Abstract
Free full text

A mammalian herpesvirus uses non-canonical expression and processing mechanisms to generate viral microRNAs
Summary
Canonical primary microRNA (pri-miRNA) precursors are transcribed by RNA polymerase II and then processed by the Drosha endonuclease to generate ~60 nt pre-miRNA hairpins. pre-miRNAs in turn are cleaved by Dicer to generate mature miRNAs. Previously, some short introns, called miRtrons, were reported to fold into pre-miRNA hairpins after splicing and debranching, and miRNAs can also be excised by Dicer cleavage of rare endogenous short hairpin RNAs. Here, we report that the miRNAs encoded by murine γ-herpesvirus 68 (MHV68) are also generated via atypical mechanisms. Specifically, MHV68 miRNAs are transcribed from RNA polymerase III promoters located within adjacent viral tRNA-like sequences. The resultant pri-miRNAs, which bear a 5′ tRNA moiety, are not processed by Drosha but instead by a cellular tRNase Z, which cleaves 3′ to the tRNA to liberate pre-miRNA hairpins that are then processed by Dicer to yield the mature viral miRNAs.
Introduction
MicroRNAs (miRNAs) are ~22 nt long regulatory RNAs that are expressed by all multicellular eukaryotes and several DNA viruses (Bartel, 2004; Umbach and Cullen, 2009). Most miRNAs are initially transcribed by RNA polymerase II (Pol II) as part of a long primary miRNA (pri-miRNA) precursor (Cai et al., 2004; Lee et al., 2004). The mature miRNA forms part of one arm of an ~80 nt long stem-loop structure, contained within the pri-miRNA, that is cleaved by the RNase III enzyme Drosha to liberate an ~60 nt pre-miRNA hairpin (Lee et al., 2003). Characteristic features of pri-mRNA stem-loops include an ~32 bp imperfect stem flanked by single-stranded RNA and bearing a large terminal loop (Han et al., 2006; Zeng et al., 2005).
Cleavage of the pri-miRNA by Drosha defines one end of the mature miRNA and leaves an ~2-nt 3′ overhang at the base of the pre-miRNA hairpin. After transport to the cytoplasm, the pre-miRNA is cleaved by a second RNase III enzyme, called Dicer (Hutvagner et al., 2001), which removes the terminal loop and leaves a second 2-nt 3′ overhang. One strand of the resultant miRNA duplex intermediate is then incorporated into the RNA- induced silencing complex RISC (Hammond et al., 2000).
A number of human and animal herpesviruses have been reported to encode viral miRNAs, and current evidence indicates that these are also transcribed by Pol II and then processed by Drosha and Dicer (Cai and Cullen, 2006; Pfeffer et al., 2005; Pfeffer et al., 2004; Umbach et al., 2008). An exception arises in the case of murine γ-herpesvirus 68 (MHV68), a relative of human Kaposi's sarcoma-associated herpesvirus. MHV68 encodes at least nine miRNAs and it has been proposed that these are initially transcribed as part of ~200 nt viral pri-miRNAs bearing a tRNA-like structure at the 5′ end and two ~60 nt RNA stem-loops toward the 3′ end (Fig. 1A) (Pfeffer et al., 2005). As the tRNA moieties contain canonical BoxA and BoxB internal Pol III promoter elements (Bowden et al., 1997), and the 3′ stem-loops end at a stretch of “U” residues, it was proposed that the MHV68 miRNAs are transcribed by Pol III (Pfeffer et al., 2005). Here, we demonstrate that the MHV68 miRNAs are indeed transcribed by Pol III to give rise to a pri-miRNA consisting of a tRNA linked to a pre-miRNA hairpin(s). These RNA structures are then separated by cleavage by the tRNA processing enzyme tRNase Z at the 3′ end of the tRNA moiety, which liberates the viral pre-miRNAs for nuclear export and Dicer processing.
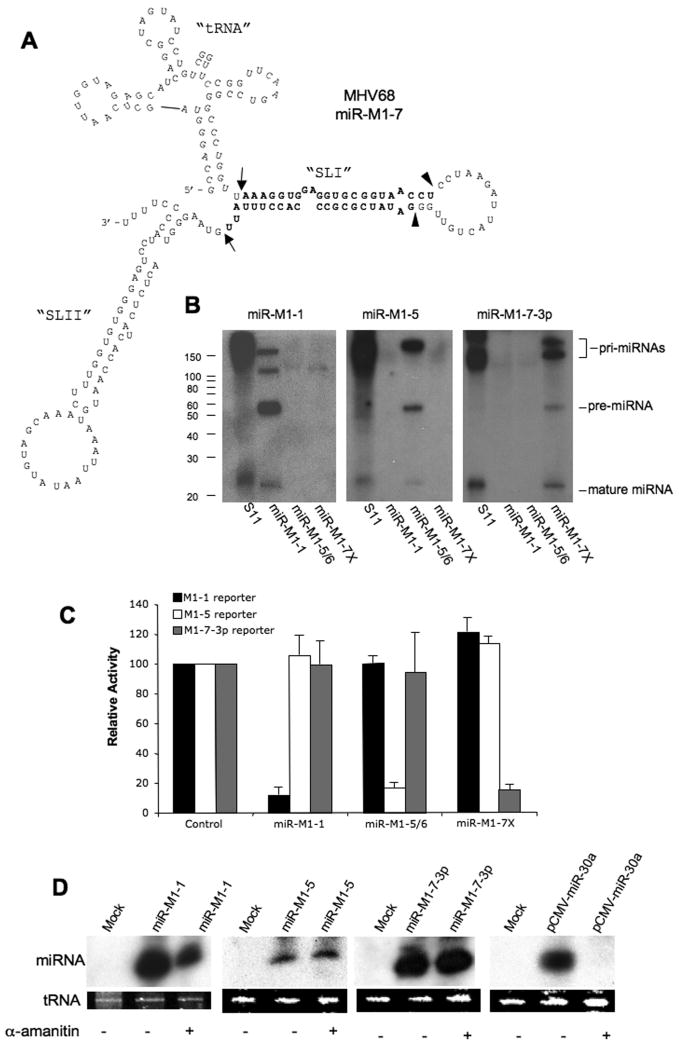
