Abstract
Free full text

A chemical and phosphoproteomic characterization of dasatinib action in lung cancer
Associated Data
Abstract
We describe a strategy to comprehend signaling pathways active in lung cancer cells and targeted by dasatinib employing chemical proteomics to identify direct interacting proteins combined with immunoaffinity purification of tyrosine phosphorylated peptides corresponding to activated tyrosine kinases. We identified nearly 40 different kinase targets of dasatinib. These include SFK members (LYN, SRC, FYN, LCK, YES), non-receptor tyrosine kinases (FRK, BRK, ACK), and receptor tyrosine kinases (Ephrin receptors, DDR1, EGFR). Using quantitative phosphoproteomics we identified peptides corresponding to autophosphorylation sites of these tyrosine kinases that are inhibited in a concentration-dependent manner by dasatinib. Using drug resistant gatekeeper mutants, we show that SFK kinases, particularly SRC and FYN, as well as EGFR are relevant targets for dasatinib action. The combined mass spectrometry based approach described here provides a system-level view of dasatinib action in cancer cells and suggests both functional targets and rationale combinatorial therapeutic strategies.
Tyrosine kinase signaling pathways play an important role in the development and progression of cancer by regulating key pathways that drive the hallmarks of cancer including cell growth, survival, invasion/metastasis and angiogenesis (1). In non-small cell lung cancer (NSCLC), a disease characterized by high propensity for metastasis and inherent drug resistance, a number of receptor and non-receptor tyrosine kinases have been implicated in the biology of the disease including the epidermal growth factor receptor (EGFR), c-MET, and SRC family kinases (SFK) amongst others (2–6). Regulation of key pathways, such as PI3K/PTEN/AKT, STATs, Ras/Raf/MAPK/ERK, and focal adhesion kinase (FAK), allows these tyrosine kinases to control cancer hallmarks.
There is a good rationale for believing that inhibitors of SFK can have anti-tumor efficacy(7). However, understanding the proper role of SFK inhibitors as anticancer therapies is hampered by a number of interacting complexities. First, SFK signaling is complex. SFK form a closely related family of non-receptor tyrosine kinases (reviewed in (8) ) that link signaling from distinct upstream cell surface receptors to downstream effector pathways. SFK cooperate with receptor tyrosine kinases (RTK), including direct phosphorylation RTK, to modulate signaling and SFK activity is required for transformation by RTK such as EGFR (9–11). These mechanisms are especially relevant given recent reports that lung cancer cell lines harboring activated EGFR mutations are sensitive to SFK inhibitors at least in part through inactivation of EGFR signaling (5, 12–15). SFK signalling also exhibits a significant amount of redundancy. In addition to SRC, FYN and YES are also activated by EGFR signaling and share a nearly ubiquitous distribution as SRC itself (16, 17). Studies with SFK knockout mouse models suggest that YES and FYN compensate for a deficiency in SRC in all tissues but osteoclasts and thus targeting one specific SFK member may be insufficient to demonstrate a phenotype in cancer (reviewed in (18)). Second, SFK inhibitors such as dasatinib (1) and bosutinib can target kinases beyond the SFK family and these significant off-target effects could be important. (19–22). Third, the uniqueness of each individual tumor cell could affect phenotypic responses from SFK inhibitors. While evidence suggests elevated SFK activity in human tumors, the consequence of SFK inhibition in individual tumor cells is difficult to discern (8). Depending on cell context, SFK signaling has been reported to activate numerous downstream signaling pathways (7, 8). A search of the HPRD database finds nearly 100 proteins interacting with SRC alone (23).
Given the complexity of SFK signaling coupled with dasatinib promiscuity, we took a multi-tiered systems level approach to understand the effect of dasatinib on signaling networks in lung cancer cells. The workflow of the strategy is shown in Supp Fig 1. We determined direct kinase targets of dasatinib by applying lung cancer cell line protein extracts to dasatinib affinity matrices and identified binding interactions using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). This approach has the advantages of examining the entire proteome expressed in the disease-relevant profile (24). To determine which of these targets are tyrosine phosphorylated in lung cancer cells, we performed immunoaffinity purification of tyrosine phosphorylated peptides and subsequent LC/MS/MS identification. Kinases that were overlapping between the two screens were evaluated for in vivo effects of dasatinib using label free quantitative phosphoproteomics using LC-MS/MS. We determined targets relevant for dasatinib activity using RNA interference and by performing dasatinib rescue studies using drug resistant forms of identified dasatinib targets. The results demonstrate wide-spread effects of dasatinib on novel tyrosine kinases and suggest a global strategy to understand multi-targeted tyrosine kinase inhibitors in cancer cells.
RESULTS
Dasatinib binds tyrosine and serine/threonine kinases
Using c-dasatinib as a bait for a previously described drug-affinity chromatography approach, we reproducibly identified more than 40 different kinases between the three cell lines investigated, H292, H441 and HCC827 with the latter displaying the E746-A750 EGFR deletion mutant (Figure 1A and Supp Table S1, Supp Fig 2 and Supp Dataset 1) (22, 25). Among the detected kinases were the cognate targets of the ABL (ABL, ARG) and SFK (SRC, LYN, YES, LCK) families of tyrosine kinases. The SRC-related kinases FRK and BRK (PTK6) as well as ACK (TNK2) were prominently found. Another strongly represented interactor of dasatinib was the C-terminal SRC tyrosine kinase (CSK), which is known to act as a negative regulator of SRC (8). Furthermore, TEC was found to bind c-dasatinib prominently in all three cell types, which is consistent with previous findings showing nanomolar inhibition by dasatinib (25).
