Abstract
Background
The ubiquitin proteasome system maintains a dynamic equilibrium of proteins and prevents accumulation of damaged and misfolded proteins, yet its role in human cardiac dysfunction is not well understood. The present study evaluated ubiquitin proteasome system function in human heart failure and hypertrophic cardiomyopathy (HCM).Methods and results
Proteasome function was studied in human nonfailing donor hearts, explanted failing hearts, and myectomy samples from patients with HCM. Proteasome proteolytic activities were markedly reduced in failing and HCM hearts compared with nonfailing hearts (P<0.01). This activity was partially restored after mechanical unloading in failing hearts (P<0.01) and was significantly lower in HCM hearts with pathogenic sarcomere mutations than in those lacking these mutations (P<0.05). There were no changes in the protein content of ubiquitin proteasome system subunits (ie, 11S, 20S, and 19S) or in active-site labeling of the 20S proteolytic subunit beta-5 among groups to explain decreased ubiquitin proteasome system activity in HCM and failing hearts. Examination of protein oxidation revealed that total protein carbonyls, 4-hydroxynonenylated proteins, and oxidative modification to 19S ATPase subunit Rpt 5 were increased in failing compared with nonfailing hearts.Conclusions
Proteasome activity in HCM and failing human hearts is impaired in the absence of changes in proteasome protein content or availability of proteolytic active sites. These data provide strong evidence that posttranslational modifications to the proteasome may account for defective protein degradation in human cardiomyopathies.Free full text

Ubiquitin Proteasome Dysfunction in Human Hypertrophic and Dilated Cardiomyopathies
Associated Data
Abstract
Background
The ubiquitin proteasome system (UPS) maintains a dynamic equilibrium of proteins and prevents accumulation of damaged and misfolded proteins, yet its role in human cardiac dysfunction is not well understood. The present study evaluated UPS function in human heart failure and hypertrophic cardiomyopathy (HCM).
Methods and Results
Proteasome function was studied in human nonfailing donor (NF) hearts, in explanted failing hearts (F), and in myectomy samples from HCM patients. Proteasome proteolytic activities were markedly reduced in F and HCM hearts vs NF hearts (P<0.01). This activity was partially restored after mechanical unloading in F hearts (P<0.01) and was significantly lower in HCM hearts with pathogenic sarcomere mutations than in those lacking these mutations (P<0.05). There were no changes in the protein content of UPS subunits (i.e. 11S, 20S, and 19S) or in active site labeling of the 20S proteolytic subunit β-5 among groups to explain decreased UPS activity in HCM and F hearts. Examination of protein oxidation revealed that total protein carbonyls and 4-hydroxynonenated proteins, and oxidative modification to 19S ATPase subunit Rpt 5, were increased in F hearts vs NF hearts.
Conclusions
Proteasome activity in HCM and F human hearts is impaired in the absence of changes in proteasome protein content or availability of proteolytic active sites. These data provide strong evidence that post-translational modifications to the proteasome may account for defective protein degradation in human cardiomyopathies.
Proteolytic degradation is critical for maintaining a dynamic equilibrium of proteins and destroying damaged or misfolded proteins. As the major pathway for intracellular protein degradation, the ubiquitin proteasome system (UPS) requires precise control to sustain most biological processes. Regulation of proteasome function may occur by altered proteasome composition (i.e. association of the 20S proteolytic core with different regulatory complexes, such as the 19S or 11S)1,2, or by post-translational modifications (i.e. phosphorylation, oxidation) that affect proteasome assembly, stability and activity3–6. Proteasome regulation thus has the potential to provide highly dynamic responses to cellular signals and stresses.
Despite recognition that UPS function is dysregulated in many diseases7, 8, 9, 10, 11, the importance of UPS function in cardiac diseases is only beginning to gain attention. Desmin-related cardiomyopathy mouse models provide compelling data for UPS dysfunction, where cardiomyocyte accumulation of protein aggregates are postulated to inhibit proteasome function by restricting entry of ubiquitinated proteins into the proteasome12, 13. Another notable example is acute cardiac ischemia, where proteasome inhibition is thought to occur as a result of accumulation of oxidized protein aggregates or oxidative injury to the proteasome14, 15. Both inhibition and activation of the UPS have been observed in models of cardiac hypertrophy16–19.
There is growing evidence for UPS dysfunction in human heart failure. Accumulation of ubiquinated proteins16, 20–22, soluble protein aggregates13, and autophagic cell death21, 22 in end-stage failing human hearts provides indirect evidence for proteasome dysfunction. However, only limited data are available on proteasome function in hypertrophied human hearts21. ATP-dependent proteasome activity has not been assessed in human cardiac hypertrophy or failure.