(A) Proposed structure of pri-miR-M1-7. Proposed cleavage sites defining the base of the pre-miR-M1-7 intermediate are indicated by open arrows and Dicer cleavage sites by closed triangles. The previously cloned (Pfeffer et al., 2005) miR-M1-7-5p and miR-M1-7-3p miRNAs are highlighted. This structured RNA can be sub-divided into a 5′ “tRNA” domain, a central stem-loop I or “SLI” domain and a 3′ “SLII” domain, as indicated. (B) Northern analysis of RNA samples derived from MHV68-infected S11 cells or 293T cells transfected with pmiR-M1-1, pmiR-M1-5/6 or pmiR-M1-7X. The end-labeled probes used are complementary to the mature miRNA and are indicated above each panel. The mobility of RNA size markers is indicated. (C) The biological activity of MHV68 miRNAs was confirmed by co-transfecting 293T cells with an Rluc-based reporter containing artificial 3′ UTR target sites for each indicated MHV68 miRNA, together with a vector encoding the MHV68 miRNAs indicated at the base of the figure. Observed Rluc activities are given relative to a culture co-transfected with a control expression vector. Average of three separate experiments with S.D. indicated. (D) 293T cells were mock-transfected or transfected with pmiR-M1-1, pmiR-M1-5/6, pmiR-M1-7X or pCMV-miR-30a and, where necessary, treated with the Pol II inhibitor α-amanitin. Total RNA was harvested at 48 h post-transfection and mature miRNA expression detected by Northern analysis.
Results
Previously, it was demonstrated that all nine miRNAs encoded by MHV68 derive from one arm of one of two proposed pre-miRNA-like stem-loops located 3′ to tRNA-like structures (Fig. 1A) (Pfeffer et al., 2005). Moreover, it was suggested that these miRNAs are initially expressed as ~200 nt pri-miRNA precursors transcribed under the control of an internal Pol III promoter present in the tRNA.
To address the validity of this hypothesis, we used PCR to isolate MHV68 genomic sequences encompassing the complete predicted pri-miRNAs for miR-M1-1, miR-M1-5/6 and miR-M1-7 and then cloned these into a vector lacking any eukaryotic promoter. After transfection into 293T cells, each plasmid gave rise to an ~22 nt mature miRNA that co-migrated with a band detected in the persistently MHV68-infected cell line S11 (Fig. 1B) (Usherwood et al., 1996). We also detected ~60 nt bands that presumably represent the pre-miRNA intermediates and finally one or two larger bands of ~130-200 nt in size that likely represent the full-length pri-miRNA and some form of processing intermediate. In the case of MHV68 miR-M1-7, we performed RNase protection analysis to map the 5′ and 3′ ends of the proposed ~60 nt pre-miRNA intermediate, and these data (Suppl. Fig. 1) confirmed that this band indeed represented the “SLI” stem-loop of miR-M1-7 as defined in Fig. 1A.
To confirm that these proposed MHV68 miRNAs are capable of programming RISCs, we co-transfected cells with each MHV68 miRNA expression vector along with a vector containing the renilla luciferase (Rluc) indicator gene linked to two artificial targets perfectly complementary to miR-M1-1, miR-M1-5 or miR-M1-7-3p (Gottwein et al., 2006). We detected the specific downregulation of each indicator plasmid by its cognate MHV68 miRNA expression plasmid (Fig. 1C).
We next addressed whether the expression of these MHV68 miRNAs was mediated by a Pol III promoter located in the tRNA-like moiety by asking if the production of these viral miRNAs was blocked by treatment with α-amanitin, a selective inhibitor of Pol II (Guialis et al., 1977). As shown in Fig. 1D, α-amanitin had no effect on the expression of MHV68 miR-M1-1, miR-M1-5 and miR-M1-7-3p in transfected 293T cells, yet blocked the expression of the cellular miRNA miR-30a transcribed under the control of a Pol II-dependent promoter. These data therefore confirm that MHV68 miRNA expression is independent of Pol II function.
Domain organization of MHV68 microRNA genes
The initial pmiR-M1-7X expression plasmid contains not only the MHV68 sequence visualized in Fig. 1A but also 104 nt of 5′ and 24 nt of 3′ viral genomic flanking sequence. To determine whether these flanking sequences are functionally relevant, we generated an expression plasmid containing only the 202 nt MHV68 sequence shown in Fig. 1A. The resultant pmiR-M1-7 construct was fully active (Fig. 2A) and produced similar, albeit slightly reduced, levels of pre-miR-M1-7 and mature miR-M1-7-3p (Fig. 2B). Deletion of the entire “SLII” domain of the proposed pri-miR-M1-7 precursor (Fig. 1A), in pmiR-M1-7ΔSLII, had no inhibitory effect on either miR-M1-7-3p function (Fig. 2A) or expression (Fig. 2B).