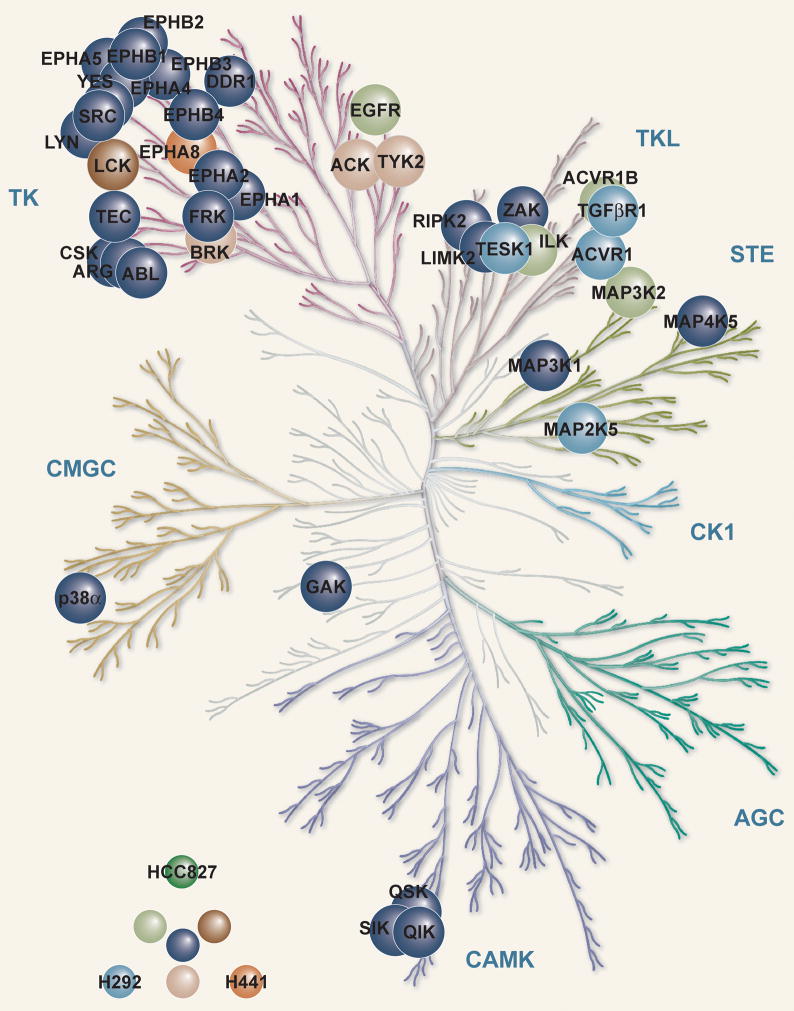
A. H292, H441 and HCC827 cell pellets were lysed and dasatinib-bound proteins were identified using c-dasatinib affinity matrixes and LC-MS/MS as described in Materials and Methods. The key for each cell line is listed in bottom left corner. Human Kinome reproduced courtesy of Cell Signaling Technology www.cellsignal.com
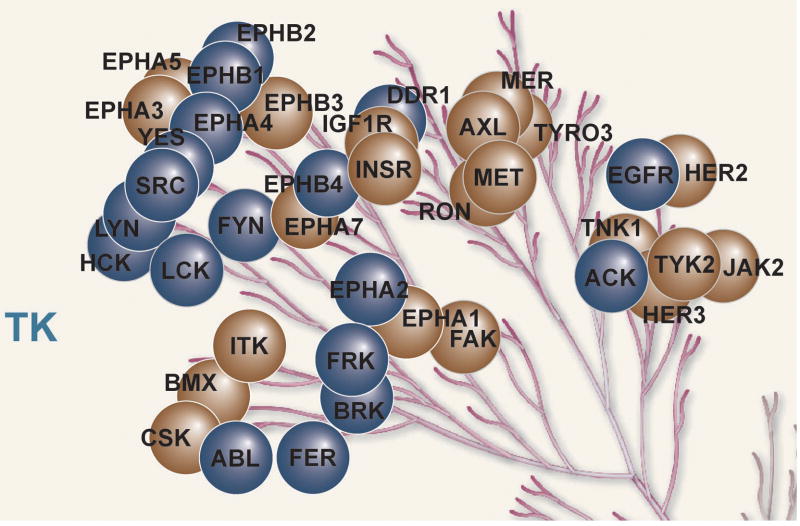
B. H292 cells were exposed to increasing concentrations of dasatinib for 3 hr after which purified pY peptides were identified and quantified. Tyrosine kinases demonstrating reduced levels of pY peptides are shown in blue circles and those showing no changes are shown in brown circles.
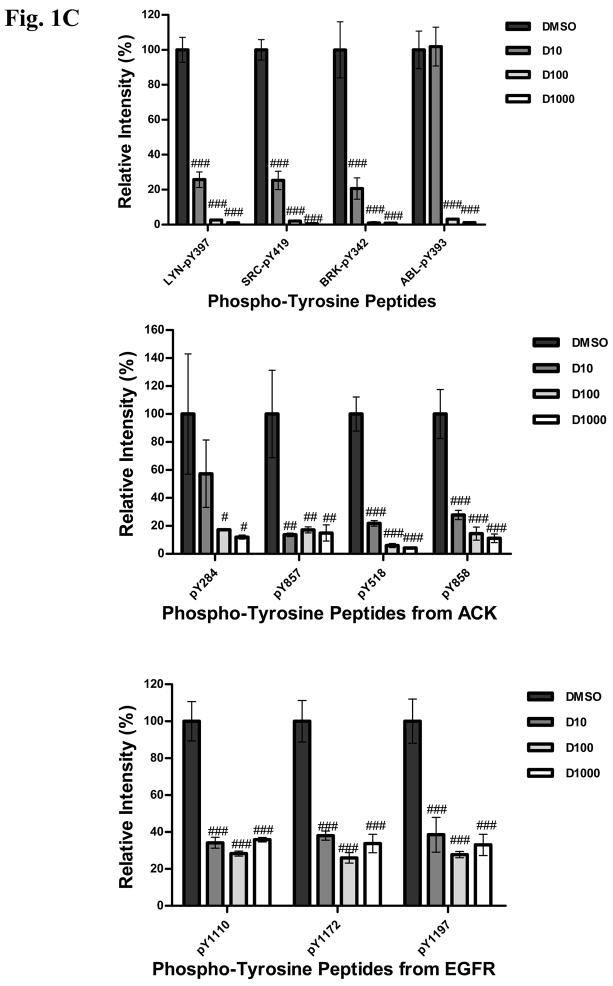
C: Effects of increasing concentration of dasatinib (10, 100, 1000 nM) on individual pY peptide amount is shown for selected targets. Y axis indicates intensity of individual peptides normalized to DMSO control. # = P<0.05; ## = P<0.01 and ### = P<0.001 using the Students T- test.
In addition to these tyrosine kinases, c-dasatinib also interacted with several serine/threonine kinases, such as cyclin G-associated kinase (GAK), the entire QIK subfamily (QIK, QSK and SIK), p38α (isoform 1 and 2), and multiple MAP kinases. Integrin-linked kinase (ILK), which is an important mediator of integrin signalling, was purified with c-dasatinib matrices from HCC827 and especially H292, but not from H441 cells (26). It is interesting to note that this pattern strongly correlates with the identification of several non-kinase proteins that are known to form a tight complex with ILK, namely PINCH, Parvin (α or β) as well as RSU1 (Supplementary Dataset 1) (26). This illustrates that the assay monitors the interaction with native molecular machines.
An important group of proteins identified as dasatinib binders are the receptor tyrosine kinases (RTKs) including the large ephrin receptor family, the members of which play various key roles in tumor biology and neurobiology. Among these, EPHA2, EPHB2 and EPHB4 are particularly strongly enriched by dasatinib in all three cell types, which correlates well with their overexpression in lung tumors (27). While EPHA2 and EPHB4 have been implicated with tumor growth, the role of EPHB2 is less clear. Noteworthy among the RTKs identified as dasatinib targets is also the discoidin domain receptor 1 (DDR1), a mediator of collagen signaling from the extracellular matrix, which we and others recently demonstrated to be potently inhibited by dasatinib (21, 22). DDR1 is highly expressed and moreover one of the most phosphorylated and thus activated RTKs in a large proportion of NSCLC tumors (28, 29). In H292 cells, we furthermore detected transforming growth factor β receptor 1 (TGFβR1) and the closely related activin A receptor 1 and 1B (also found in HCC827) to bind c-dasatinib.
