premethylated Ptcra enhancer–promoter–reporter–insulator plasmid (F2R2-M2 and F2R2-M3). GAPDH was analyzed as a loading control. (D) Bisulfite sequencing was used to examine DNA methylation at the integrated Ptcra enhancer 4 d after retroviral transduction of the F2R2-M2 and F2R2-M3 VL3-3M2 clones. DNA was examined from untransduced cells (WT) or cells transduced with the control retrovirus (Control) or the retroviruses expressing the Mi-2β or Brg1/Brm (BBM2) shRNAs. DNA methylation was examined at the integrated Ptcra enhancer and promoter, as well as at CpGs located upstream of (UP) or downstream from (DN) the enhancer. (E) EGFP mRNA expression from the F2R2-M2 and F2R2-M3 clones was examined by real-time RT–PCR. As a control, EGFP mRNA levels were monitored in two different clones (5 and 34) transfected with the unmethylated Ptcra enhancer–promoter–reporter–insulator plasmid. Gapd mRNA levels were monitored in each sample as a control.
To determine whether repressive chromatin was responsible for the resistance of differentiated cells to the establishment of enhancer marks, we asked whether this resistance was dependent on protein complexes that contribute to the assembly of repressive chromatin. For this analysis, we focused on the repressive Mi-2β/NuRD complex. This complex, which contains an ATP-dependent nucleosome remodeling protein (Mi-2β), a methyl-CpG-binding protein (MBD2), and histone deacetylases (HDAC1 and HDAC2), has been implicated in the generation of repressive chromatin at methylated promoters (for review, see Denslow and Wade 2007).
Strikingly, knockdown of Mi-2β by retroviral delivery of an shRNA in VL3-3M2 thymocytes containing the stably integrated and repressed Ptcra enhancer–promoter–EGFP–insulator plasmid resulted in susceptibility to establishment of an unmethylated window at the integrated Ptcra enhancer (Fig. 6B–D). However, the Ptcra promoter region within the same reporter plasmid (CpGs −386 and −241) remained more heavily methylated (Fig. 6D; data not shown). In parallel experiments, the Ptcra enhancer remained heavily methylated in cells that were left untransduced (Fig. 6D, WT) or were transduced with retroviruses lacking shRNA sequences (Fig. 6D, Control) or expressing an shRNA that targets the Brg1 and Brm subunits of the SWI/SNF remodeling complexes (Fig. 6D, BBM2). Similar results were obtained with three additional independent VL3-3M2 clones containing the stably integrated Ptcra reporter plasmid, although the precise enhancer CpGs that lost their methylation following Mi-2β knockdown varied from clone to clone (Supplemental Fig. 2). Collectively, these experiments demonstrate that CpG methylation at the stably integrated Ptcra enhancer is maintained in differentiated cells by repressive complexes like the Mi-2β/NuRD complex.
Interestingly, despite the significant loss of CpG methylation at the Ptcra enhancer following Mi-2β knockdown, the EGFP reporter gene remained silent (Fig. 6E, left), in contrast to the abundant EGFP expression observed in two VL3-3M2 clones (clones 5 and 34) stably transfected with an unmethylated Ptcra reporter plasmid (Fig. 6E, right). Thus, although VL3-3M2 thymocytes contain all of the transcription factors required for transcription of the endogenous Ptcra gene, and although Mi-2β knockdown confers susceptibility to the establishment of an unmethylated window at the integrated Ptcra enhancer, these differentiated cells must possess additional repressive chromatin features that prevent transcriptional activation.
Establishment of an unmethylated window at the Ptcra enhancer in ES cells is subject to both positive and negative regulation
Previous studies identified several DNA motifs that are important for Ptcra enhancer function in thymocytes (Fig. 7A; Reizis and Leder 2001, 2002). ChIP experiments revealed that DNA-binding proteins such as Sp1, E47, and CSL bind the endogenous Ptcra enhancer in ES cells (Supplemental Fig. 3). To determine which DNA motifs are functionally responsible for establishing the unmethylated window at the Ptcra enhancer in ES cells, we used the premethylation/stable transfection assay to analyze a series of five substitution mutants and a 30-bp deletion mutant (Fig. 7A). Ptcra enhancer–promoter–reporter–insulator plasmids containing these mutant sequences were premethylated and stably transfected into ES cells. Several clones containing each mutant were expanded and analyzed by bisulfite sequencing.