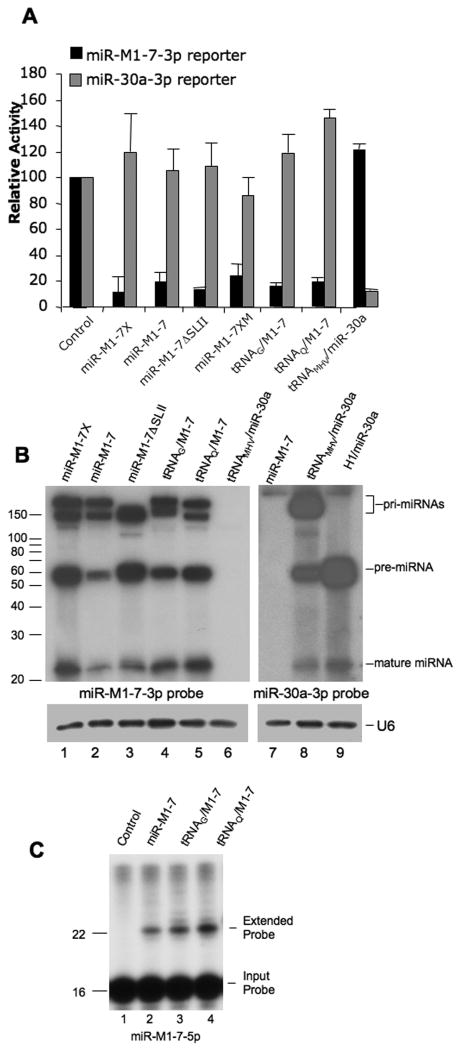
(A) This functional analysis of the indicated pri-miR-M1-7 mutants was performed in 293T cells, as described in Fig. 1C, except that a similar miR-30a-3p indicator construct was included as a control. Average of three experiments with S.D. indicated. (B) Northern analysis of mature miRNA and miRNA precursor expression in 293T cells using the same plasmids analyzed in panel A. The 32P-labeled oligonucleotide probes used were complementary to miR-M1-7-3p (lanes 1-6) or miR-30a (lanes 7-9). RNA size markers are indicated. U6 was used as a loading control. (C) Primer extension analysis to map the 5′ end of miR-M1-7-5p expressed in 293T cells transfected with the indicated expression vectors. The end-labeled primer is 16 nt in length, while mature miR-M1-7-5p is 22 nt (Fig. 1A). Mock-transfected 293T cells were used as a control.
To address whether the proposed MHV68 tRNA moiety (Fig. 1A) is uniquely able to drive expression of the linked MHV68 miR-M1-7-3p miRNA, we substituted the first 72 nt of the MHV68 sequence shown in Fig. 1A, as defined by the 5′ arrow, with a human glycine tRNA, to give ptRNAG/M1-7, or a human glutamine tRNA, to give ptRNAQ/M1-7. Remarkably, both human tRNAs proved capable of generating active, mature miR-M1-7-3p, as shown both by indicator assay (Fig. 2A) and Northern analysis (Fig. 2B). To confirm that the resultant miRNAs are processed at the same sites as the parental miRNA, we performed primer extension analysis to map the 5′ end of mature miR-M1-7-5p. All three expression vectors produced miRNAs with identical 5′ ends (Fig. 2C).
We next asked if SLI is indeed functioning as a pre-miRNA intermediate by substituting an authentic cellular pre-miRNA, derived from human miR-30a (Zeng et al., 2002), in place of the SLI sequence in miR-M1-7ΔSLII, to give ptRNAMHV/miR-30a. This vector inhibited the expression of an Rluc indicator gene linked to two artificial miR-30a target sites, but did not affect expression of a vector bearing miR-M1-7-3p sites (Fig. 2A). Moreover, RNAs that co-migrated with authentic pre-miR-30a and mature miR-30 were expressed in tRNAMHV/miR-30 transfected cells (Fig. 2B, lanes 8 and 9).
To determine whether the tRNA-based Pol III promoter present in miR-M1-7 (Fig. 1A) could be replaced by the heterologous, external Pol III-dependent H1 promoter (Baer et al., 1990), we replaced the tRNA moiety present in wild-type miR-M1-7 with the H1 promoter, such that transcription would initiate at the first nucleotide of “SLI”, defined by the 5′ arrow present in Fig. 1A. The pH1/M1-7 vector selectively downregulated an indicator construct bearing M1-7-3p target sites (Suppl. Fig. 2A) and expressed a high level of both pre- and mature miR-M1-7-3p in transfected cells (Suppl. Fig. 2B). As expected, deletion of SLII, to give pH1/M1-7ΔSLII, did not affect the expression of mature, active miR-M1-7-3p (Suppl. Fig. 2).
pri-miR-M1-7 is processed by tRNase Z but not by Drosha
tRNA genes are transcribed to give precursor tRNAs that are processed at the 5′ end by RNase P and at the 3′ end by tRNase Z (Hartmann et al., 2009). In particular, tRNase Z cleaves pre-tRNAs 3′ to the first nucleotide after the acceptor stem—the discriminator nucleotide—followed by the addition of the sequence 5′-CCA-3′. Previous analysis of the MHV68 tRNAs has demonstrated that these are also cleaved 3′ to the discriminator nucleotide and modified by the addition of CCA, although these viral tRNAs do not appear to be aminoacylated (Bowden et al., 1997). Moreover, the 5′ end of miR-M1-7-5p has been shown both by sequencing (Pfeffer et al., 2005) and here by primer extension (Fig. 2C) to coincide with the second nucleotide 3′ to the base of the acceptor stem (Fig 1A). Finally, we have shown that the viral tRNA moiety present in miR-M1-7 can be functionally substituted by wild-type human tRNAGly or tRNAGlu (Fig. 2B) without affecting miRNA processing. Together, these results argue that the cleavage between the tRNA and SLI in miR-M1-7, as well as in other MHV68 pri-miRNAs, is likely performed by tRNase Z and not by Drosha.