Epidermal growth factor receptor (EGFR) as the hallmark of aberrant tyrosine kinase activity in NSCLC presented one of the most interesting findings of our study. Whereas c-dasatinib purified only low levels of wild-type EGFR as expressed in H292 and H441 cells, the activated mutant form expressed in HCC827 cells was detected as the dasatinib target with the highest number of unique peptides within this cell type (38 peptides). Consistent with this EGFR identification pattern, we also detected the well-known EGFR adaptor protein GRB2 only in HCC827 cells suggesting a co-purification of GRB2 with EGFR (Supplementary Dataset 1).
Dasatinib effects on target phosphorylation
We next turned to identifying which of these kinases, particular the tyrosine kinases, are (i) expressed in the active phosphorylated state in lung cancer cells and (ii) are inhibited by dasatinib. We used immunoaffinity capture of tyrosine phosphorylated peptides in conjunction with LC-MS/MS to produce a catalog of tyrosine phosphorylated proteins that could be screened against the dasatinib binding data and validated as targets inhibited by dasatinib. In addition to the H292, H441, and HCC827 cells used for the chemical targets screen, we included additional lung cancer cell lines including PC9, H1975, H460, H1299, and A549 cells to produce a more complete picture of tyrosine phosphorylation in lung cancer. Full data is presented in Supp. Dataset 2. Overall our analysis identified tyrosine phosphorylated peptides corresponding to 43 tyrosine kinases. Results from the chemical proteomics experiments and phosphotyrosine peptide/protein catalog were searched to identify proteins both bound to dasatinib and tyrosine phosphorylated and therefore possibly functionally important. Of the 40 kinases bound to dasatinib, we identified tyrosine phosphorylated peptides corresponding to 23. These kinases included ABL, ARG, DDR1, EGFR, EPHA1, EPHA2, EPHA4, EPHB1, EPHB2, EPHB3, EPHB4, FRK, LCK, LYN, p38α, BRK, RIPK2, SNF1LK, SRC, TEC, ACK, TYK2, and YES. These results indicate that the majority of the kinases bound to dasatinib are tyrosine phosphorylated and possibly important in signal transduction in lung cancer cells.
We then exposed H292 cells to increasing concentrations of dasatinib (10nM, 100nM, and 1000nM) for 3 hr, collected total proteins and digested these with trypsin. Tyrosine phosphorylated peptides were purified using anti-pY antibodies and identified by LC-MS/MS. For each run we generated extracted ion chromatograms to quantify the amounts of individual peptides (Supp Fig S3). Of the 38 tyrosine kinases identified through immunoaffinity pY purification in H292 cells, we found inhibition by dasatinib in 18 (Supp Table S2A, Figure 1B). We observed concentration-dependent reductions in the amount of tyrosine phosphorylated LYN, SRC, ACK, BRK, FRK, and ABL (Supp Table S2A, Fig. 1C). Dasatinib had dramatic effects on all Ephrin receptors evaluated. Finally, similar to what we and others have observed, dasatinib reduced the amount of tyrosine phosphorylated peptides corresponding to different sites on EGFR although, interestingly, no concentration-dependent effects were observed (5, 12). These findings with EGFR could be explained by inhibition of SFK-dependent tyrosine phosphorylation on EGFR and/or direct inhibition of EGFR autophosphorylation, as EGFR was found in our dasatinib pulldowns in H292 cells (30, 31). These experiments validate these tyrosine kinases as targets of dasatinib that can be inhibited at low nM concentrations.
In addition to the direct targets of dasatinib, we were able to examine indirect targets that change in response to inhibiting tyrosine kinase activity of the direct targets. We examined peptides corresponding to proteins reported to be SRC substrates for changes in tyrosine phosphorylation in response to dasatinib, such as FAK, CASL, paxillin, cortactin, and CDCP1 (Supp Table S2B). From these results we conclude that this approach can identify dasatinib-induced changes in both direct binding partners of dasatinib (tyrosine kinases) as well as downstream substrate proteins.
Phenotypic effect of RNAi knockdown of dasatinib targets
We initially attempted to evaluate a subset of these dasatinib targeted tyrosine kinases for evidence in their role in lung cancer cell growth using siRNA. Using pools of individual siRNA, we initially knocked down SRC, LYN, BRK, and ACK in H292 cells and examined the effects on cellular viability (Figure 2). The choice of targets was made based on evidence cited from the literature that particular tyrosine kinases, such as SRC, BRK, and ACK, can be involved in cellular growth pathways and can potentially interact with EGFR. Similar to effects seen with SRC, BRK has been reported to physically interact with EGFR and BRK overexpression can lead to sensitization of cells to EGF and can enhance Erb-mediated activation of PI3K (32). BRK has also been shown to affect breast cancer proliferation through ERK5 and p38 signaling pathways (33). ACK has also been reported to interact with EGFR and has been implicated as an important downstream substrate in Ras-transformed cells (34, 35). While LYN is typically thought to be more important in hematological tissues, our results along with more recent reports suggests a possible role for LYN in epithelial cancers such as lung and prostate cancer (36). We felt that it was unlikely that inhibition of a single target would result in substantial changes in cell growth absent evidence of genomic alterations or “oncogene addiction”. This is especially relevant for SFK where functional redundancy is known to exist. Indeed, to our knowledge, H292 cells have no genomic alteration in a tyrosine kinase that renders them addicted to this particular protein.
