Abstract
Free full text

Genomic gems: SINE RNAs regulate mRNA production
Summary
Mammalian short interspersed elements (SINEs) are abundant retrotransposons that have long been considered junk DNA; however, RNAs transcribed from mouse B2 and human Alu SINEs have recently been found to control mRNA production at multiple levels. Upon cell stress B2 and Alu RNAs bind RNA polymerase II (Pol II) and repress transcription of some protein-encoding genes. Bidirectional transcription of a B2 SINE establishes a boundary that places the growth hormone locus in a permissive chromatin state during mouse development. Alu RNAs embedded in Pol II transcripts can promote evolution and proteome diversity through exonization via alternative splicing. Given the diverse means by which SINE encoded RNAs impact production of mRNAs, this genomic junk is proving to contain hidden gems.
Introduction
Generating an mRNA is a complex process with numerous points of control. The enzyme Pol II synthesizes mRNA transcripts in eukaryotes, and a number of additional general transcription factors (GTFs) are necessary for transcription to occur [1]. Transcription is both positively and negatively regulated by a host of factors including promoter specific activators and repressors, chromatin and histone associated proteins, and co-regulator complexes that can bridge the general transcription machinery to regulatory proteins [1-4]. The transcription reaction undergoes a series of steps including formation of the preinitiation complex at the promoters of genes, initiation of transcription, promoter escape and transcript elongation. Pre-mRNA splicing is another key regulatory step in generating a mature mRNA. Splicing occurs co-transcriptionally and patterns of alternative splicing can be affected by transcription [5, 6]. Although the majority of factors that are known to regulate transcription or alternative splicing are proteins, a comparably small number of non-coding RNAs (ncRNAs) have also been found to function as regulators of these processes [7].
The mobile retroelements Alu, B1 and B2 are part of a family known as short interspersed elements (SINEs). An astounding 11% of the human genome is comprised of Alu SINEs [8]. By comparison, the mouse genome contains approximately 550,000 B1 and 350,000 B2 SINEs [9, 10]. SINEs, which have an internal RNA polymerase III (Pol III) promoter, encode a small RNA and propagate non-autonomously by using the L1 long interspersed element (LINE) machinery to incorporate into their host genome [11]. SINE elements are ubiquitous, and are located throughout their host genome from intergenic regions to being embedded in protein encoding genes [12-14]. Consequently, many SINEs are also transcribed as part of larger Pol II transcripts.
Historically, SINEs were thought of as “junk DNA”, useful primarily for determining phylogenetic relationships between organisms and probing mammalian speciation [15-17]. However, a number of labs found Pol III SINE transcripts to increase under a variety of cell stresses [18-22]. Furthermore, bioinformatic analysis showed thousands of SINEs were present in constrained nonexonic elements, suggesting that SINEs serve a biological and perhaps evolutionary role [23].
What was long thought of as genomic junk is turning out to contain hidden treasure; SINEs have been found to possess diverse and evolutionarily important biological functions from altering gene expression, to localizing mRNAs, to serving as mobile Pol II promoters [24-27]. This review will concentrate on recent studies investigating the regulation of mRNA production by RNAs transcribed from Alu, B1, and B2 SINEs.
B2 and Alu RNAs act as transregulators of mRNA transcription
Pol III transcribes Alu RNA, B2 RNA, and B1 RNA in a regulated fashion (Figure 1A) [28-30]. Human Alu RNA is ~300 nt in length and is composed of two 7SL derived arms linked by an A-rich region [31]. The ~140 nt mouse B1 RNA is similar to the arms of Alu RNA [32]. The ~180 nt B2 RNA originated from tRNA and is not related to Alu and B1 RNA in sequence or secondary structure [33]. The observation that upon heat shock, and other cellular stresses, levels of Pol III transcribed Alu, B1, and B2 RNAs increase led to the hypothesis that they play a functional role during the response to stress [22, 34].