To determine whether tRNase Z and/or Drosha contribute to miR-M1-7 processing, we used RNA interference (RNAi) to knock down each of these two endonucleases in human cells. Two distinct synthetic small interfering RNAs (siRNAs) were used for each target analyzed, and each siRNA proved capable of strongly and specifically downregulating the expression of either tRNase Z or Drosha in HeLa cells (Suppl. Fig. 3). A complication that emerged was that while Drosha knock-down was well tolerated by HeLa cells, knockdown of tRNase Z, which is essential for tRNA biogenesis (Hartmann et al., 2009), proved toxic. As a result, our analysis of miRNA expression had to be performed 72 hr after transfection with the tRNase Z-specific siRNAs, i.e., before a full knock-down of tRNase Z could be achieved, in order to avoid confounding cytopathic effects. Representative knock-down experiments are shown in Figs. 3A and 3B, while a compilation of data derived from replicate experiments is presented in Fig. 3C.
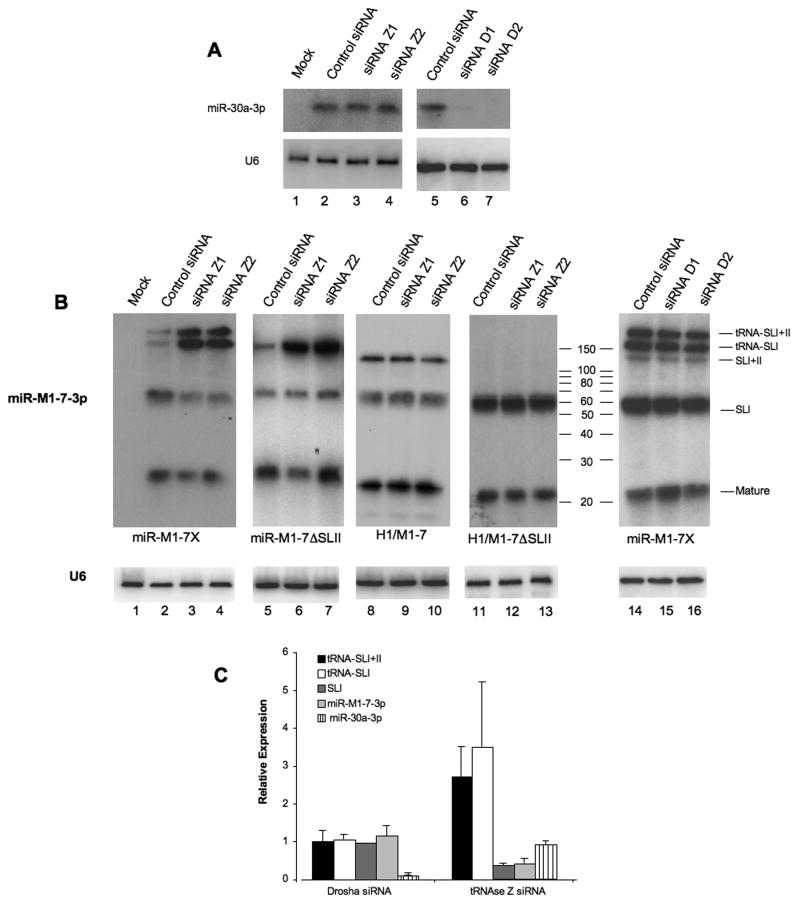
(A) Northern analysis of the expression of mature miR-30a-3p in HeLa cells treated with a control siRNA or two distinct siRNAs specific for tRNase Z (Z1 and Z2) or Drosha (D1 and D2) and then transfected with pCMV-miR-30a. U6 RNA was used as a loading control. (B) Similar to panel A, except that these blots analyze HeLa cells co-transfected with the indicated siRNAs and either pmiR-M1-7X (lanes 1-4 and 14-16), pmiR-M1-7ΔSLII (lanes 5-7), pH1/M1-7 (lanes 8-10) or pH1/M1-7ΔSLII (lanes 11-13). Size markers are indicated and U6 was used as a loading control. (C) Quantitation of the level of expression of full-length wild-type pri-miR-M1-7 (tRNA-SLI+SLII), the miR-M1-7 tRNA-SLI intermediate, the SLI pre-miR-M1-7 intermediate and mature miR-M1-7-3p in HeLa cells transfected with pmiR-M1-7 and depleted for either RNase Z or Drosha by RNAi. Human miR-30a was used as a control. Average of four experiments with S.D. indicated. Data are presented relative to the level of expression of each RNA species in cells treated with a control siRNA, which was set at 1.
Knock-down of Drosha by either siRNA tested, both of which efficiently depleted endogenous Drosha expression (Suppl. Fig. 3B), blocked the processing of human miR-30a (Fig. 3A, lanes 5-7), which is known to be Drosha dependent (Zeng et al., 2005), but had no detectable effect on the processing of full-length miR-M1-7 (Fig. 3B, lanes 14-16). In contrast, knock down of tRNase Z had no effect on miR-30a processing (Fig. 3A, lanes 1-4), but did inhibit processing of miR-M1-7X (Fig. 3B, lanes 1-4). Specifically, Northern analysis of miR-M1-7 processing in HeLa cells in which tRNase Z expression had been attenuated using one of two different siRNAs revealed an ~3-4 fold increase in the level of the full-length, ~200 nt pri-miR-M1-7 transcript and of the ~132 nt tRNA-SLI premature termination product (Fig. 3C). The identity of these RNAs was confirmed using both a miR-M1-7-3p-specific probe (Fig. 3B) and a probe specific for the tRNA moiety (Suppl. Fig. 4). In contrast, expression of pre-miR-M1-7 and mature miR-M1-7-3p decreased by ~2-3 fold (Fig. 3B, lanes 1-4 and Fig. 3C). Analysis of HeLa cells transfected with pmiR-M1-7ΔSLII also revealed a marked increase in the expression of the truncated tRNA-SLI pri-miRNA and a slight decrease in the expression of the pre-miRNA and mature miR-M1-7-3p (Fig. 3B, lanes 5-7). However, we did not detect any effect of loss of tRNase Z function on the processing of the SLI+SLII transcript produced by pH1/M1-7 or the SLI pre-miRNA stem-loop produced by pH1/M1-7ΔSLII (Fig. 3B, lanes 8-13).