The current study examines UPS function in cardiac tissue obtained from patients with end-stage heart failure or hypertrophic cardiomyopathy (HCM). Measurement of peptidase-specific fluorogenic substrate cleavage over a range of ATP concentrations revealed marked reductions in proteasome activity for both HCM and failing hearts and inhibition of activity in response to ATP compared to non-failing controls. Proteasome subunit content, configuration, and access to the active sites within the proteolytic core were not different among groups. These findings suggest post-translational events are key contributors to proteasome dysfunction. Importantly, decreased proteasome activity in end-stage heart failure is observed at a point when total protein oxidation, and oxidative modification of the 19S ATPase subunit, Rpt 5, are increased. These results provide strong evidence that oxidative stress contributes to proteasome dysregulation in end-stage heart failure.
Methods
Expanded methods are available in the on-line Data Supplement.
Human heart tissue procurement
Tissue from non-failing, failing and HCM hearts was collected as described in supplemental methods. Tissue collection from human hearts used in this study has the approval of the University of Michigan Institutional Review Board (IRB) and subjects gave informed consent.
Proteasome activity assay
An optimized method was used for determining heart tissue chymotrypsin-like and caspase-like activities23 (see supplemental methods).
Active-site labeling and in-gel detection of the 20S proteasome
Active site labeling of proteasome catalytic sites was performed as described by Verdoes et al24, 25 (see supplemental methods)
Measurement of protein oxidation products
Protein carbonyls and 4-Hydroxynonylated proteins (4HNE) were analyzed as described14 (see supplemental methods).
Preparation of enriched 26S proteasome fraction, detection of protein carbonylation of proteasome subunits and 2D gel electrophoresis
An enriched 26S-proteasome fraction was prepared as described26. Oxidation of proteasome subunits was detected using the protein carbonyl assay following separation of DNPH-tagged proteins on a 4 – 20% gel. Assignment of carbonyl reactive bands to specific subunits was made by 2D gel electrophoresis26 followed by membrane stripping and recognition by antibodies specific for proteasome subunits (see supplemental methods).
Statistical analysis
Data are expressed as mean ± SEM unless otherwise indicated. Continuous variables were analyzed using 1-way ANOVA. 26S proteasome activity over a range of ATP concentrations was analyzed using 2-way repeated-measures ANOVA with Bonferroni adjustment for multiple comparisons. Comparison of chymotrypsin-like activity between pre- and post-LVAD patients was assessed using paired t-tests. Race and gender comparisons among patient groups (Table 1) were analyzed by chi-square. Where appropriate, data were log transformed prior to analysis. All statistical analysis was performed using Sigma Stat 3.0 with P<0.05 indicating significant differences.
Table
Demographics and Clinical Characteristics of Patients for Myocardial Tissue Analysis
| Nonfailing (NF) (n=8) | HCM (n=29) | Failing (F) (n=33) | P value | |
|---|---|---|---|---|
| Age | 59 ± 4 (40–73) | 49 ± 3 (26–76) | 54 ± 2 (18–68) | <0.01 * |
| Male gender | 4 (50%) | 14 (48%) | 26 (79%) | 0.03 § |
| White | 7 (88%) | 28 (97%) | 27 (82%) | 0.2 |
| Number of years since diagnosis | - | 5.4 ± 1.4 (0.2–40) | 10.2 ± 1.4 (0.4–30) | 0.02 |
| Non-ischemic etiology of heart failure | - | - | 17 (52%) | - |
| LV assist device | - | - | 16 (48%) | - |
| Ejection fraction (%) | 58 ± 2 | 71 ± 2 | 15 ± 1 | <0.001 ** |
| Maximum LV wall thickness (mm) | 11 ± 0.8 | 23 ± 1 | 10 ± 0.4 | <0.001 * |
| Known sarcomere gene mutation ‡ | - | 12/22 | - | - |
Values represent mean ± SEM, with range from minimum to maximum in parentheses.
Note: Each set of experiments used tissue from a subset of the patients listed above. Most tissue samples were used in more than one experiment.