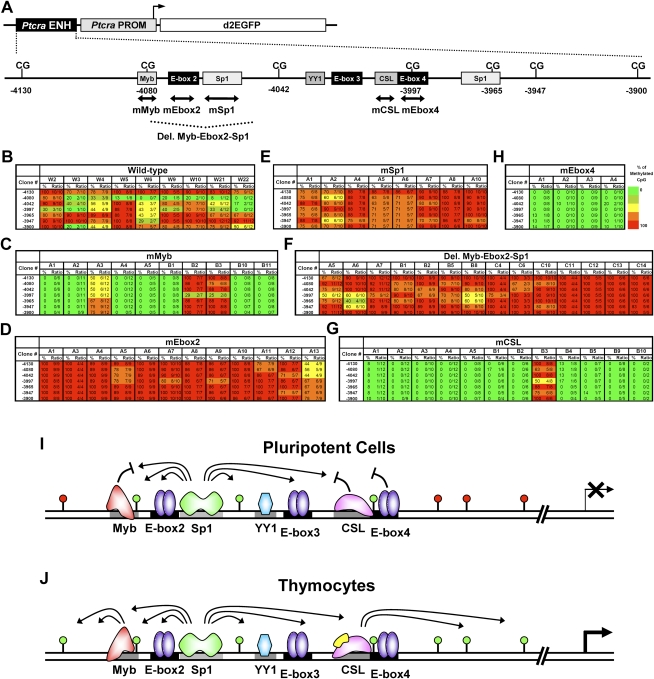
Dynamic regulation of the unmethylated window within the Ptcra enhancer in ES cells. (A) A diagram of the Ptcra enhancer–promoter–EGFP–insulator plasmid is shown. The 0.37-kb Ptcra enhancer and the 0.5-kb promoter were inserted upstream of a destabilized EGFP reporter (pd2EGFP). The cloned Ptcra enhancer contains seven CpG dinucleotides (−4130 to −3900). Putative transcription factor-binding sites are shown as shaded boxes. Regions analyzed by deletion and substitution mutation are indicated by a dashed line and double-headed arrows, respectively. (B–H) Ptcra enhancer methylation was monitored by bisulfite sequencing in several independent stable clones containing the wild-type enhancer (B) or six different mutant enhancers (C–H). Other mutations that had no effect on establishment of an unmethylated window are shown in Supplemental Figures 4 and 5. (I,J) A working model for the positive and negative regulation of DNA methylation at the Ptcra enhancer mark in pluripotent cells and thymocytes is shown.
Susceptibility to establishment of the unmethylated window was eliminated by mutations in an Sp1 site or in E-box 2 (Fig. 7D,E). Consistent with the hypothesis that proteins bound to these elements play a central role in establishment of the unmethylated window, the window was also absent in 13 of 14 clones containing the Myb-Ebox2-Sp1 deletion (Fig. 7F). Importantly, Sp1 was shown previously to play a role in preventing DNA methylation (Brandeis et al. 1994; Macleod et al. 1994).
Surprisingly, mutation of the Myb site, CSL site, or E-box 4, which flank the Sp1/E-box 2 cassette, resulted in greatly enhanced loss of methylation following stable transfection of the premethylated plasmids into ES cells (Fig. 7C,G,H). Dramatic loss of methylation was also observed with a Ptcra deletion mutant, F2R5, in which the sequences encompassing the CSL site and E-box 4 were removed (Supplemental Fig. 4C). In contrast, mutation of a YY1 site or E-box 3 had no effect on establishment of the unmethylated window (Supplemental Fig. 5). Although most clones containing the Myb and CSL mutations showed dramatic loss of methylation, a few clones were resistant (Fig. 7C,G), perhaps because they fortuitously integrated into heterochromatin. Importantly, all of the premethylated mutant plasmids were strongly resistant to the establishment of an unmethylated window following transfection into VL3-3M2 thymocytes (Supplemental Figs. 6, 7). This finding is consistent with previous evidence that the Myb and CSL sites play critical positive roles in Ptcra enhancer activity in thymocytes (Deftos et al. 2000; Reizis and Leder 2001, 2002). These data demonstrate that establishment of an unmethylated window at the Ptcra enhancer in ES cells is subject to both positive and negative regulation. Moreover, different tissue-specific enhancers are marked by different transcription factors in ES cells, with the Alb1 enhancer marked by FoxD3 and the Ptcra enhancer most likely marked by Sp1 and an E protein.