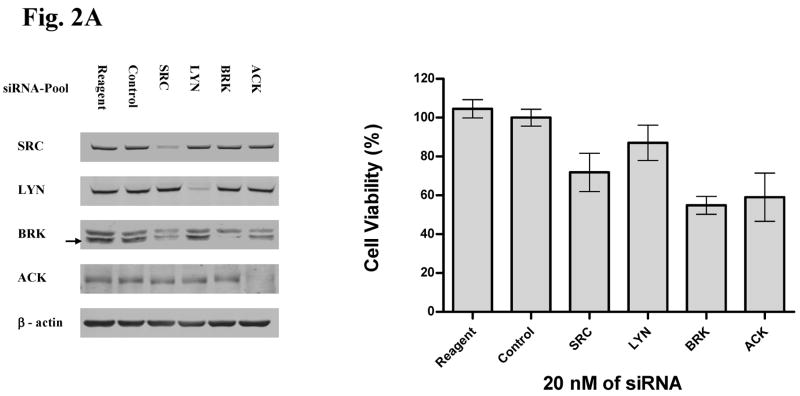
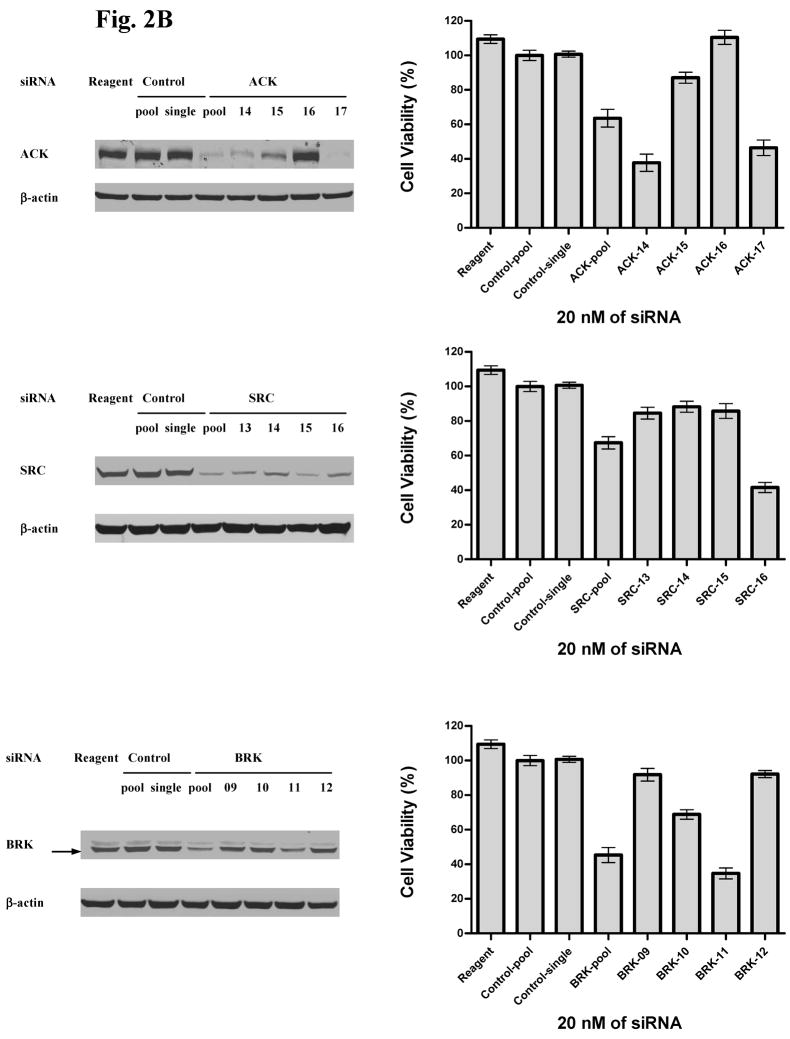
A. In left panel, H292 cells were exposed to indicated siRNA molecules for 72 hr and western analysis used to determine target modulation. Reagent = transfection reagents without siRNA, control siRNA = mismatch siRNA with transfection reagents. Arrow indicates BRK band at appropriate molecular weight compared to non-specific upper band. In right panel, effects of indicated siRNA on cell viability were assessed after 12 days. Results were normalized to reagent-only treated cells and error bars represent standard deviation.
B. H292 cells were exposed to indicated siRNA molecules and effects on target modulation and cell viability assessed. Reagent = transfection reagents without siRNA, pool = 4 individual siRNA combined with final concentration of 20 nM, single = single siRNA molecule, numbers represent individual siRNA against indicated targets, control = mismatch siRNA. β-actin was used to ensure equal loading for western blotting. In right panel, effects of indicated siRNA on cell viability was assessed after 12 days. Results were normalized to reagent-only treated cells and error bars represent standard deviation.
As expected, knockdown of these kinases had modest effects on the growth of H292 cells. However, both ACK and BRK knockdown resulted in more inhibition of cell growth compared to SRC or LYN (Figure 2A). We performed further validation of these initial experiments with individual siRNA against ACK, BRK, and SRC (Figure 2B and Supp. Fig S4). These results suggest good correlation between the degree of ACK knockdown and effects on cell growth further supporting the role of ACK in controlling cell growth. SRC knockdown had consistent effects on target inhibition and effects on cell growth but were modest compared to effects seen with ACK. Effects of individual siRNA on BRK knockdown paralleled effects seen on cell viability although only one of four siRNA had obvious effects on BRK protein, yielding less confidence in functional relevance of this target. We attempted similar studies with HCC827 cells but found no significant effects of any of the tested siRNA on cell viability (data not shown). These results in HCC827 cells suggest that the combinatorial effects of multiple target inhibition by dasatinib is necessary for the anti-growth properties of dasatinib or alternatively other dasatinib targets that we did not evaluate are more important in regulating growth and survival in this particular cell line driven by mutant EGFR.
Functional characterization of dasatinib targets
We next addressed which of these dasatinib targets identified by chemical proteomics and phosphoproteomics contributes to anti-proliferative and/or pro-apoptotic effects of dasatinib in lung cancer cells. Since cell context is important in response to dasatinib, we initially examined both HCC827 cells with activating EGFR mutation and H292 cells with wildtype EGFR. We used drug resistant forms of kinases to delineate their roles in dasatinib induced apoptosis and/or growth arrest (37). Cells were infected with lentivirus expressing either wildtype kinases or kinase alleles with gatekeeper mutations and cell viability examined in response to increasing concentrations of dasatinib (Figure 3 and Supp. Fig S5, S6 and S7). We included in this analysis 13 tyrosine kinases: SRC, FYN, LYN, LCK, FRK, DDR1, ABL, EPHB1, EPHA2, EPHA4, ACK, BRK, and EGFR. Protein expression of the lentiviral constructs was confirmed in 10 of the 13 targets (3 were not evaluated, Supp. Fig S7).
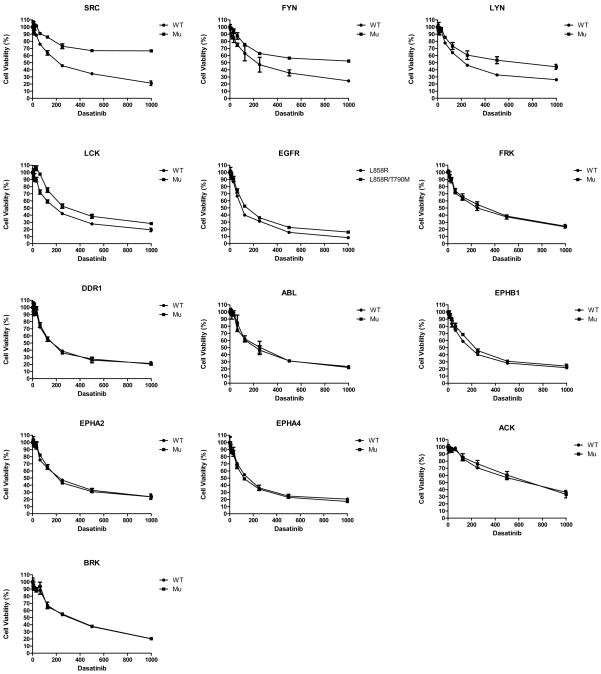
HCC827 cells were infected with lentivirus expressing wildtype and mutant gatekeeper form of each indicated target for 48 hr. Subsequently cells were exposed to increasing concentrations of dasatinib for 120 hr after which cell viability was assessed. Cell viability as a function of dasatinib concentration is plotted with error bars representing standard deviation and results normalized to DMSO treated cells.
First, we find that SRC, FYN, and to some extent LYN and LCK are able to rescue HCC827 cells from dasatinib (Figure 3). No effects were observed with the remaining dasatinib targets except some minor effects of EGFR. Similar studies were performed in H292 cells, where dasatinib results in cell cycle arrest (Supp Fig S5A and Supp Fig S8B). Similar to that of HCC827 cells, we found evidence that SFK rescue effects of dasatinib, as SRC and FYN led to dasatinib rescue; this compares to LYN and LCK that had minimal effects. None of the other targets examined, including ACK, demonstrated rescue of dasatinib activity in these cells. Thus, while our siRNA results suggests that levels of ACK protein expression can be important in control of cellular growth in H292 cells, ACK kinase activity is not critical in the context of dasatinib-induced growth arrest.