Results
Distinct characteristics of hypertrophic and dilated cardiomyopathies
The mean age of HCM patients was significantly younger compared to NF and failing groups, and there were more males in the failing group (Table). The duration of symptomatic disease was longer in the failing compared to the HCM patients (Table). As expected, systolic function and LV wall thickness (measured by echocardiography or magnetic resonance imaging), were markedly different among groups. Patients with end-stage heart failure had severely decreased systolic function and normal wall thickness, while patients with HCM displayed hyperdynamic function and marked hypertrophy. Twenty-two HCM patients had clinical genetic testing, of whom 12 carried a pathogenic sarcomere mutation, consistent with genotype results in large HCM cohorts27, 28. Mutations were identified in MYBPC (n=10, E542Q, R495Q, D1076fs, W1098X, IVS27+1 G>A, G1248_C1253 dup, IVS30+2 T>G [2 related probands], and E258K [2 unrelated probands]), TNNT2 (n=1, D86A), and TPM1 (n=1, I284V) within the HCM group. All HCM patients exhibited severe left ventricular outflow tract obstruction and New York Heart Association (NYHA) Class III–IV symptoms while patients undergoing LVAD implantation or transplantation exhibited NYHA Class IV symptoms. Ninety percent of HCM patients and 55% of patients with heart failure were taking beta blockers at the time of tissue procurement. Forty five percent of heart failure patients were on intravenous inotropic therapy and 52% were on anti-arrhythmic therapy.
Ubiquitin proteasome peptidase activity is markedly impaired in HCM and failing hearts compared to controls
Proteasome peptidase activity in whole heart protein extracts was measured over a range of ATP concentrations using synthetic fluorogenic peptides as substrates for chymotrypsin-like and caspase-like activities. Overall, ATP-dependent chymotrypsin-like activity was markedly decreased in failing and HCM hearts compared to non-failing control hearts (P<0.001, Figure 1A). In non-failing hearts, there was a small, non-statistically significant rise in chymotrypsin-like activity from 0 to 14 µM (p=0.37). In contrast, chymotrypsin-like activity decreased significantly in both HCM and failing hearts in response to ATP (P<0.001, range from 3.5 µM to 1 mM, data not shown for entire concentration curve). Basal and ATP-dependent caspase-like activities were similarly reduced in failing and HCM hearts compared to non-failing hearts (P<0.001, Figure 1B), with neither ATP-dependent activation nor inhibition in any of the 3 patient groups.
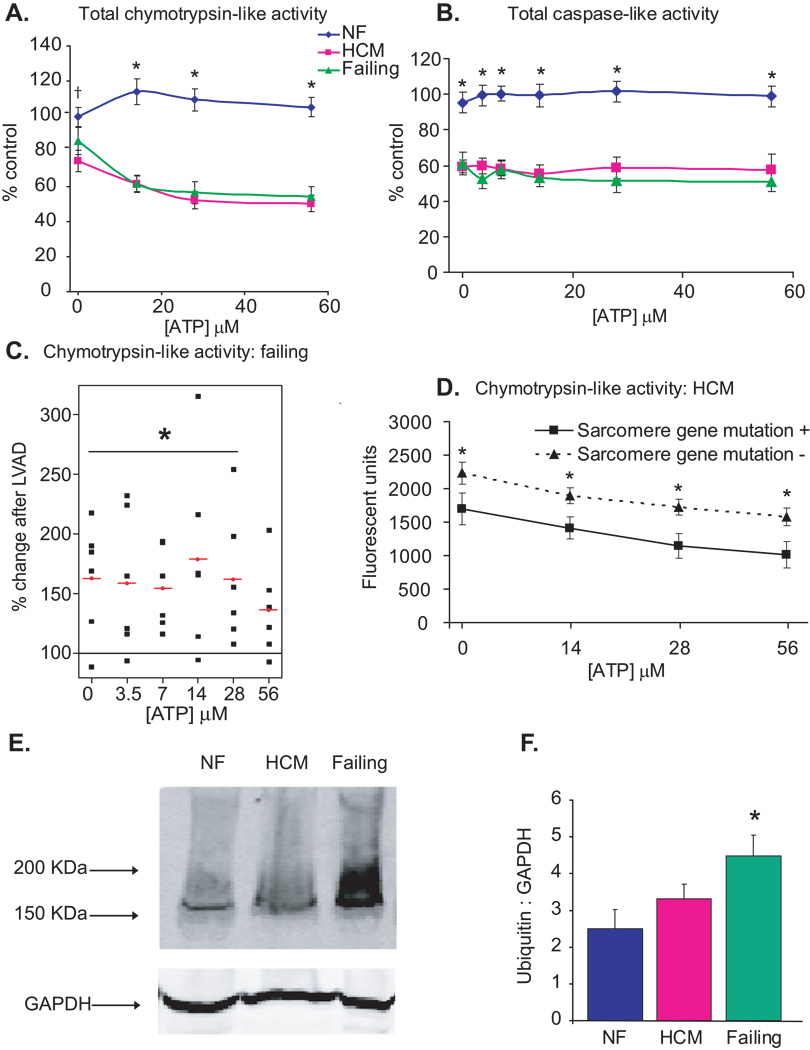
NF=nonfailing, HCM=hypertrophic cardiomyopathy. All values are expressed as a mean ± SEM. A. Heart tissue homogenates (60 µg total cytosolic protein) were assayed for proteasome peptidase activity in the presence of ATP. Total chymotrypsin-like activity represents the difference in fluorescence in the presence and absence of the specific inhibitor, lactacystin (18 µmol/L). †P<0.05 for NF (n=6) vs HCM (n=17). *P<0.01 for NF vs HCM and NF vs Failing (n=14). B. Total caspase-like activity