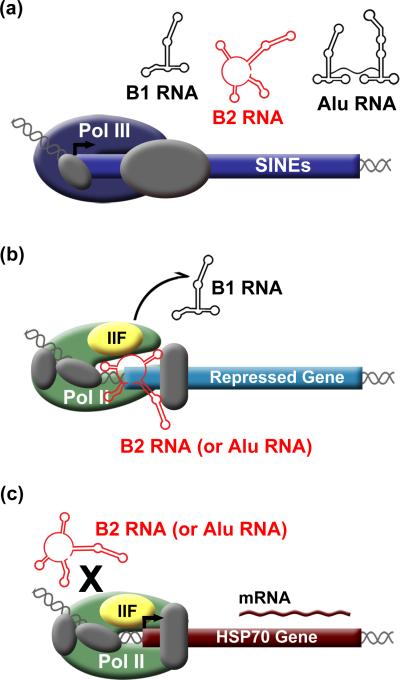
SINE RNAs control the heat shock response in mouse and human cells. A) Pol III transcribed SINE RNAs increase upon heat shock. B) During heat shock, mouse B2 RNA or human Alu RNA enters complexes at the promoters of repressed genes. TFIIF facilitates dissociation of B1 RNA from Pol II. C) Heat shock activated genes are resistant to repression by B2 RNA and Alu RNA.
Experiments have determined that B2 and Alu RNAs function as repressors of mRNA transcription during heat shock (Figure 1B) [35, 36••]. As cells respond to heat shock, transcription of some genes is upregulated (e.g. hsp70, Figure 1C), while transcription of others is repressed (e.g. actin, and hexokinase II). Antisense oligonucleotides against Alu RNA or B2 RNA, introduced into human or mouse cells, abrogated transcriptional repression at several genes upon heat shock, indicating that B2 and Alu RNAs function to inhibit transcription [35, 36••]. In a purified transcription system, Alu RNA and B2 RNA were potent repressors, whereas B1 RNA was not [36••, 37]. Electrophoretic mobility shift assays (EMSAs) showed that B2 RNA, Alu RNA, and unexpectedly B1 RNA, bind directly to core Pol II [36••, 37]. B2 and Alu RNAs assemble with Pol II, GTFs, and the promoter into complexes that can be resolved by EMSA. Consistent with this finding, in heat shocked mouse or human cells, B2 RNA or Alu RNA co-localize with Pol II at the promoters of transcriptionally repressed genes [36••]. Together these observations led to the model that Alu RNA and B2 RNA are up-regulated upon heat shock, bind Pol II, enter complexes at promoters, and block transcription (Figure 1B).
Analogous to protein transcriptional regulators, B2 and Alu RNAs possess multiple functional domains that can be mixed and matched to create chimeric ncRNAs with new functions [36••]. The first hints that these ncRNAs have distinct functional domains came from deletion studies with B2 RNA. Nucleotides 81-130 of B2 RNA were fully functional for binding Pol II and repressing transcription in vitro; however, further truncation to 99-130 yielded an RNA domain capable of binding Pol II, but lacking the ability to repress transcription [38]. Similarly, deletion analysis of Alu RNA demonstrated that it had two separable “Pol II binding” domains and two different “transcriptional repression” domains [36••]. B1 RNA, because it binds Pol II but does not repress transcription, can be considered a Pol II binding domain. Fusing the Alu RNA repression domains individually to B1 RNA created chimeric ncRNAs that functioned as transcriptional repressors [36••]. Perhaps other ncRNAs exist that contain Pol II binding domains and possess different functional domains to regulate transcription in diverse ways.
Recently, a series of in vitro studies were performed to determine the molecular mechanism by which Alu RNA and B2 RNA repress transcription. Given that these ncRNAs co-occupy promoters with Pol II, they likely repress transcription after complexes bind DNA but prior to initiation. B2 RNA and Alu RNA were found to inhibit transcription from a template designed to bypass open complex formation, suggesting that they repress transcription prior to promoter melting [39•]. UV crosslinking and DNase I footprinting assays using complexes containing ncRNA, Pol II, and GTFs, showed significant disruption of contacts between Pol II and promoter DNA, as well as ncRNA-induced conformational changes [39•]. These data support a model in which B2 RNA and Alu RNA repress transcription by preventing Pol II from properly engaging the DNA after assembling into complexes with promoter-associated GTFs (Figure 1B).