Discussion
The molecular features of ES cells and iPS cells responsible for their pluripotency and capacity for self-renewal have been examined in considerable depth, primarily through analyses of key regulatory proteins and networks, and through analyses of histone modification patterns at genes involved in developmental decisions. The results presented here suggest that pluripotency may also require protein–DNA interactions at enhancers for typical tissue-specific genes. During differentiation and the accompanying transition to a repressive chromatin environment, these enhancer marks may be essential for maintaining competence for transcriptional activation. Furthermore, we find that establishment of these marks in ES cells is under both positive and negative control.
Our studies of the Alb1 and Ptcra enhancers suggest that various transcription factors may mark tissue-specific enhancers in pluripotent cells. FoxD3 is responsible for the unmethylated CpG at the Alb1 enhancer, with binding sites for Sp1 and E proteins required at the Ptcra enhancer. The only important property of the bound transcription factors may be an ability to prevent assembly of repressive chromatin during differentiation. However, it is noteworthy that FoxD3 but not FoxA1 was capable of promoting the loss of CpG methylation at the Alb1 enhancer when ectopically expressed in MEFs. This finding suggests that FoxD3 may possess unique properties that are important for marking the Alb1 enhancer in pluripotent cells.
At the Ptcra enhancer, the stable transfection assay with premethylated plasmids was used to identify an Sp1 site and an E-box that are critical for establishment of the unmethylated window. An unexpected finding was the dramatic enhancement of the unmethylated window when either the Myb site or CSL/E-box 4 cassette was disrupted. Because a similar enhancement was observed upon mutation of any of these three sites, factors bound to the sites appear to act in synergy to limit the action of factors bound to the intervening Sp1 and E-box 2 sites. B-Myb and the CSL protein are both expressed in ES cells, but additional experiments will be needed to determine whether they are responsible for this negative activity. Although the significance of this negative activity remain unknown, the results raise the possibility that broader loss of methylation in the vicinity of the Ptcra enhancer in ES cells may be detrimental for proper regulation of the Ptcra gene.
The striking differences in the fate of premethylated plasmids transfected into pluripotent ES and iPS cells in comparison with a variety of differentiated cell types strengthens the hypothesis that chromatin in pluripotent cells is fundamentally different from that in differentiated cells. It is formally possible that the differences we observe are entirely due to the expression of transcription factors in pluripotent cells that are uniquely capable of gaining access to repressive chromatin. However, we favor the view that our findings are related to the general differences in chromatin structure between pluripotent and differentiated cells reported by others (see above; Meshorer and Misteli 2006; Meshorer et al. 2006; Guenther et al. 2007; Efroni et al. 2008). It has been proposed that a hyperdynamic chromatin environment is beneficial for developmental plasticity, presumably by facilitating diverse gene activation and repression programs that accompany cell fate decisions. The results shown here suggest that hyperdynamic chromatin could be equally beneficial for allowing transcriptional competence to be established at tissue-specific genes that remain silent until long after commitment to a specific lineage.
The failure to establish unmethylated windows on premethylated plasmids stably transfected into differentiated cells suggests that the absence of a pre-existing enhancer mark at an endogenous tissue-specific gene would prevent transcriptional activation in differentiated cells. However, a previous study of the immunoglobulin μ heavy-chain enhancer revealed active transcription from a premethylated plasmid directly transfected into B cells when the core μ enhancer was flanked by native matrix attachment regions, whereas the premethylated core enhancer by itself was resistant to activation (Forrester et al. 1999). This result raises the possibility that some enhancers may not require a mark established in pluripotent cells for their activation in differentiated cells. However, in our experiments but not in the previous experiments, the enhancer–promoter–reporter cassettes were flanked by β-globin insulator sequences; these insulators may have been important for maintaining the premethylated Ptcra and Il12b enhancers in their methylated and silent states following transfection into differentiated cells, by protecting the enhancer–promoter–reporter cassette from surrounding chromatin influences and from the cointegrated drug resistance gene.