We next examined downstream signaling in these cells expressing dasatinib resistant kinases with focus on SRC and EGFR (Figure 4). We examined effects of dasatinib on EGFR and SRC phosphorylation as well as effects on downstream AKT and ERK phosphorylation in three cell lines (HCC827, PC9, and H292). In these experiments, we included PC9 cells as a drug insensitive SRC mutation protected cells from dasatinib similar to that in HCC827 cells (Supp Fig S5B). In both the HCC827 and PC9 cells infected with lentivirus expressing wildtype SRC, dasatinib resulted in progressive concentration-dependent decreases in tyrosine 1068 and 845 EGFR, AKT, and ERK phosphorylation consistent with prior observations (Figure 4A) (5). Cells expressing the SRC gatekeeper mutant exposed to dasatinib retain phosphorylation at the autophosphorylation site of SFK (pY 419) yet demonstrate similar reduced tyrosine 1068 phosphorylation of EGFR as cells infected with wildtype SRC. This contrasts to lack of effect on tyrosine 845 phosphorylation of EGFR consistent with this site being a major SRC phosphorylation site. A gatekeeper of ABL was similarly unable to rescue effects on EGFR tyrosine phosphorylation (Supp. Fig S9). In HCC827 cells, we found reductions in AKT and ERK phosphorylation, although not to the extent seen in wildtype SRC infected cells, suggesting some compensation by SRC to maintain signaling despite EGFR inhibition. More pronounced effects of SRC on maintenance of AKT and ERK phosphorylation are observed in the PC9 cells. Consistent with the idea that SRC can partially compensate for EGFR inhibition in the context of dasatinib treatment, we find that dasatinib-induced apoptosis is lower in both HCC827 and PC9 cells expressing the SRC gatekeeper as opposed to wildtype SRC (Figure 4B). The combined findings that (i) inhibition of EGFR 1068 phosphorylation (a known autophosphorylation site) by dasatinib despite persistent SRC activity and (ii) EGFR identified in our dasatinib pulldowns strongly suggest that EGFR is also a direct target of dasatinib. The results of dasatinib on in vivo phosphorylation also agree with our recently reported results finding dasatinib inhibits EGFR kinase activity with more pronounced effects seen with activating EGFR mutants compared with wildtype EGFR (38). These results also suggest that, to some extent, persistent SRC compensates for inhibition of EGFR depending on cell context.
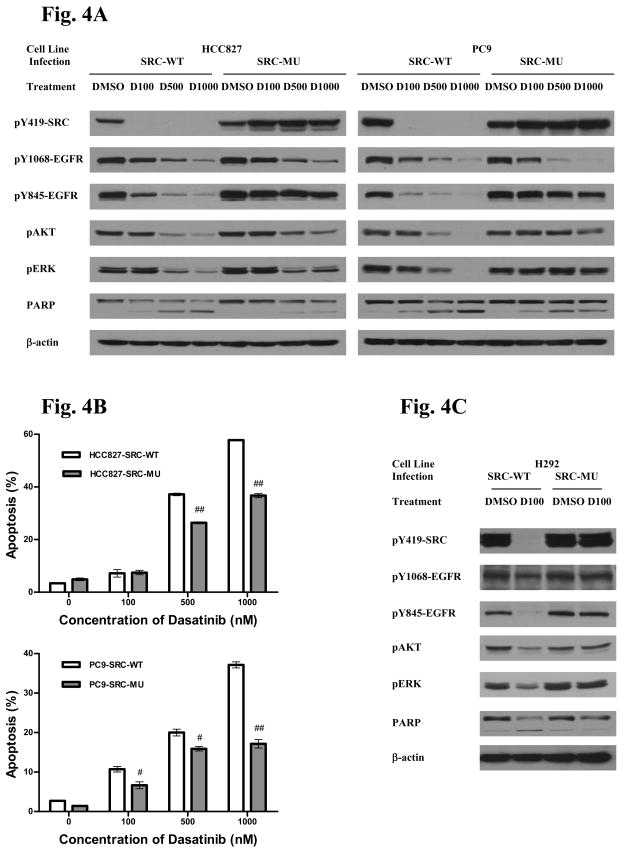
A. HCC827 and PC9 cells were infected with lentiviruses expressing either wildtype SRC or SRC gatekeeper mutant. Cells were exposed to indicated concentrations of dasatinib(D), for example D100 = 100 nM of dasatinib, for 24 hr and protein lysates used for western analysis with indicated antibodies. DMSO = solvent control.
B HCC827 and PC9 cells were infected with lentiviruses expressing either wildtype SRC or SRC gatekeeper. Cells were exposed to indicated concentrations of dasatinib for 60 hr and apoptosis assayed using cleaved-caspase-3. Results indicate mean ± SD. # = P<0.01 and ## = P<0.001 using the Students T-test.
C. H292 cells were infected with lentiviruses expressing either wildtype SRC or SRC gatekeeper mutant. Cells were exposed to 100 nM of dasatinib for 24 hr and protein lysates used for western analysis with indicated antibodies.
We also examined signaling in the H292 cells (Figure 4C). Tyrosine 1068 is difficult to assess in these cells given very low levels of tyrosine phosphorylated EGFR but some subtle reduction is apparent with dasatinib and stronger more obvious effects are seen on tyrosine 845 phosphorylation. In cells expressing the SRC gatekeeper, tyrosine 845 is clearly maintained while it is difficult to assess effects on tyrosine 1068. Dasatinib inhibits AKT and ERK phosphorylation in cells infected with wildtype SRC while the gatekeeper SRC infected cells have no changes in AKT or ERK phosphorylation in the presence of dasatinib. These results also implicate SRC in control of distal signaling in lung cancer cells with wildtype EGFR.
We also included in this rescue analysis (Figure 3 and Supp Fig S5A) an EGFR allele with activating mutation (L858R) along with a T790M gatekeeper mutation (T790M) since our chemical proteomics results strongly implicated mutant EGFR in the HCC827 cells as a direct target of dasatinib. Interestingly, while expression of L858R/T790M resulted in minimal rescue effects in HCC827, more pronounced effects were seen in H292 cells. As HCC827 have been recently reported to have 35 copies of EGFR, we considered whether the low levels of lentiviral-driven L858R/T790M EGFR may have been insufficient to rescue cells from effects of dasatinib (39). We therefore performed similar studies in PC9 cells that contain an E746-A750 EGFR deletion mutation, are highly sensitive to EGFR TKI, and contain a total of 5 copies of EGFR (39).