represents the difference in fluorescence in the presence and absence of the specific inhibitor, ZPNAC (10 µmol/L). *P<0.01 for NF (n=5) vs HCM (n=8) and NF vs Failing (n=8). C. Comparison of peak chymotrypsin-like activity in samples from the same patients (n=6) before and after implantation of a left ventricular assist device (LVAD). Each black square represents a single observation and red circle with line represents the mean % change in post-LVAD compared to pre-LVAD samples (*P<0.05). D. Comparison of chymotrypsin-like activity in samples from HCM patients with or without sarcomere gene mutations. *P<0.05 for mutation positive (n=6) vs mutation negative (n=7). E. Representative immunoblot of human total protein homogenates (50 µg) probed with an antibody against polyubiquinated proteins. F. Densitometric analysis for polyubiquinated proteins (>100KDa), standardized to GAPDH as a protein loading control. N=5 (NF), 11 (HCM), 10 (failing). *P<0.05 for NF vs failing.
Paired heart tissue samples collected prior to and after left ventricular unloading were analyzed from heart failure patients treated with left ventricular assist devices (LVADs) as a bridge to cardiac transplantation. The mean time from LVAD implantation to transplantation was 30.78 ± 11 (range 12.3–74) weeks and the mean increase in LV ejection fraction was 11 ± 4% (P<0.05). Proteasome activity increased significantly after mechanical unloading of the left ventricle (P<0.05 for [ATP] 0–28 µM, Figure 1C), consistent with recently published data29, and indicating upregulation of the UPS machinery in response to unloading and partial functional recovery of the left ventricle. In the genotyped HCM subgroup, mean chymotrypsin-like activity was significantly lower in hearts from patients with vs those without pathogenic sarcomere gene mutations (P<0.05, Figure 1D) despite their younger age (37 ± 4 vs 54 ± 5 yrs, P<0.05)..
Proteasome dysfunction can result in intracellular accumulation of ubiquinated proteins12, 30. Polyubiquinated proteins were increased in failing (P<0.05) compared to non-failing hearts (Figure 1E and F). However, ubiquinated protein content was not different between HCM and non-failing hearts despite a similar magnitude reduction in proteasome activity in HCM and failing hearts.
Akt and p53 levels are increased in HCM and failing hearts
In the next set of studies, potential pathways that could be affected by proteasome dysfunction were examined. Akt, a serine/threonine kinase, is the major mediator of mTOR activation and a key regulator of protein synthesis and cardiac hypertrophy. Akt is a direct target for degradation by the UPS, and Akt signaling is decreased in skeletal muscle atrophy31, 32. We found Akt protein expression to be significantly increased in failing and HCM hearts compared to controls (Figure 2A), consistent with the observed decrease in proteasome activity in these 2 groups.
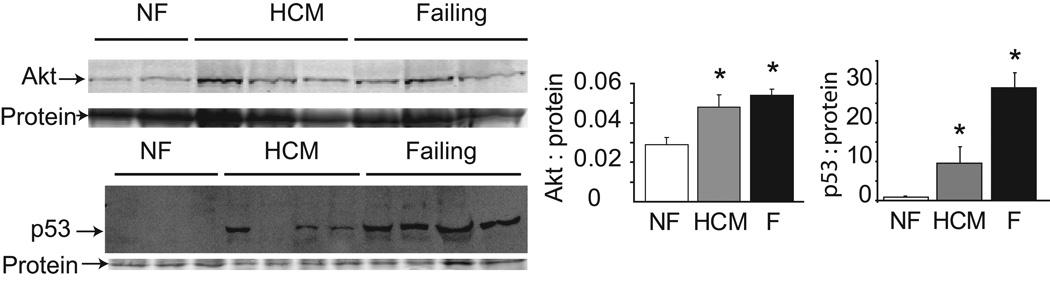
Representative immunoblots (left) and densitometric analysis (right) for Akt and p53. * P<0.05 for Akt and P<0.001 for p53 comparing both HCM and failing groups to non-failing.