B1 RNA and B2 RNA bind Pol II competitively and with similarly high affinity, which raised the intriguing question of whether B1 RNA could block B2 RNA from binding Pol II and repressing transcription. This possibility was investigated using an in vitro transcription system, and surprisingly, B2 RNA was found to repress transcription when B1 RNA had been pre-bound to Pol II [40•]. Further experiments elucidated that TFIIF facilitates the dissociation of B1 RNA from Pol II (Figure 1B). Moreover, fusing a transcriptional repression domain from Alu RNA onto B1 RNA created a chimeric ncRNA that remained stably bound to Pol II in the presence of TFIIF, showing that repression domains make Pol II-ncRNA complexes resistant to the destabilizing effects of TFIIF [40•]. Identifying the precise location of the ncRNA docking site on Pol II and how it interacts with ncRNAs will provide a better understanding the interplay of ncRNA repression domains, Pol II, and TFIIF.
A B2 SINE serves as a boundary element to regulate transcription during organogenesis
Recently, a B2 SINE was found to regulate mRNA transcription in a developmental and tissue specific manner [41••] (Figure 2). The murine growth hormone (GH) locus has differential expression profiles during the development of the pituitary gland. In specific cells of the pituitary, the GH gene transitions from being silenced to being transcriptionally active at embryonic stage 17.5. Fluorescence in situ hybridization experiments found that the GH gene localization changed from regions of heterochromatin to euchromatin just prior to the stage of pituitary development at which the GH gene became activated. Moreover, ChIP analysis showed that the movement to euchromatin was temporally coupled to a change in the methylation state of lysine 9 on histone H3 at the GH promoter. Specifically, the promoter started in a state in which H3K9 was trimethylated, which is a mark for condensed heterochromatin, and changed to a state in which H3K9 was dimethylated. More extensive ChIP analysis across the GH locus revealed that there was a localized transition between H3K9 trimethyl and H3K9 dimethyl in a region 10-14 kb upstream of the GH transcription start site. This observation led to the hypothesis that a boundary element existed at this location [41••].
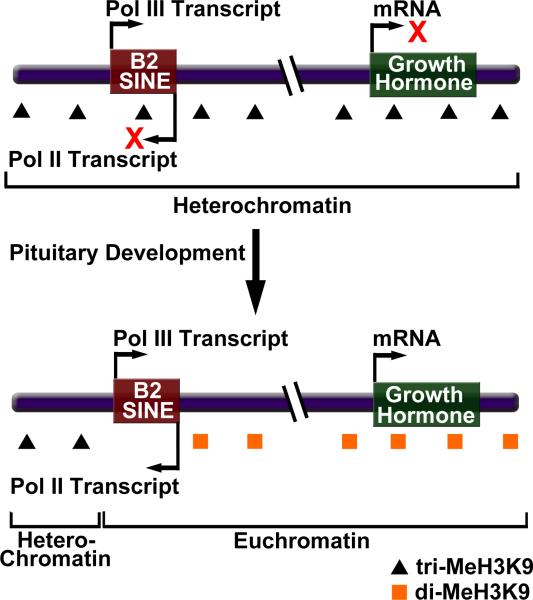
A B2 SINE serves as a boundary element to regulate transcription during pituitary development in mouse cells. Bidirectional transcription of a B2 SINE upstream of the growth hormone locus facilitates a change in chromatin structure from a repressive heterochromatic state to a permissive euchromatic state.
To test the boundary element hypothesis different portions of the region containing the putative boundary element were tested for enhancer blocking activity in a transient transfection assay. The results showed that a 1.1 kb region strongly blocked reporter gene expression when placed between the enhancer and the core promoter [41••]. Intriguingly, this region contained a B2 SINE, and deletion of the B2 SINE eliminated the enhancer blocking activity. Strand specific RT-PCR detected the presence of both Pol III (sense) and Pol II (anti-sense) B2 transcripts, with the Pol II transcript appearing coincident with the change in H3K9 methylation. Transgene experiments containing the GH gene and the upstream B2 element lacking the Pol II or Pol III promoter showed that both were important for enhancer blocking activity. It appeared that ongoing transcription from these promoters was required for enhancer blocking activity since supplying the B2 transcripts in trans had no effect on enhancer function. Together these results suggest that a mouse B2 SINE serves as a boundary element; its bi-directional transcription causes a developmentally important change in chromatin structure (from heterochromatin to euchromatin), which establishes a permissive environment that allows transcription of the GH gene [41••]. Interestingly, in human cells over 9000 Alu SINEs are found within 1 kb upstream of transcription initiation sites, raising the possibility that some Alu SINEs might also affect mRNA transcription by serving as boundary elements [42].