Changes in dasatinib and erlotinib(2) induced cell viability were assessed in PC9 cells stably expressing either a mutant drug sensitive EGFR allele (L858R) or a drug resistant allele (L858R/T790M)(Figure 5A). Here expression of the L858R/T790M allele has a strong rescue effect on dasatinib viability suggesting that EGFR is an important target in the context of dasatinib-induced death. Signaling studies show again that cells expressing the L858R allele have reduced EGFR, AKT, and ERK phosphorylation in response to increasing concentrations of dasatinib (Figure 5B). This is especially apparent at concentrations of dasatinib greater than 100 nM. In cells expressing L858R/T790M, while dasatinib had some effect on EGFR phosphorylation, the effect was incomplete even up to 1000 nM, and minimal changes were observed in phosphorylated AKT and ERK. In agreement with the effects on EGFR phosphorylation and AKT/ERK signaling, cells expressing the T790M form of L858R EGFR had less apoptosis when exposed to dasatinib (Figure 5C).
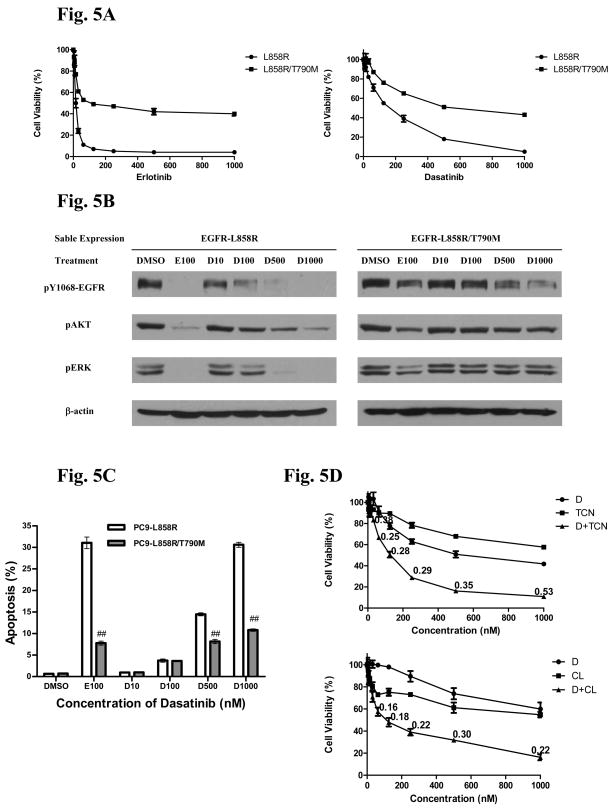
A. PC9 cells expressing either L858R EGFR or L858R/T790M EGFR were exposed to indicated concentrations of dasatinib (D) or erlotinib (E) for 120 hr and cell viability was accessed. Cell viability was normalized to untreated cells and error bars represent standard deviation.
B PC9 cells expressing either L858R EGFR (left panel) or L858R/T790M EGFR (right panel) were exposed to indicated concentrations of erlotinib (E) or dasatinib (D) for 24 hr. Protein lysates were transferred to membranes and probed with indicated antibodies.
C. PC9 cells expressing either L858R EGFR or L858R/T790M EGFR were exposed to indicated concentrations of dasatinib for 60 hr and apoptosis assayed using cleaved-caspase-3. Results indicate mean ± SD. # = P<0.01 and ## = P<0.001 using the Students T-test.
D. PC9 cells expressing L858R/T790M EGFR were exposed to indicated concentrations of dasatinib (D), tricirabine (TCN), or the combination for 120 hr after which cell viability was assessed. A parallel group of cells were exposed to indicated concentrations of dasatinib (D), CL-387,785 (CL), or the combination for 120 hr after which cell viability was assessed. Values at each data point indicate combination index calculated by Chou-Talaly approach.
The results suggest that full activity of dasatinib in lung cancer cells with mutant EGFR results from dual SRC and EGFR inhibition. As acquired T790M mutations render cells resistant to dasatinib similar to that observed with erlotinib, this would limit effects of dasatinib in the resistant state (40). As the above results demonstrate an important role for SRC, we considered if dual SRC inhibition in conjunction with inhibition of mutant EGFR-driven AKT signaling could be effective in cells with EGFR gatekeeper mutations. Therefore, we examined dasatinib in combination with two strategies to inhibit the EGFR-AKT pathways: direct AKT inhibition or indirect AKT pathway inhibition using a compound able to overcome T790M-mediated resistance such as an irreversible EGFR kinase inhibitor. PC9 cells expressing the T790M allele were exposed to increasing concentrations of dasatinib, the AKT inhibitor tricirabine(3), or the combination; a parallel group of cells received dasatinb, the irreversible EGFR inhibitor CL-387,785(4), or the combination (41, 42). The results in Figure 5D show strong synergistic combination effects of dual SRC and AKT inhibition (either by direct AKT inhibition with tricirabine or indirect through CL-387,785) suggesting this as a rationale strategy to overcome acquired EGFR resistance by the T790M gatekeeper.
DISCUSSION
We describe a system wide, multi-tiered MS-based approach to discern mechanisms of dasatinib in lung cancer cells using drug affinity purification for identifying tyrosine kinase binding partners coupled with immunoaffinity purification of tyrosine phosphorylated peptides to identify and measure drug-sensitiving of active forms of these kinases and their substrates. This approach is able to surmount the complex function of SFK in signal transduction and the highly promiscuous nature of dasatinib and thus could be important in discerning tyrosine kinase inhibitor mechanism of action in diverse tumor types. Overall we believe this translational strategy could be important for candidate target prioritization and along with more complex algorithms to detect changes in peptide amount could be used to interrogate pathway effects of tyrosine kinase inhibitors.
Our results define a set of signaling nodes modulated by dasatinib that can be considered targets in lung cancer and possibly other epithelial cancers. This includes a large set of non-receptor tyrosine kinases including multiple SFK members, ACK, BRK, FRK as well as receptor tyrosine kinases such as EGFR and Eph members. It is notable that the dasatinb affinity matrix identified SRC kinase family members in lung cancer cells beyond SRC including LYN, LCK, and YES. A number of dasatinib targets appear to be not only expressed in the active state in cell lines but also in tumor specimens, such as EGFR, DDR1, SRC family members, BRK, FRK, and ACK (Supp Figure S10) (29). How these kinases influence lung cancer biology, treatment sensitivity and overall prognosis remains to be understood. Given the role of genetic alterations in tyrosine kinases and sensitivity to tyrosine kinase inhibition it will be interesting to examine these dasatinib tyrosine kinase targets for evidence of genetic alterations. It is also possible that dasatinib targets play important roles in signal transduction in lung cancer cells “addicted” to upstream tyrosine kinases such as EGFR, ALK, and PDGFR. ALK and PDGFR, which is also a direct target of dasatinib, are known to signal through SRC kinases and may require the function of other dasatinib kinase targets for signal transduction (9). Thus, these studies suggest combination strategies of dasatinib with other TKI or signal transduction inhibitors that attack critical signaling pathways important for tumor maintenance.