As noted above, tRNase Z cleavage at the 3′ end of a pre-tRNA is followed by the addition of the sequence 5′-CCA-3′, which is an anti-determinant for tRNase Z cleavage (Mohan et al., 1999). We therefore inserted the CCA sequence immediately 3′ to the predicted tRNase Z cleavage site in miR-M1-7 (Fig. 1A) and examined whether this would affect miR-M1-7 processing. In fact, this mutation blocked the production of pre- and mature miR-M1-7-3p and of the free tRNA, yet had little effect on the production of the full-length miR-M1-7 transcript or of the tRNA-SLI intermediate (Suppl. Fig. 5). In contrast, mutational insertion of the sequences GGT, GAT or AGT had no inhibitory effect on miR-M1-7 processing (data not shown). These results again indicate that tRNase Z mediates the cleavage event that separates the tRNA moiety present in pri-miR-M1-7 from SLI.
If the SLI subdomain liberated by processing of miR-M1-7 (Fig. 1A) is indeed acting as a pre-miRNA, as implied by the fact that substitution of miR-M1-7 SLI with authentic human pre-miR-30a results in both pre- and mature miR-30a expression (Fig. 2B), then mature miR-M1-7-3p expression should be Dicer dependent. Indeed, knock-down of Dicer using RNAi strongly inhibited the production of mature miR-M1-7-3p without affecting the expression of pre-miR-M1-7 (Suppl. Fig. 6).
Specific cleavage of pri-miR-M1-7 by tRNase Z in vitro
To address whether Drosha or tRNase Z is capable of processing pri-miR-M1-7 appropriately in vitro, we used an affinity matrix to isolate FLAG-tagged Drosha or tRNase Z from overexpressing 293T cells. We then tested the ability of these partially purified proteins to accurately process a pri-miRNA known to be a substrate for Drosha, i.e., KSHV pri-miR-K5 (Gottwein et al., 2006), or one of two miR-M1-7 derived precursors, i.e., the 132 nt tRNA-SLI transcript that results from premature termination at the end of SLI (Fig. 4A), or the full-length, 200 nt pri-miR-M1-7 (Fig. 4B).
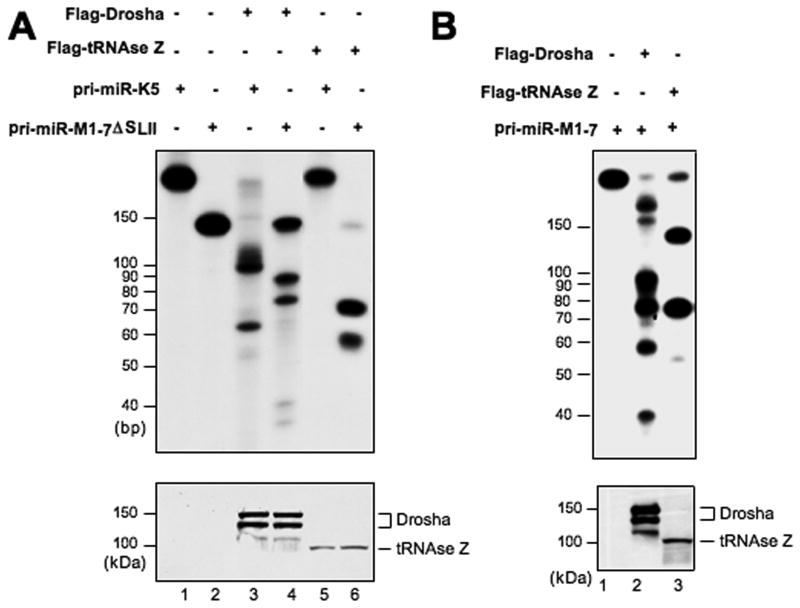
(A) Upper panel: In vitro processing of a 132 nt transcript, consisting of the tRNA and SLI domains of MHV68 pri-miR-M1-7, and of the 260 nt pri-miR-K5 transcript. 32P-labeled transcripts were incubated in the presence of recombinant FLAG-tRNase Z or FLAG-Drosha isolated from overexpressing 293T cells, and any resultant cleavage products separated by denaturing gel electrophoresis and visualized by autoradiography. Size markers are indicated at left. Lower panel: The protein preparations used in this experiment were analyzed by Western blot using a FLAG-specific antiserum. (B) Similar to panel A, except that a 200 nt 32P-labeled transcript, consisting of the predicted full-length pri-miR-M1-7 (Fig. 1A) was analyzed.
Incubation of the 132 nt miR-M1-7 tRNA-SLI probe with FLAG-tRNase Z resulted in the complete digestion of this labeled transcript to give the predicted tRNA (72 nt) and SLI (60 nt) products (Fig. 4A, lane 6). In contrast, tRNase Z did not detectably process the 260 nt pri-miR-K5 transcript (Fig. 4A, lane 5).