Transcription factor p53 is a major mediator of apoptosis in response to various stimuli33 including oxidative stress, and its stability is regulated by UPS degradation. p53 protein expression was markedly increased in both HCM and failing hearts compared to controls, with more variable levels seen in the HCM group (Figure 2). However, the expression of one of p53’s downstream targets, proapoptotic factor Bax, was not significantly different among groups (data not shown).
Proteasome subunit composition and availability of active sites are not altered in human cardiomyopathies
Downregulation of certain proteasome populations or alteration of the proteasome particle configuration are potential mechanisms for proteasome dysfunction. Protein content of representative subunits from the 20S, 19S and 11S proteasomes was therefore evaluated. No differences were observed in the content of α-7 (20S), Rpt 1, 3 and 4 (19S ATPases), Rpn 10 (19S ubiquitination recognition site), and PA28α (11S activator) among non-failing, HCM and failing hearts (Supplemental Figure 1A). Availability of the active site for the 20S subunit β-5, the peptidase with intrinsic chymotrypsin-like activity, was assessed using the novel cell permeable fluorescent probe, MV151, that irreversibly binds to the active sites on proteasome peptidases24, 25. No differences in β-5 active-site labeling were observed among groups (Supplemental Figure 1B). These data suggest that proteasome dysfunction in HCM and failing hearts cannot be explained by alterations in proteasome subunit content, configuration or active site accessibility within the proteolytic core.
Oxidative protein damage and oxidative modification of 19S proteasome subunit Rpt 5 are increased in failing hearts
Lack of a difference in the absolute or relative content of proteasome species in heart tissue from patients with HCM or heart failure suggests a post-translational mechanism(s) to explain proteasome dysfunction. Oxidative injury to the proteasome is one such event that could interfere with protein substrate delivery to the 20S catalytic core. Accordingly, we assessed the degree of protein oxidation in human non-failing, HCM and failing hearts by measuring protein carbonyls and 4-hydroxynonenylated proteins. These oxidized derivatives were significantly increased in failing hearts (P<0.05), but not in HCM hearts, compared to non-failing hearts (Figure 3A and B). There was a non-significant reduction in total protein carbonyls after mechanical unloading with LVAD (Supplemental Figure 2), that correlates with improved functionality as shown above. To determine if the proteasomes themselves were oxidatively modified in failing hearts, we analyzed protein carbonylation of proteasome subunits in highly enriched 26S fractions prepared from non-failing and failing heart samples. Sufficient tissue was not attainable from any single HCM sample for this analysis. A single 53 KDa band was more reactive with the DNPH antibody in the failing compared to non-failing samples (Figure 3C). The identity of this band was confirmed to be Rpt5 by 2-D gel electrophoresis and immunodetection (Figure 3D). These data support the hypothesis that oxidative modification to the proteasome itself could account in part for the proteasome dysfunction observed in failing human hearts.