ACK results are particularly interesting and worth some discussion. ACK has been found to be amplified in some patients with lung cancer and this correlates with advanced stage of disease (43). We identified ACK in dasatinib pulldowns in H292 and H441 cells. The major site of autophosphorylation on ACK is Y284 which was found to be reduced by dasatinib treatment (44). In addition to this site, Y518 phosphorylation was reduced as well as Y857/858. In addition to its kinase activity, the C-terminal domain of ACK can form complexes with SH2 and SH3 domains of other important signaling proteins including SRC, HCK, NCK, and GRB2 (44–46). Our observation that siRNA mediated knockdown of ACK inhibited cell growth, while no rescue was observed in dasatinib treated cells expressing ACK with a gatekeeper mutation, was surprising at first. These results however may suggest that scaffold function of ACK, rather than kinase activity, could be important in the context of cell growth control in these cells. Similar seemingly paradoxical findings were also found in EGFR, where kinase dead forms of EGFR could still elicit downstream signaling (47). In addition to inhibition of kinase activity by dasatinib, the inhibition of ACK tyrosine phosphorylation may reflect indirect effects of SRC inhibition and could be important in deregulating ACK scaffold functions, proper protein localization, or formation of protein-protein complexes important for downstream signaling. Further studies are necessary to examine ACK interactions as well as evaluate functions of distinct tyrosine phosphorylation sites. These results also highlight the ability of the combined chemical proteomics, quantitative proteomics, and dual validation strategy to glean novel insights into kinase function.
Compared to previous studies in leukemia, we observed a high prevalence of RTKs and serine/threonine receptor kinases present in the NSCLC dasatinib profiles. (22). This applies particularly to EGFR and here mainly to the deletion mutant present in HCC827 cells, which might have a higher affinity for dasatinib in addition to being overexpressed as compared to wild-type EGFR found in H292 and H441. This observation however correlates well with the different susceptibility of this cell line to dasatinib as compared to H292 and H441. Also the ephrin-type A receptor tyrosine kinases, and within these particularly EPHA2, are as a family strongly represented throughout the investigated NSCLC cell lines while not identified in K562. ILK (H292 and HCC827) and TGFβR1 (H292) are absent from the K562 profile as well, although they are also only identified in certain NSCLC cell lines. Validation of ILK and TGFβR1 as targets of dasatinib could be interesting given the important roles these proteins play in metastasis of lung cancer (48, 49). Likewise, the contribution of MAPK binding of dasatinib to changes in signalling and physiology remains to be determined.
The correlation of the generated drug target profiles with quantitative phosphoproteomics analyses and subsequent validation of functional targets in this study may offer additional explanations for the pharmacological effects seen in these cells with dasatinib and thus highlight the potential of a multi-targeted drug such as dasatinib for the application in NSCLC. The dual EGFR and SFK activity of dasatinib raises important considerations for how these agents are used clinically. Our experience with dasatinb in lung cancer patients finds a peak concentration of ~600 nM with daily dosing and a short half life of ~3 hours (unpublished). These pharmacokinetic properties may be effective for SFK inhibition but may be insufficient for durable and potent EGFR inhibition, both critical factors for killing tumors cells driven by oncogenic kinases (50). Our results also suggest that gatekeeper mutations in EGFR, found in nearly 50% of tumors with acquired resistance to EGFR TKI, are likely to be resistant to dasatinib. Nonetheless, our results suggest that pairing of dasatinib, which effectively eliminates SFK signaling, with AKT pathway inhibition with either irreversible EGFR inhibitors or direct AKT inhibitors, could be effective in this patient subset.
In general, our results suggest that dasatinib, besides inhibiting downstream signaling by SFK, could simultaneously target NSCLC at many different nodes and shut down multiple relevant signaling pathways originating from the extracellular matrix (integrins, collagens), ephrins and growth factors such as EGF and TGFβ. As suggested also by our functional data, dasatinib could therefore present an important component for combination therapy with other pathway inhibitors.
MATERIALS AND METHODS
Full descriptions of all materials and methods can be found under “Supplemental Methods and Materials”.
Cell Lines, Plasmids and Reagents
All cells were maintained in RPMI-1640 medium supplemented with 10% newborn calf serum (NCS) from Sigma (St. Louis, MO) except H292 and H441 cells which were grown in RPMI-1640 medium containing glucose. cDNA for L858R EGFR and L858R/T790M EGFR were provided by Dr. William Pao (Vanderbilt University, Nashville, TN), ACK was provided by Dr. Nupam Mahajan (Moffitt Cancer Center, FL), and Human BRK was purchased from ORIGENE (Rockville, MD). Lentiviral plasmids containing WT and gatekeeper SRC, FYN, LYN, LCK, ABL, DDR1, FRK, EphB1, EphA2 and EphA4 were generously provided by Dr. Jinyan Du and Dr. Todd Golub (The Broad Institute, MA). Erlotinib was provided by OSI Pharmaceuticals (Melville, NY) and dasatinib by Bristol-Myers Squibb (New Brunswick, NJ). Tricirabine were provided by Dr. Jin Cheng (Moffitt Cacner Center, FL) and CL-387,785 was purchased from AXXORA (San Diego, CA). Stock solutions of dasatinib, erlotinib, tricirabine and CL-387, 785 in 100% DMSO were diluted directly into the media to indicated concentrations.
Drug Affinity Purification, Data Processing, and Analysis
Drug affinity experiments were performed in triplicate and analyzed by nano-LC tandem mass spectrometry as described previously using total cell lysates from H292, H441 and HCC827 cells (22).
Phosphopeptide Immunoprecipitation
Phosphopeptides were purified according to manufacturer’s recommendations of Cell Signaling PhosphoScan pTyr100 Kits (Beverly, MA). Briefly, 1×10^8 cells were lysed in urea buffer; extracted proteins were reduced by dithiothreitol, alkylated by iodoacetamide, and then digested by trypsin. Peptide purification was carried out using Sep-Pak C18 columns and anti-phosphotyrosine antibody beads.
Analysis of Phosphopeptides by LC-MS/MS
The eluted peptide mixtures were analyzed using a nanoflow liquid chromatograph (U3000, Dionex, Sunnyvale, CA) coupled to an electrospray ion trap mass spectrometer (LTQ-Orbitrap, Thermo, San Jose, CA) in a data-dependent manner for tandem mass spectrometry peptide sequencing experiments. Both MASCOT and SEQUEST search results were summarized in Scaffold 2.0. The integrated peak areas for phosphotyrosine peptide quantification were calculated from extracted ion chromatograms (EIC) using QuanBrowser from Xcalibur 2.0.
Cell Viability, Proliferation and Apoptosis Assays
Cell viability assays (CellTiter-Glo) were employed according to the manufacturer’s recommendations of CellTiter-Glo Luminescent Cell Viability Assay from Promega (Madison, WI). Cell cycle profiles were analyzed in a flow cytometer (FACS 420, Becton Dickinson) after propidium iodide staining (Roche (Indianapolis, IN)). Apoptosis assays were performed according to manufacturer’s recommendations using PE-conjugated Monoclonal Active Caspase-3 Antibody Apoptosis Kit (BD Pharmingen, San Diego, CA). Significant differences between values obtained in DMSO control groups and different treatment groups were determined using the Students t test. P values of less than0.05 were assigned significance.