Although we found that knockdown of Mi-2β expression in differentiated cells eliminates the barrier to establishment of enhancer marks, Mi-2β is not necessarily responsible for the differences in chromatin structure between pluripotent cells and differentiated cells. Mi-2β expression levels are comparable in pluripotent and differentiated cells (Supplemental Fig. 2B), and the NuRD complex that contains Mi-2β as its nucleosome remodeling ATPase carries out critical functions in maintaining the undifferentiated state of stem cells (Kaji et al. 2006; Yoshida et al. 2008). Furthermore, we found that Mi-2β knockdown in thymocytes was not sufficient for transcriptional activation from the premethylated Ptcra enhancer and promoter. We suspect that the Mi-2β/NuRD complex is one of a variety of protein complexes that contribute to the repressive chromatin structure observed in differentiated cells.
It is noteworthy that CpGs at the Ptcra and Il12b enhancers were unmethylated, not only in pluripotent cells and expressing cells, but also in most other differentiated cell types we examined (Xu et al. 2007), possibly as a remnant of the unmethylated window established during early embryogenesis. At these enhancers, the broad maintenance of the unmethylated windows would eliminate the need for its re-establishment during the generation of iPS cells from somatic cells. In contrast, the Alb1 enhancer was fully methylated in most differentiated cell types, with the unmethylated window largely restricted to ES cells and hepatocytes (Xu et al. 2007). Thus, the re-establishment of a mark at this enhancer and at other enhancers with similar properties may be necessary for reprogramming to a pluripotent state.
Although the enhancer marks we studied at the Alb1, Ptcra, and Il12b enhancers were identified originally as unmethylated windows, the bound factors may be more important for transcriptional competence than the unmethylated state. The bound factors may prevent both CpG methylation and the assembly of silent chromatin structures at the enhancers during differentiation. Notably, when premethylated plasmids were stably transfected into differentiated cells, the CpG methylation presumably led to the rapid assembly of repressive chromatin via the action of methyl-CpG-binding proteins (Bird and Macleod 2004), promoting resistance to transcription factor binding and transcriptional activation.
After the completion of this study, the first genome-wide bisulfite sequencing analysis with human ES cells was reported (Lister et al. 2009). Efforts to determine whether unmethylated CpG dinucleotides are commonly found at enhancers for typical tissue-specific genes have not been completed. However, the DNA methylation properties of the human homologs of the mouse Ptcra, Il12b, and Alb1 enhancers are consistent with our findings. Specifically, a window of low CpG methylation was observed at the human PTCRA enhancer in human ES cells (Lister et al. 2009). The low CpG methylation was accompanied by low levels of active histone modifications (Lister et al. 2009), consistent with our published data with the mouse enhancer (Xu et al. 2007). Also consistent with our findings (Xu et al. 2007), histone modifications associated with active or inactive chromatin were not observed at the human IL12B and ALB1 enhancers (Lister et al. 2009). Unfortunately, the CpGs that are unmethylated in the mouse Il12b and Alb1 enhancers are not conserved in the human enhancers. The human IL12B enhancer contains no CpGs at all, with surrounding CpGs heavily methylated. CpGs in the human ALB1 enhancer were fully methylated, similar to our finding that the mouse enhancer is fully methylated except for the CpG that coincides with the FoxD3-binding site (which is not conserved in the human enhancer). In the future, it will be important to determine whether FoxD3 is bound to the human ALB1 enhancer in ES cells, and whether common sets of factors are bound to the mouse and human Il12b (IL12B) and Ptcra (PTCRA) enhancers. Future work will also be needed to determine whether the unmethylated status of CpGs in these enhancers is a contributing effector or simply a mark of important pioneer factor interactions.
In summary, these findings may contribute to an understanding of the molecular features of ES cells and iPS cells that are essential for their pluripotency, and of the initial events leading to the activation of tissue-specific genes. The reprogramming of somatic cells to a pluripotent state involves numerous molecular events, including the silencing of genes that define differentiated lineages, the activation of genes associated with pluripotency, and broad transitions in chromatin structure. The barriers that limit the efficiency and success of reprogramming remain poorly understood. Efforts to identify and overcome these barriers may benefit from increased knowledge of the properties of tissue-specific genes in pluripotent cells and of the reliance of tissue-specific gene expression on events that take place prior to lineage commitment.