Analysis of Drosha function revealed efficient processing of pri-miR-K5 to yield the expected ~60 nt pre-miRNA and the two ~100 nt 5′ and 3′ flanking sequences (Fig. 4A, lane 3). Incubation of the 132 nt miR-M1-7 tRNA-SLII transcript with Drosha resulted in partial processing that gave rise to cleavage products of ~90 nt, ~76 nt, ~42 nt and ~37 nt in length (Fig. 4A, lane 4). While we do not know the exact origin of these cleavage products, it is apparent that these do not represent the predicted ~72 nt tRNA and ~60 nt SLI RNA products expected upon accurate processing of the miR-M1-7 tRNA-SLI precursor.
Analysis of the processing of the full-length, ~200 nt pri-miR-M1-7 transcript by tRNase Z resulted in the production of RNAs that migrate at the sizes predicted for the tRNA moiety (~72 nt) and the SLI+SLII intermediate (~128 nt) (Fig. 4B, lane 3). In contrast, incubation with Drosha again gave rise to a complex pattern of products of unclear origin. Overall, these data therefore indicate that tRNase Z is necessary and sufficient to cleave the predicted pri-miR-M1-7 transcript at a specific site located between the tRNA moiety and SLI (Fig. 1A). In contrast, tRNase Z was not able to detectably cleave at the predicted processing site located between SLI and SLII. Although Drosha proved able to efficiently process the pri-miR-K5 transcript to generate the expected ~60 nt pre-miRNA (Fig. 4A). Drosha is not able to cleave pri-miR-M1-7 at either of the predicted processing sites.
Alternative Pol III transcription termination of MHV68 pri-miRNAs
The in vitro data shown in Fig. 4 indicate that tRNase Z cleaves between the tRNA and SLI moieties present in MHV68 pri-miR-M1-7 but also suggest that neither tRNase Z nor Drosha is able to cleave between SLI and SLII (Fig. 1A). One possibility is that a third, as yet undefined, cellular endonuclease cleaves at this location. However, we noted that the sequences near the 3′ end of SLI in miR-M1-7, and also in all six other MHV68 miRNA genes, are U-rich. In fact, all seven MHV68 miRNA genes contain the sequence 5′-CUUU-3′ at the junction of the predicted SLI and SLII sequences (Suppl. Fig. 7A and B). Pol III transcription in vertebrate cells generally terminates at the sequence 5′-UUUU-3′ (Baer et al., 1990), but termination at variant sequences, such as 5′-UUAUU-3′, has also been reported (Gunnery et al., 1999) and we therefore wondered if this might represent a leaky Pol III transcription termination signal. Indeed, as shown in Suppl. Fig. 7C, insertion of this 13-nt sequence element from miR-M1-1 or miR-M1-7 into an ~200 nt RNA driven by the Pol III-dependent H1 promoter resulted in partial termination of transcription, and insertion of an even shorter consensus sequence (5′-CCUUCUUU-3′) had the same effect. Moreover, insertion of this element from miR-M1-5/6 resulted in almost complete termination of Pol III transcription. Mutated forms of these U-rich sequences, in contrast, did not induce premature termination (Suppl. Fig. 7C). The efficient termination of Pol III transcription induced by the sequence derived from miR-M1-5/6 presumably reflects the presence of four tandem “U” residues, rather than the three seen in miR-M1-1 and miR-M1-7 (Suppl. Fig. 7A) and correlates with the observation that miR-M1-5/6 differs from both miR-M1-1 and miR-M1-7 in giving rise to a single >100 nt transcript in transfected cells (Fig. 1B).
We next engineered these same mutations into the pmiR-M1-7X and pmiR-M1-5/6 expression plasmids and examined their effect on miRNA processing. Consistent with the hypothesis that this 5′-CUUU-3′ element induces partial transcription termination in wild-type miR-M1-7, we observed a significant (~5 fold) decrease in the level of the tRNA-SLI processing intermediate and a concomitant ~3 fold increase in the expression of an ~120 nt band that is likely to represent a miR-M1-7 processing intermediate consisting of SLI linked to SLII (Suppl. Fig. 7D). Nevertheless, the expression (Suppl. Fig. 7D) and function (Fig. 2A) of the mature miR-M7-3p miRNA was largely unaffected by mutation of this U-rich sequence in pmiR-M1-7XM.
While miR-M1-7 does not give rise to a detectable miRNA derived form the SLII sub-domain (Suppl. Fig. 2B and Pfeffer et al., 2005), miR-M1-5/6 gives rise to miR-M1-5 from SLI and miR-M1-6 from SLII. This is surprising, as transcription termination at the end of SLI is apparently efficient (Suppl. Fig. 7C), thus presumably precluding transcription of SLII. We therefore mutated the 5′-UUUU-3′ sequence present in miR-M1-5/6 (Suppl. Fig. 7A) and analyzed the effect on miR-M1-5 and miR-M1-6 expression. While this mutation resulted, as expected, in the production of an RNA that migrated at the predicted size for tRNA-SLI+SLII, it also inhibited the production of both pre- and mature miR-M1-5 (Suppl. Fig. 7E, lane 3). However, this same mutation also induced the expression of both pre- and mature miR-M1-6 (Suppl. Fig. 7E, lane 7). It therefore appears that the full-length tRNA-SLI+SLII precursor can be processed to yield miR-M1-6 from SLII but is not efficiently processed to yield miR-M1-5, which instead derives from the prematurely terminated tRNA-SLI RNA (Suppl. Fig. 7E, lane 4). Together, these data indicate that the MHV68 miRNA genes undergo alternative transcription termination at the 3′ end of SLI and at the 3′ end of SLII to produce two distinct pri-miRNA species that can, in the case of miR-M1-5 and miR-M1-6, give rise to two distinct mature viral miRNA species.