Protein Expression Analysis
Western blotting was performed as previously described (5). Odyssey Infrared Imaging System detection of proteins was accomplished using IRDye™ 800CW or IRDye™ 680 - labeled secondary antibody from LI-COR (Lincoln, NE) and then imaged using the LI-COR Odyssey Infrared Imaging Scanner.
Transfection of Small Interfering RNA
The small interfering RNAs (siRNA) used were ON-TARGET plus SMART pool along with mismatch as a negative control obtained from Dharmacon (Chicago, IL). Transfection was performed with lipofectamine™ RNAiMAX from Invitrogen (Carlsbad, CA) using reverse transfection procedure as recommended by manufacturer.
Dasatinib resistant alleles and rescue experiments
Open reading frames (ORFs) of BRK, EGFR L858R and EGFR L858R/T790M were PCR amplified from the relevant plasmids using Pfu DNA Polymerase from Stratagene (La Jolla, CA) and AccuPrime Pfx DNA Polymerase from Invitrogen for ACK amplification, and then the inserted entry vector for the gateway system using pENTR D-TOPO Cloning Kits from Invitrogen (Carlsbad, CA). ACK and BRK gatekeeper residues were identified by previous methods (37). The ATP kinase domain was identified (http://www.expasy.org/prosite/) and all selected kinases domain were clustered via ClustalW2 from EMBL-EBI (http://www.ebi.ac.uk/Tools/clustalw2/index.html). From these results, the threonine gatekeeper position in ACK and BRK were identified and mutated to isoleucine.
Mutagenesis was performed by QuikChange II Site-Directed Mutagenesis Kit from Stratagene in the entry vector. Wild-type and gatekeeper mutants ACK, BRK and EGFR were inserted into pLenti6/UbC/V5-DEST gateway vector from Invitrogen. Each lentivirus expressing either wild type or gatekeeper mutant tyrosine kinase was produced in 293FT cells according to the manufacturer’s recommendations of ViraPower Lentiviral Expression Systems (Invitrogen).
For rescue experiments, HCC827 or H292 cells were seeded in black wall 96-well plate from NUNC (Rochester, New York). After overnight incubation, cells were infected with 30μl of viruses per well for 48 hours, and then treated with a series dilution of dasatinib for 5 days for CellTiter-Glo cell viability assay. Stable cell lines such as EGFR T790M or L858R PC9 cell lines were generated by blasticidin selection two weeks after EGFR T790M or L858R lentivirus infection.
Acknowledgments
We thank Frank Lee (BMS Oncology) for providing dasatinib, Genentech & OSI Pharmaceuticals for providing erlotinib, Klarisa Rikova, (Cell Signaling Technology, Danvers, MA) for providing NSCLC tumor spectral count data, Cell Signaling Technology (Danvers, MA) for allowing reproduction of the kinome map, Gerhard Durnberger for data uploading to PRIDE, Drs. Jinyan Du and Todd Golub for providing lentiviral constructs, Takeshi Yoshida M.D. for helpful discussions, and Patricia Johnston for administrative assistance. The work was partially funded by grants from the National Functional Genomics Center, the Moffitt Cancer Center SPORE in Lung Cancer (P50-CA119997), Joan’s Legacy/LUNGevity Foundation (E.B.H), the Austrian federal ministry of science and research bm.wf within the GEN-AU program (GZ200.142/1-VI/I/2006 and GZ 200.145/1-VI/1/2006) and by the Austrian Academy of Sciences (ÖAW). This work has been supported in part by the Proteomics Core, Molecular Biology and Sequencing Core, and the Flow Cytometry Core at the H. Lee Moffitt Cancer Center & Research Institute (Tampa, FL).
Footnotes
AUTHORSHIP
J.L. designed and performed the experiments, analyzed and interpreted the data, performed statistical analyses, made the figures and wrote the manuscript; U.R. performed all chemical proteomics experiments, analyzed and interpreted the data, performed statistical analyses, made the figures and wrote the manuscript; B.F. performed all extracted ion chromatogram experiments, analyzed and interpreted the data, performed statistical analyses, made the figures and wrote the manuscript in the US; A.E. provided statistical and bioinformatics support for interpreting the LC-MS/MS data in the US; S.E. advised on the research, performed statistical and bioinformatics support for interpreting the LC-MS/MS data and critically read the manuscript; J.G. helped perform pY peptide purification from lung cancer cell lines; Y.B. provided technical assistance with cell lines; L.S. provided technical help with mutagenesis; K.B. analyzed chemical proteomics experiments and operated mass spectrometers in Austria; J.C. analyzed chemical proteomics experimental data and performed the bioinformatics analyses in Austria; J.K. advised on the research, supervised all proteomics experiments in the US, and critically read the manuscript; G.S.F. supervised the chemical proteomics experiments, advised on the research and critically read the manuscript; E.B.H. was overall responsible for this research and edited the manuscript.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nchembio.332
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2842457
Citations & impact
Impact metrics
Article citations
Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14(1):9923, 30 Apr 2024
Cited by: 0 articles | PMID: 38688959 | PMCID: PMC11061198
Prognostic value of RNA methylation-related genes in gastric adenocarcinoma based on bioinformatics.
PeerJ, 12:e16951, 29 Feb 2024
Cited by: 2 articles | PMID: 38436027 | PMCID: PMC10909369
Characterization of gastric cancer-stimulated signaling pathways and function of CTGF in cancer-associated fibroblasts.
Cell Commun Signal, 22(1):8, 02 Jan 2024
Cited by: 3 articles | PMID: 38167009 | PMCID: PMC10763493
CSF3R T618I Collaborates With RUNX1-RUNX1T1 to Expand Hematopoietic Progenitors and Sensitizes to GLI Inhibition.
Hemasphere, 7(10):e958, 11 Oct 2023
Cited by: 0 articles | PMID: 37841755 | PMCID: PMC10569757
Differential network analysis of ROS1 inhibitors reveals lorlatinib polypharmacology through co-targeting PYK2.
Cell Chem Biol, 31(2):284-297.e10, 16 Oct 2023
Cited by: 0 articles | PMID: 37848034
Go to all (193) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: possible combinations in solid tumors.
Clin Cancer Res, 17(17):5546-5552, 13 Jun 2011
Cited by: 184 articles | PMID: 21670084
Review
Dasatinib (BMS-354825) tyrosine kinase inhibitor suppresses invasion and induces cell cycle arrest and apoptosis of head and neck squamous cell carcinoma and non-small cell lung cancer cells.
Clin Cancer Res, 11(19 pt 1):6924-6932, 01 Oct 2005
Cited by: 202 articles | PMID: 16203784
Dasatinib inhibits site-specific tyrosine phosphorylation of androgen receptor by Ack1 and Src kinases.
Oncogene, 29(22):3208-3216, 12 Apr 2010
Cited by: 68 articles | PMID: 20383201 | PMCID: PMC2880659
Transforming growth factor β signaling overcomes dasatinib resistance in lung cancer.
PLoS One, 9(12):e114131, 11 Dec 2014
Cited by: 5 articles | PMID: 25501935 | PMCID: PMC4263601
Funding
Funders who supported this work.
NCI NIH HHS (3)
Grant ID: R01 CA123174-02
Grant ID: P50 CA119997
Grant ID: R01 CA123174