Materials and methods
Cell culture and reagents
The VL3-3M2 murine thymocyte line was maintained in RPMI 1640 medium (Cellgro) containing 10% fetal bovine serum (FBS; Omega Scientific). The J774 macrophage line was maintained in Dulbecco's modified Eagle's medium (DMEM; Cellgro) with 10% low-endotoxin FBS (Omega Scientific). ES cell line CCE was maintained on gelatin-coated petri dishes in standard ES medium (DMEM with 15% FBS, 10−4 M nonessential amino acids, 2 μM L-glutamine, 1% pen/strep, 0.05 μM β-mercaptoethanol, 1000 U/mL LIF). ES cell line R1 was maintained on mitomycin C-arrested MEFs (feeders). ES cells were differentiated into macrophages as described (Keller et al. 1993). Macrophages were activated with LPS (10 μg/mL) and recombinant murine IFN-γ (10 U/mL; PharMingen). LPS/IFN-γ activation of ES cell-derived macrophages results in efficient induction of the endogenous Il12b gene and many other proinflammatory genes, with expression levels comparable with those found in primary bone marrow-derived macrophages (SD Pope, unpubl.).
Primary MEFs were isolated from day 13.5 to day 14.5 embryos from C57BL/6J mice and maintained in DMEM with 10% FBS and 0.05 μM β-mercaptoethanol. iPS cell lines (Maherali et al. 2007) were maintained on mitomycin C-arrested MEFs (feeders) in standard ES medium. It is noteworthy that certain primary MEF lines exhibited reduced methylation at the −10,695 site of the Alb1 enhancer, whereas other MEF lines contained a fully methylated enhancer. The Alb1 enhancer was fully methylated in the MEFs used to generate the iPS cells and in the MEFs used for other experiments in this study.
Endoderm cells were isolated from mouse ES cells by growing the cells to induce differentiation, followed by sequential FACS using CD4-FoxA2 as a marker and, subsequently, CD25-FoxA3. This procedure has been shown to routinely generate definitive endoderm capable of differentiating into hepatic cells (Gouon-Evans et al. 2006; Gadue et al. 2009). Each preparation of cells was tested by the expression of endogenous endoderm markers.
Antibodies were from Chemicon (FoxD3, AB5678; CSL, AB5790; β-actin, MAB1501R), Santa Cruz Biotechnologies (Oct-4, sc-5279; Brg1, sc-10768; GAPDH, sc-59540; B-Myb, sc-13028; c-Myb, sc-7874; Sp1, sc-59; TBP, sc-273; E47, sc-763; HEB, sc-357; RNA pol II, sc-899), BD Pharmingen (HMG1, 556528), and Abcam (histone H3, ab8898), or were prepared in our laboratory (Mi-2β, GST).
Bisulfite sequencing, RT–PCR, and ChIP
Bisulfite sequencing was performed as described (Xu et al. 2007). PCR primers are listed in Supplemental Table 1.
For RT–PCR, RNA was extracted using TRI-reagent (MRC), treated with RNase-free DNase I, and purified with RNeasy kit (Qiagen). Quantified RNA (2 μg) was reverse-transcribed using SuperScript II (Invitrogen) and random hexamer primers. cDNA samples were analyzed in triplicate with the iQ SYBR Green Supermix (Bio-Rad) using iCycler iQ Real-Time PCR Detection System (Bio-Rad). PCR amplification parameters were 3 min at 95°C and 45 cycles of 15 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C. ChIP assays were performed as described (Su et al. 2004), with the ChIP samples analyzed by real-time PCR.
Plasmids and stable transfection
Mutations in the Ptcra enhancer–promoter–EGFP–insulator plasmid (Xu et al. 2007) were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The Il12b enhancer–promoter–EGFP plasmid was generated by subcloning a 1.1-kb Il12b enhancer fragment (−10,258 to −9182) and a 0.4-kb promoter fragment (−356 to +54) into the KpnI/MluI and MluI/EcoRI sites, respectively. Twenty-five micrograms to 40 μg of each plasmid were linearized using XmnI (Ptcra) or NotI (Il12b), followed by in vitro methylation with SssI (CpG) methylase (New England Biolabs) as described (Xu et al. 2007). The methylated DNA was extracted with phenol/chloroform, precipitated with ethanol, and dissolved in 50 mM Tris-HCl (pH 7.4). The Il12b-EGFP BAC was created by homologous recombination in E. coli. The EGFP cDNA was introduced into the second exon of the Il12b gene. Short DNA sequence tags adjacent to the Il12b promoter and enhancer were introduced by site-directed mutagenesis, followed by homologous recombination in E. coli.