Discussion
Previously, two classes of Drosha-independent/Dicer-dependent miRNAs have been identified. These are the miRtrons, in which a pre-miRNA intermediate is generated after debranching a short intron (Berezikov et al., 2007; Ruby et al., 2007), and natural shRNAs, which mimic the pre-miRNA structure (Babiarz et al., 2008). In this manuscript, we identify a third class of Drosha-independent miRNAs, encoded by the murine γ-herpesvirus MHV68, that is initially expressed fused to a tRNA that provides the Pol III promoter required for pri-miRNA transcription. The resultant pri-miRNA is then processed by tRNase Z, which cleaves 3′ to the tRNA moiety to liberate a canonical pre-miRNA stem-loop that is processed by Dicer to give a mature miRNA. The MHV68 miRNAs are therefore distinct from previously reported Drosha-independent miRNAs in that their biogenesis is dependent on the action of a distinct endonuclease, i.e., tRNase Z. As all nine MHV68 miRNAs are predicted to be initially transcribed as tRNA fusions (Pfeffer et al., 2005), it is likely that they all require this RNase Z-dependent processing pathway. At present, no other viral or cellular miRNAs that are initially transcribed as tRNA fusions have been identified, although a small number of human miRNAs occupy genomic locations adjacent to known tRNAs (Borchert et al., 2006).
A surprising feature of the MHV68 tRNA:miRNA fusions is that they do not appear to contain any unique features; i.e., the viral tRNAs can be substituted by cellular tRNAs and the viral SLI sequence can be replaced with a known cellular pre-miRNA without any inhibition of mature miRNA biogenesis being observed (Fig. 2). Therefore, artificial tRNA:shRNA fusions could be used to express artificial shRNA or pre-miRNAs in culture or in vivo. Indeed, it has been previously reported that a tRNA:shRNA fusion can repress target mRNA expression effectively, although the mechanism underlying the processing of this fused transcript was not analyzed (Scherer et al., 2007). Remarkably, we now not only demonstrate that highly active tRNA:miRNA fusions exist in nature but also that these are specifically processed by a tRNA processing enzyme instead of by the canonical cellular pri-miRNA processing machinery.
Experimental Procedures
RNA expression analysis
miRNA expression was analyzed by Northern blot, as previously described (Cai et al., 2004). Briefly, 2×106 293T cells were transfected with 10 μg of miRNA expression plasmid using calcium phosphate. 48 hr later, total RNA was isolated (TriReagent, Sigma) and 25 μg fractionated on a 15% TBE-Urea gel (Bio-Rad). RNA was transferred to HyBond-N membrane (Amersham) and UV-crosslinked (Stratalinker, Stratagene). Membranes were pre-hybridized in ExpressHyb (Clontech) and then incubated at 37°C with a 32P-end labeled oligonucleotide (Suppl. Table 1). Membranes were washed with 2× SSC/0.1%SDS at room temperature and miRNAs visualized by autoradiography.
To determine if MHV68 miRNA expression is Pol III dependent, we transfected 1.2×106 293T cells with 800 ng of each miRNA expression construct. 50 μg/ml of α-amanitin (Sigma) was added into the medium at 2 hr post-transfection, if necessary. Cells were harvested 48 hr later and Northern analyses performed as described above. Primer extension assays were performed as described (Zeng et al., 2005), using total RNA isolated from 293T cells transfected with pmiR-M1-7, ptRNAG/M1-7 or ptRNAQ/M1-7. The end-labeled probe used had the sequence 5′-AGGTTACCGCACCTCC-3′.
RNA interference assays
Silencer siRNA duplexes specific for human Drosha or tRNase Z were purchased from Applied Biosystems. The Drosha siRNAs targeted nucleotides 2136-2156 and 3404-3424 of Drosha mRNA, where the first residue of the open reading frame is designated position 1. The tRNase Z-specific siRNAs targeted residues 286-306 and 2228-2248 of tRNase Z mRNA. A control siRNA (Block-It Alexa Fluor Red, Invitrogen) was used to assay for non-specific siRNA transfection effects. All siRNAs were transfected at a final concentration of 20 nM.
To analyze the role of tRNase Z in MHV68 miRNA processing, HeLa cells were transfected with tRNase Z-specific siRNAs using Lipofectamine RNAiMax (Invitrogen). 24 hr post-transfection, cells were transfected with 20 μg of pmiR-M1-7X (or derivatives) or pCMV-miR-30a (Zeng et al., 2002), using Lipofectamine2000. 48 hr later, total RNA was isolated using TriReagent (Sigma-Aldrich). To analyze the role of Drosha in MHV68 miRNA processing, HeLa cells were transfected with Drosha-specific siRNAs using RNAiMax (Invitrogen). 48 hr post-transfection cells were re-transfected with each Drosha siRNA along with 20 μg of pmiR-M1-7X or pCMV-miR-30a, using Lipofectamine2000 (Invitrogen). After an additional 48 hr, total RNA was isolated as described above and analyzed by Northern blot. Membranes were also imaged and bands quantitated using the Typhoon 9200 Variable Mode Imager and ImageQuant software. (GE Healthcare Life Sciences). Finally, blots were stripped and re-probed for U6 RNA, as a loading control.
In vitro miRNA processing
Radiolabeled substrates for in vitro processing assays were generated by transcription of PCR products containing the T7 promoter (Gottwein et al., 2006; Zeng and Cullen, 2006). To generate recombinant, FLAG-tagged tRNase Z and Drosha, 293T cells were transfected either with the expression plasmid pcDNA3.1/FLAG-tRNase Z or with both pCK/FLAG-Drosha and pCMV-HA-DGCR8. At 48 h post-transfection, cells were sonicated in lysis buffer (10 mM Tris-HCl, pH 7.4, 250 mM NaCl, and protease inhibitor cocktail (Roche)), and the supernatant incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 2 hr, followed by 5 washes with lysis buffer. For in vitro processing by tRNase Z, the immunoprecipitate was mixed with radiolabeled transcript in reaction buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1.5 mM DTT, 3.2 mM MgCl2) and incubated at 50 °C for 15 min (Takaku et al., 2004). Drosha cleavage assays were performed as previously described (Gottwein et al., 2006; Zeng and Cullen, 2006). Cleavage products were analyzed by electrophoresis on 15 % denaturing polyacrylamide gels.