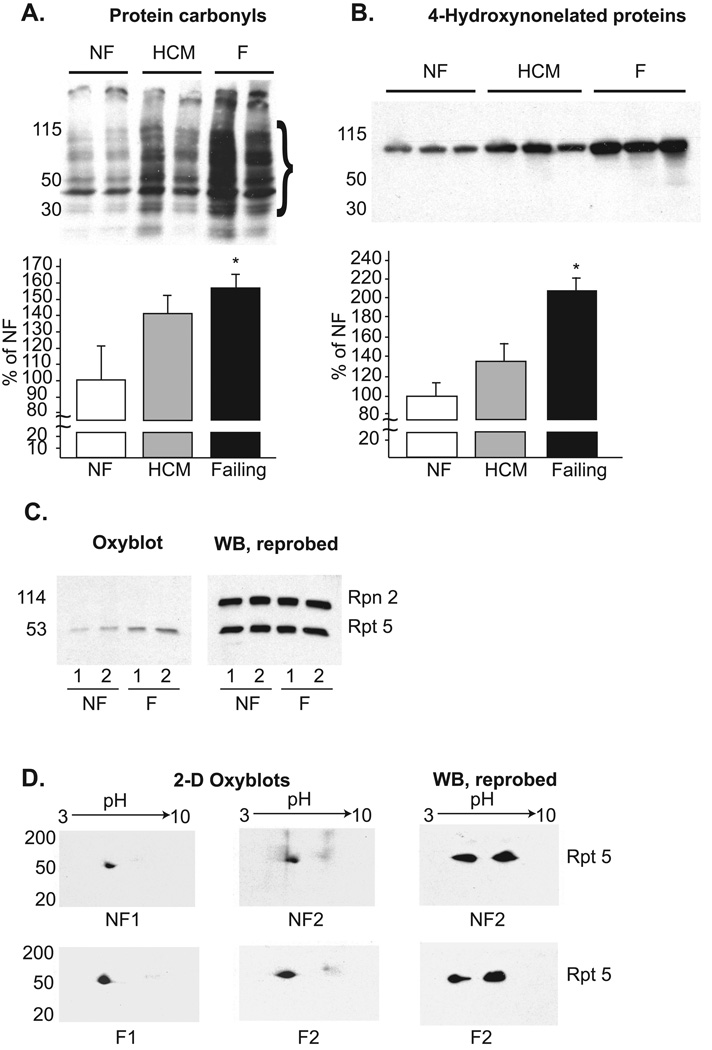
Samples were analyzed for (A) protein carbonyls after exposure to 2,4-dinitrophenylhydrazine (DNPH) and (B) 4-hydroxynonelated proteins. The depicted membranes are representative of 5 to 6 different samples in each group. Densitometric analysis is presented in the graphs beneath the membranes. In the protein carbonyl samples, overall average density was calculated over the range of 30 to 115 kDa. *P<0.05 for NF vs failing hearts. C. (Left) Proteasome subunit proteins from highly enriched 26S proteasome fractions were reacted with DNPH and probed with a DNPH-specific antibody. (Right) The membrane was stripped and reprobed with antibodies specific for Rpt5 and Rpn2. D. (Left and middle) 2D-gel electrophoresis of proteasome subunits previously reacted with DNPH. These membranes were probed with an antibody specific for DNPH. (Right) The membranes were stripped and reacted with an antibody to Rpt 5.
Discussion
In this report, we show that two distinct human cardiac pathologies, HCM and heart failure, are associated with marked proteasome dysfunction compared to non-failing controls. One earlier study described highly variable levels of proteasome peptidase activities in human hearts, with no significant difference between failing and non-failing hearts22. The use of freshly prepared protein homogenates from hearts preserved with cardioplegia prior to freezing, and inclusion of ATP in our assays may explain the observed differences23. An additional key finding in the present study is the identification of post-translational proteasome oxidation, suggesting that oxidation may be an important mechanism involved in proteasome dysfunction in human heart failure.
Our studies of proteasome peptidase activity yielded several unique and reproducible observations in human hearts. A small, but statistically insignificant rise in proteasome activity is observed in non-failing hearts in response to ATP, a magnitude lower than expected given the known dependence of the 26S proteasome on ATP for assembly and protein processing34. This suggests a high level of basal activation in human heart tissue due to either residual activating ATP and/or a lower required activating concentration of ATP for the human compared to the rodent 26S proteasome. Additional possibilities include dissociation of the 19S from 20S during tissue processing34 or diffusion of small fluorogenic substrates without the need for ATP-dependent processing by the 19S complex35.
The most striking observation with regard to proteasome activity was the marked inhibition of activity at low concentrations of ATP in HCM and failing human hearts compared to non-failing hearts. A biphasic response of proteasome activity to ATP has been reported in rat hearts previously23, but the reason for inhibition with increasing concentrations of ATP is currently unknown. Inhibition of activity by ATP in human HCM and failure is particularly notable as it distinguishes these disease states from the non-failing control hearts. We speculate that the addition of ATP could promote the assembly of a damaged or dysfunctional 19S to the 20S core, resulting in restricted substrate entry. The observation of oxidative damage to the 19S subunit Rpt5 supports this notion.
Mechanical circulatory support is a major advance in the treatment of refractory heart failure. LVAD support has many favorable effects on the myocardium, including increased myocyte contractile force36, improved β-adrenergic responsiveness36, normalization of Ca2+ cycling37, and global changes in gene expression38. We propose an additional benefit of mechanical unloading of the failing heart to be activation of the proteasome, analogous to findings with skeletal muscle unloading in which there is a shift in the balance from protein synthesis to degradation9, 39. Activation of the proteasome in heart failure by mechanical unloading may reduce the toxic accumulation of oxidized or damaged proteins and contribute to left ventricular reverse remodeling.