Stable transfection of ES cells, primary MEFs, VL3-3M2 thymocytes, and J774 macrophages with the premethylated plasmids was performed as described (Xu et al. 2007). Stable transfection of ES cells with the premethylated Il12b-EGFP BAC was performed using Lipofectamine 2000 (Invitrogen). Stable clones were selected with G418 (150–250 μg/mL) or puromycin (1–1.5 μg/mL), expanded, and screened as described (Xu et al. 2007).
Retroviral transduction and RNAi
Foxa1 and Foxd3 cDNAs were cloned into the retroviral pQCXIP vector (Clontech) and transfected into HEK 293T cells with the packaging vector pCL-10A1 (Clontech) using Lipofectamine 2000. Virus supernatants were collected 48 and 72 h after transfection, filtered through a 0.45-μm syringe filter, and stored at 4°C. Primary MEFs (4 × 105 cells per well) were seeded in six-well plates, and 2 mL of virus supernatant with 20 μL of 1 M HEPES (pH 7.5) and 5 μL of polybrene (8 mg/mL in 20 mM HEPES at pH 6.2) were added. Spin infections were performed at 2500 rpm for 1.5 h at 30°C. Cells were selected with puromycin (1.5 μg/mL) 24 h post-infection. FoxA1 and FoxD3 expression was monitored by Western blot using anti-Flag antibody (Sigma, F-3165).
Retroviral RNAi knockdown of Mi-2β and Brg1/Brm was performed as described (Ramirez-Carrozzi et al. 2006). RNAi knockdown of FoxD3 in ES cells was performed by transfection of siRNA oligonucleotides against Foxd3 (Dharmacon) into ES cells using the manufacturer's protocol.
Acknowledgments
This work was supported by NIH grants R01 CA127279 and R21 CA137278 (to S.T.S.), R37 GM36477 (to K.S.Z.), and T32 AI07323 (to S.D.P.), and predoctoral training grant T1-00005 from the California Institute for Regenerative Medicine (to J.X.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1861209.
Supplemental material is available at http://www.genesdev.org.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. [Abstract] [Google Scholar]
- Bird A, Macleod D. Reading the DNA methylation signal. Cold Spring Harb Symp Quant Biol. 2004;69:113–118. [Abstract] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. [Abstract] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. [Europe PMC free article] [Abstract] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. [Abstract] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. [Abstract] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. [Abstract] [Google Scholar]
- Clevidence DE, Overdier DG, Tao W, Qian X, Pani L, Lai E, Costa RH. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc Natl Acad Sci. 1993;90:3948–3952. [Europe PMC free article] [Abstract] [Google Scholar]
- Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. [Europe PMC free article] [Abstract] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. [Abstract] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. [Europe PMC free article] [Abstract] [Google Scholar]
- Forrester WC, Fernandez LA, Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer–promoter interactions. Genes & Dev. 1999;13:3003–3014. [Europe PMC free article] [Abstract] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci. 2006;103:16806–16811. [Europe PMC free article] [Abstract] [Google Scholar]
- Gadue P, Gouon-Evans V, Cheng X, Wandzioch E, Zaret KS, Grompe M, Streeter PR, Keller GM. Generation of monoclonal antibodies specific for cell surface molecules expressed on early mouse endoderm. Stem Cells. 2009;27:2103–2113. [Europe PMC free article] [Abstract] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. [Abstract] [Google Scholar]
- Gouon-Evans V, Boussemart L, Gadue P, Nierhoff D, Koehler CI, Kubo A, Shafritz DA, Keller G. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24:1402–1411. [Abstract] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes & Dev. 1996;10:1670–1682. [Abstract] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. [Europe PMC free article] [Abstract] [Google Scholar]
- Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes & Dev. 2002;16:2650–2661. [Europe PMC free article] [Abstract] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. [Abstract] [Google Scholar]
- Kaji K, Cabellero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. [Abstract] [Google Scholar]
- Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993;13:473–486. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. [Abstract] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009 10.1038/nature08514. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. [Abstract] [Google Scholar]
- Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes & Dev. 1994;8:2282–2292. [Abstract] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. [Abstract] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. [Abstract] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. [Europe PMC free article] [Abstract] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. [Europe PMC free article] [Abstract] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. [Abstract] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes & Dev. 2006;20:282–296. [Europe PMC free article] [Abstract] [Google Scholar]
- Reizis B, Leder P. The upstream enhancer is necessary and sufficient for the expression of the pre-T cell receptor α gene in immature T lymphocytes. J Exp Med. 2001;194:979–990. [Europe PMC free article] [Abstract] [Google Scholar]
- Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes & Dev. 2002;16:295–300. [Europe PMC free article] [Abstract] [Google Scholar]
- Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Dynamic assembly of silent chromatin during thymocyte maturation. Nat Genet. 2004;36:502–506. [Abstract] [Google Scholar]
- Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada DA, Fletcher CF, Jenkins NA, Copeland NG, Klemsz M, et al. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem. 1996;271:23126–23133. [Abstract] [Google Scholar]
- Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol Cell Biol. 2005;25:1804–1820. [Europe PMC free article] [Abstract] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. [Abstract] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. [Abstract] [Google Scholar]
- Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. [Abstract] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. [Abstract] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. [Abstract] [Google Scholar]
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci. 2007;104:12377–12382. [Europe PMC free article] [Abstract] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, et al. The role of the chromatin remodeler Mi-2β in hematopoietic stem cell self-renewal and multilineage differentiation. Genes & Dev. 2008;22:1174–1189. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. [Abstract] [Google Scholar]
- Zhou L, Nazarian AA, Smale ST. Interleukin-10 inhibits interleukin-12 p40 gene transcription by targeting a late event in the activation pathway. Mol Cell Biol. 2004;24:2385–2396. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou L, Nazarian AA, Xu J, Tantin D, Corcoran LM, Smale ST. An inducible enhancer required for Il12b promoter activity in an insulated chromatin environment. Mol Cell Biol. 2007;27:2698–2712. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.1861209
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/23/24/2824.full.pdf
Free to read at www.genesdev.org
http://www.genesdev.org/cgi/content/abstract/23/24/2824
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/content/full/23/24/2824
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/reprint/23/24/2824
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102655240
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/gad.1861209
Article citations
RUNX1 C-terminal mutations impair blood cell differentiation by perturbing specific enhancer-promoter networks.
Blood Adv, 8(10):2410-2423, 01 May 2024
Cited by: 0 articles | PMID: 38513139 | PMCID: PMC11112616
Molecular Basis of Cell Reprogramming into iPSCs with Exogenous Transcription Factors.
Results Probl Cell Differ, 72:193-218, 01 Jan 2024
Cited by: 0 articles | PMID: 38509259
A gene regulatory network for neural induction.
Elife, 12:e73189, 03 Mar 2023
Cited by: 4 articles | PMID: 36867045 | PMCID: PMC10038663
G&D vignettes.
Genes Dev, 37(1-2):63-68, 01 Jan 2023
Cited by: 0 articles | PMID: 37061958 | PMCID: PMC10046443
Epigenetic Regulation of Inflammatory Signaling and Inflammation-Induced Cancer.
Front Cell Dev Biol, 10:931493, 08 Jun 2022
Cited by: 15 articles | PMID: 35757000 | PMCID: PMC9213816
Review Free full text in Europe PMC
Go to all (126) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells.
Proc Natl Acad Sci U S A, 104(30):12377-12382, 18 Jul 2007
Cited by: 82 articles | PMID: 17640912 | PMCID: PMC1941477
Transcriptional competence in pluripotency.
Genes Dev, 23(24):2793-2798, 01 Dec 2009
Cited by: 25 articles | PMID: 20008929 | PMCID: PMC2800094
X Chromosome Dosage Influences DNA Methylation Dynamics during Reprogramming to Mouse iPSCs.
Stem Cell Reports, 10(5):1537-1550, 19 Apr 2018
Cited by: 23 articles | PMID: 29681539 | PMCID: PMC5995367
Epigenetic modifications in the embryonic and induced pluripotent stem cells.
Gene Expr Patterns, 29:1-9, 04 Apr 2018
Cited by: 25 articles | PMID: 29625185
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R21 CA137278
Grant ID: R01 CA127279-01A2
Grant ID: R01 CA127279
Grant ID: R21 CA137278-01
NIAID NIH HHS (2)
Grant ID: T32 AI07323
Grant ID: T32 AI007323
NIGMS NIH HHS (2)
Grant ID: R37 GM036477
Grant ID: R37 GM36477