Acknowledgments
The authors thank Drs. E. Lees, S. Efstathiou, V. N. Kim and E. J. Underwood for reagents used in this research, which was supported by NIH grant AI067968 to BRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. [Europe PMC free article] [Abstract] [Google Scholar]
- Baer M, Nilsen TW, Costigan C, Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97–103. [Europe PMC free article] [Abstract] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. [Abstract] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. [Europe PMC free article] [Abstract] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. [Abstract] [Google Scholar]
- Bowden RJ, Simas JP, Davis AJ, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78(Pt 7):1675–1687. [Abstract] [Google Scholar]
- Cai X, Cullen BR. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J Virol. 2006;80:2234–2242. [Europe PMC free article] [Abstract] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. [Europe PMC free article] [Abstract] [Google Scholar]
- Gottwein E, Cai X, Cullen BR. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol. 2006;80:5321–5326. [Europe PMC free article] [Abstract] [Google Scholar]
- Guialis A, Beatty BG, Ingles CJ, Crerar MM. Regulation of RNA polymerase II activity in alpha-amanitin-resistant CHO hybrid cells. Cell. 1977;10:53–60. [Abstract] [Google Scholar]
- Gunnery S, Ma Y, Mathews MB. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J Mol Biol. 1999;286:745–757. [Abstract] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. [Abstract] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. [Abstract] [Google Scholar]
- Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A. The making of tRNAs and more - RNase P and tRNase Z. Prog Mol Biol Transl Sci. 2009;85:319–368. [Abstract] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. [Abstract] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. [Abstract] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. [Europe PMC free article] [Abstract] [Google Scholar]
- Mohan A, Whyte S, Wang X, Nashimoto M, Levinger L. The 3′ end CCA of mature tRNA is an antideterminant for eukaryotic 3′-tRNase. RNA. 1999;5:245–256. [Europe PMC free article] [Abstract] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. [Abstract] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. [Abstract] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. [Europe PMC free article] [Abstract] [Google Scholar]
- Scherer LJ, Frank R, Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. [Europe PMC free article] [Abstract] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M. The N-terminal half-domain of the long form of tRNase Z is required for the RNase 65 activity. Nucleic Acids Res. 2004;32:4429–4438. [Europe PMC free article] [Abstract] [Google Scholar]
- Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. [Europe PMC free article] [Abstract] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. [Europe PMC free article] [Abstract] [Google Scholar]
- Usherwood EJ, Stewart JP, Nash AA. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J Virol. 1996;70:6516–6518. [Europe PMC free article] [Abstract] [Google Scholar]
- Zeng Y, Cullen BR. Recognition and cleavage of primary microRNA transcripts. Methods Mol Biol. 2006;342:49–56. [Abstract] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. [Abstract] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2009.12.016
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276509009204/pdf
Subscription required at www.molecule.org
http://www.molecule.org/cgi/content/reprint/37/1/135
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.molcel.2009.12.016
Article citations
Evaluation of immune sensor responses to a viral small noncoding RNA.
Front Cell Infect Microbiol, 14:1459256, 08 Oct 2024
Cited by: 0 articles | PMID: 39450336 | PMCID: PMC11499242
Exonuclease-enhanced prime editors.
Nat Methods, 21(3):455-464, 01 Feb 2024
Cited by: 4 articles | PMID: 38302659 | PMCID: PMC10927552
Pluripotency-Associated microRNAs in Early Vertebrate Embryos and Stem Cells.
Genes (Basel), 14(7):1434, 12 Jul 2023
Cited by: 1 article | PMID: 37510338 | PMCID: PMC10379376
Review Free full text in Europe PMC
microRNAs in action: biogenesis, function and regulation.
Nat Rev Genet, 24(12):816-833, 28 Jun 2023
Cited by: 98 articles | PMID: 37380761
Review
Shaping the host cell environment with viral noncoding RNAs.
Semin Cell Dev Biol, 146:20-30, 28 Dec 2022
Cited by: 1 article | PMID: 36581481 | PMCID: PMC10101873
Review Free full text in Europe PMC
Go to all (143) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Intronic microRNA precursors that bypass Drosha processing.
Nature, 448(7149):83-86, 24 Jun 2007
Cited by: 895 articles | PMID: 17589500 | PMCID: PMC2475599
Dicer cleaves 5'-extended microRNA precursors originating from RNA polymerase II transcription start sites.
Nucleic Acids Res, 46(11):5737-5752, 01 Jun 2018
Cited by: 31 articles | PMID: 29746670 | PMCID: PMC6009592
The nuclear RNase III Drosha initiates microRNA processing.
Nature, 425(6956):415-419, 01 Sep 2003
Cited by: 2988 articles | PMID: 14508493
MicroRNA biogenesis: isolation and characterization of the microprocessor complex.
Methods Mol Biol, 342:33-47, 01 Jan 2006
Cited by: 96 articles | PMID: 16957365
Review
Funding
Funders who supported this work.
NIAID NIH HHS (5)
Grant ID: AI067968
Grant ID: R01 AI067968
Grant ID: R01 AI067968-04
Grant ID: R56 AI067968
Grant ID: R01 AI067968-03