Prior studies of protein degradation, aggregation, and autophagic cell death in human heart tissue have been largely limited to end-stage heart failure13, 20, 22, with little direct evidence for proteasome dysfunction. Data on human cardiac hypertrophy is limited to one previous report demonstrating an increase in ubiquitin-related autophagy with aortic stenosis, only when concomitant systolic dysfunction was present21. Our study is the first to examine proteasome function in human HCM. In contrast to the present work, proteasome activation occurs in response to hypertrophic stimuli in animal and cellular models17–19 Differences in the nature of the inciting stimulus, rate of hypertrophy development, and disease duration may not adequately reflect the chronic and insidious nature of human hypertrophic heart disease. Over half the cases of familial HCM are linked to mutations in sarcomere genes27, 40, and this held true in the patients in this study. Proteasome activity was lower in patients with sarcomere gene mutations, compared to those without mutations. Along with previous observations of the effects of myosin binding protein C truncation mutant expression on UPS function in neonatal cardiac myocytes41, our results support the hypothesis that sarcomere mutant protein expression contributes to proteasome dysfunction in HCM. Decreased proteasome activity in human HCM also challenges the applicability of proteasome inhibition as a therapeutic strategy for suppressing cardiac hypertrophic growth.
Consistent with previous reports, polyubiquitinated proteins were increased in failing hearts compared to non-failing controls. Somewhat surprisingly, this was not the case for the HCM hearts, despite a comparable magnitude of reduction in proteasome activity to the failing hearts. The reason for this finding is not clear, but possible explanations include differences in the duration of proteasome dysfunction or in the rate or magnitude of accumulation of damaged or misfolded proteins in HCM compared to failing hearts.
Increased expression of key mediators of hypertrophy and apoptosis pathways, Akt and p53 respectively, is notable since both proteins are direct targets of the UPS and proteasome dysfunction would therefore be expected to increase their stability and steady state expression. Elevated p53 expression has been observed previously in failing human hearts42, but has not been assessed in human hypertrophy. The importance of the finding of increased p53 expression in human HCM patients is highlighted by previous work showing that p53 is essential for the transition from hypertrophy to heart failure in an animal model of pressure overload by inhibition of Hif-1 and subsequent impairment of angiogenesis43.
What is the mechanism for proteasome dysfunction in human heart failure and HCM? Studies on the cardiac proteasome reveal significant heterogeneity in expression of different subunits and isoforms26. Different proteasome subpopulations confer proteolytic substrate specificity, and may fluctuate in response to environmental stresses. For example, upregulation of the 11S activated proteasome was recently described in an experimental rat model of diabetic cardiomyopathy44. However, in our study, lack of a change in proteasome subunit expression or in the availability of proteolytic active sites invokes a post-translational mechanism to explain proteasome dysfunction in the human diseased heart. One attractive hypothesis is that excess production of oxidized proteins could inhibit proteasome function by interfering with protein substrate delivery to the 20S catalytic core14, 15, 45, and/or by oxidative modification of the proteasome itself15. Increases in total protein oxidation and oxidative modification of 19S subunit, Rpt 5, with heart failure support this idea. These data are particularly intriguing, given the recent observation that Rpt 5 plays a critical role in assembly and activation of the 26S proteasome46. The present observations suggest that strategies to reduce proteasome oxidation may hold therapeutic value for the treatment of heart failure.
Unlike failing hearts, total protein oxidation was not significantly increased in HCM hearts, although a trend was observed. Other post-translational modifications, such as phosphorylation, may play a more important role in this subgroup of patients. Phosphorylation of proteasome subunits in vitro promotes assembly of the 26S and increases proteolytic activity3–6. However, the functional significance of proteasome phosphorylation in vivo is largely unknown, and will be important to address in future studies on HCM and failing hearts.
The use of human heart tissue to identify potential disease mechanisms has obvious scientific value, although certain limitations should be acknowledged. In most cases, data is obtained at a single time point at an advanced stage of disease, which precludes tracking of disease pathways or determination of causality. In addition, statistical power is reduced due to limited availability of non-failing donor heart tissue and heterogeneity of clinical disease.
In conclusion, a marked and consistent decrease in proteasome activity in HCM and failing human hearts is observed in the absence of changes in proteasome protein content or availability of proteolytic active sites. A post-translational defect in regulation of protein substrate processing, and/or oxidative damage to accessible proteasome subunits, appears to be responsible for impaired protein degradation in these human disease states. Defective protein quality control has significant implications for disease pathogenesis in promoting the toxic accumulation of protein aggregates47 and increasing steady state levels of pro-hypertrophic and pro-apoptotic factors. Future work in human tissue, complemented by animal models, will be necessary to define the precise mechanisms for proteasome dysfunction, and to establish a causal link to cardiomyopathy progression.