How bidirectional transcription of the B2 SINE results in boundary element function is not yet understood. It seems unlikely that the means by which a boundary is established is directly related to the mechanism by which Pol III transcribed B2 RNA represses transcription during heat shock. In the heat shock situation, B2 RNA (a Pol III transcript) binds Pol II in trans and blocks transcription initiation, whereas in the HG locus, boundary element function requires ongoing Pol II and Pol III transcription. tRNA genes have been found to serve as boundary elements in yeast, but the mechanism of function also appears to be somewhat different from that of the GH B2 SINE (as discussed in [43]). Although ongoing Pol III transcription of tRNA genes is required for boundary element function in yeast, there is no evidence that anti-sense Pol II transcription through the tRNA genes is required. Moreover, Pol III transcription through a yeast tRNA gene can disrupt a nucleosomal array, which results in the inability of silent chromatin to propagate through the tRNA gene. By contrast, transcription of the B2 SINE does not appear to change the localization of nucleosomes [43]. Additional experiments will be needed to uncover the mechanism by which bidirectional transcription of the HG B2 SINE establishes a boundary in chromatin.
Alu RNA contained in primary mRNA can change patterns of alternative splicing
Genomic complexity is increased by the ability to produce multiple different mature mRNAs from a single primary mRNA through alternative splicing. Bioinformatic studies suggest that greater than 70% of human genes are alternatively spliced [44, 45]. Recent studies have found that Alu RNAs embedded within Pol II transcripts can influence alternative splicing. Alu SINEs are thought to be present in over 5% of alternatively spliced internal exonic regions, hence a large portion of mature mRNAs contain pieces of Alu RNA [46, 47••, 48]. Rather than being contained in vital, conserved regions of mRNAs, the majority of the Alu sequences present in mature mRNAs occur in non-conserved alternatively spliced exons, allowing new protein products to be sampled and selected [45, 49, 50•].
Both sense and antisense strands of Alu contain a number of potential 5’ and 3’ splice sites, enabling previously intronic Alu elements to be alternatively exonized in mRNAs (Figure 3A), and exonization of Alu RNA occurs primarily using portions of the antisense strand of the right arm of Alu [49, 50•, 51•, 52, 53]. In the adenosine deaminase acting on RNA 2 (ADAR2) gene, exonization occurs such that a portion of Alu sequence adds 40 amino acids to an alternatively spliced gene product [54, 55]. Studies investigating the determinants of Alu exonization of the ADAR2 and putative glucosyltransferase (PGT) minigenes revealed multiple mechanisms by which Alu regulates alternative splicing [50•, 54]. For example, mutations in the left arm of Alu can dramatically shift the splicing profile of right arm derived Alu exons from alternatively spliced to constitutively spliced [50•]. Additionally, single point mutations in a 5’ splice site within an Alu embedded in ADAR2 mRNA switched an alternatively spliced site to a constitutively spliced site [51•]. A study of 330 Alu-derived exons demonstrated Alu exonization in multiple tissue specific transcripts [47••]. The importance of Alu exonization in human evolution and in enhancing human genomic diversity is just beginning to be uncovered.