Acknowledgements
Herman Overkleeft and Martijn Verdoes (Leiden University, The Netherlands) for their kind gift, Bodipy TMR-Ah3L3VS (MV151). Kenneth Margulies (University of Pennsylvania) for providing samples of non-failing human heart tissue. John Younger (University of Michigan) and Ananda Sen (University of Michigan) for statistical consultation.
Funding sources: NIH HL093338 (SMD), NIH HL67254 (MVW), and NIH HL68936 (SRP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: There are no conflicts to disclose
Reference list
Full text links
Read article at publisher's site: https://doi.org/10.1161/circulationaha.109.904557
Read article for free, from open access legal sources, via Unpaywall:
https://www.ahajournals.org/doi/pdf/10.1161/CIRCULATIONAHA.109.904557
Free after 12 months at intl-circ.ahajournals.org
http://intl-circ.ahajournals.org/cgi/reprint/121/8/997.pdf
Free to read at intl-circ.ahajournals.org
http://intl-circ.ahajournals.org/cgi/content/abstract/121/8/997
Free after 12 months at intl-circ.ahajournals.org
http://intl-circ.ahajournals.org/cgi/content/full/121/8/997
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Integrating Clinical Phenotype With Multiomics Analyses of Human Cardiac Tissue Unveils Divergent Metabolic Remodeling in Genotype-Positive and Genotype-Negative Patients With Hypertrophic Cardiomyopathy.
Circ Genom Precis Med, 17(3):e004369, 10 Jun 2024
Cited by: 0 articles | PMID: 38853772 | PMCID: PMC11188634
Potential limitations of microdystrophin gene therapy for Duchenne muscular dystrophy.
JCI Insight, 9(11):e165869, 07 May 2024
Cited by: 3 articles | PMID: 38713520 | PMCID: PMC11382885
Genetic blockade of the activation of 26S proteasomes by PKA is well tolerated by mice at baseline.
Am J Cardiovasc Dis, 14(2):90-105, 15 Apr 2024
Cited by: 0 articles | PMID: 38764549 | PMCID: PMC11101957
Stress-Induced Proteasome Sub-Cellular Translocation in Cardiomyocytes Causes Altered Intracellular Calcium Handling and Arrhythmias.
Int J Mol Sci, 25(9):4932, 30 Apr 2024
Cited by: 1 article | PMID: 38732146 | PMCID: PMC11084437
Genetic ablation of Lmp2 increases the susceptibility for impaired cardiac function.
Front Mol Biosci, 11:1148948, 07 Mar 2024
Cited by: 0 articles | PMID: 38516190 | PMCID: PMC10955435
Go to all (160) article citations
Other citations
Wikipedia (Showing 10 of 34)
- https://en.wikipedia.org/wiki/PSMD10
- https://en.wikipedia.org/wiki/PSMA5
- https://en.wikipedia.org/wiki/PSMA6
- https://en.wikipedia.org/wiki/PSMA7
- https://en.wikipedia.org/wiki/PSMB1
- https://en.wikipedia.org/wiki/PSMB10
- https://en.wikipedia.org/wiki/PSMB2
- https://en.wikipedia.org/wiki/PSMB3
- https://en.wikipedia.org/wiki/PSMB4
- https://en.wikipedia.org/wiki/PSMB5
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Impaired assembly and post-translational regulation of 26S proteasome in human end-stage heart failure.
Circ Heart Fail, 6(3):544-549, 20 Mar 2013
Cited by: 32 articles | PMID: 23515276 | PMCID: PMC3864674
Energetics and function of the failing human heart with dilated or hypertrophic cardiomyopathy.
Eur J Clin Invest, 29(6):469-477, 01 Jun 1999
Cited by: 61 articles | PMID: 10354207
Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy.
Cardiovasc Res, 79(3):472-480, 28 Mar 2008
Cited by: 83 articles | PMID: 18375498
Proteasome dysfunction in cardiomyopathies.
J Physiol, 595(12):4051-4071, 16 Mar 2017
Cited by: 42 articles | PMID: 28181243
Review
Funding
Funders who supported this work.
NHLBI NIH HHS (5)
Grant ID: R01 HL068936-07
Grant ID: R01 HL093338-01A1
Grant ID: R01 HL068936
Grant ID: R01 HL067254
Grant ID: R01 HL093338