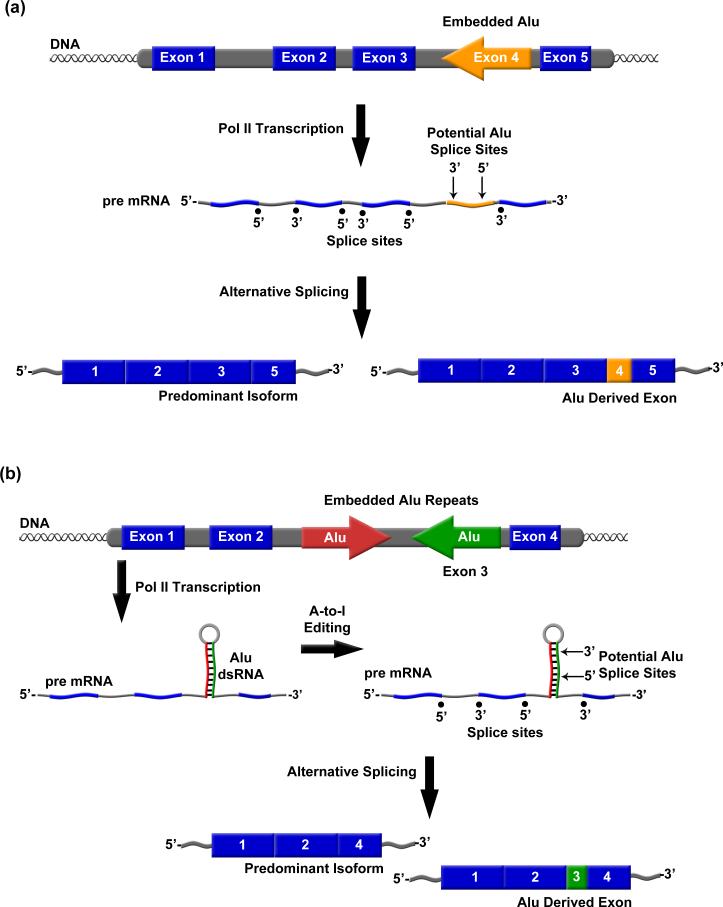
Alu RNA exonization promotes proteome diversity. A) Shown is an example in which embedded Alu RNA is exonized via alternative splicing into an mRNA. B) Alu RNA indirect repeats embedded in an mRNA can become double stranded and undergo A-to-I editing, resulting in Alu exonization.
Embedded Alu RNA also influences gene expression via adenosine-to-inosine (A-to-I) editing of double stranded Alu RNA by ADARs (Figure 3B). Studies suggest that 90% of all A-to-I editing occurs in Alu RNA [56, 57]. Approximately 45% of Alu elements that reside within intronic sequences are present in multiple copies per intron and in both orientations [42, 58•]. In these situations Alu RNA inverted repeats can form double stranded structures [56, 57, 58•, 59]. Subsequent editing of these Alu dsRNAs can influence gene expression through varying mechanisms including deleting premature stop codons, altering triplet codons, and creating new splice sites [58•, 60, 61•, 62]. Examples of Alu regulated gene expression via A-to-I editing include the creation of a 3’ splice site in the ADAR gene, and both creation of a 3’ splice site and deletion of a stop codon in the NARF gene [61•, 62, 63]. Together these studies demonstrate another means by which embedded Alu RNA can influence gene expression and the diversity of proteomes.
Conclusion
It is becoming apparent that SINE encoded RNAs can perform vital roles in regulating transcription and mRNA splicing, thereby controlling processes as diverse as the stress response, development, and proteome diversity. During cell stress, Pol III transcribed Alu and B2 RNAs repress mRNA transcription. Transcription of B2 RNA can establish a boundary and thereby regulate gene expression via controlling the state of chromatin in a developmentally specific manner. Embedded Alu RNAs can affect alternative mRNA splicing, which enables the sampling of new and potentially advantageous proteins. At this juncture it is not obvious that these three regulatory events share common mechanistic features, beyond the involvement of SINE RNAs. Indeed, the vast numbers of SINEs that inhabit mammalian genomes might have established a situation in which different SINEs could evolve distinct and unrelated functions. Both the B2 SINE that serves as a boundary element and the cases of Alu SINEs facilitating alternative splicing likely resulted from mammalian evolution taking advantage of SINEs that inserted at specific locations. In the case of Pol III transcribed B2 and Alu RNAs binding Pol II in trans and repressing transcription, it is likely that mammalian evolution took advantage of expressed, structured ncRNAs to play a valuable role in responding to cellular stress. The effects that SINE RNAs have on gene expression are just beginning to be revealed. We imagine that the discoveries of the functions of the ncRNAs expressed from these gems of the genome will delight and astound for many years to come.
Acknowledgements
This work was supported by a Public Health Service grant (R01 GM068414) from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
[This study shows that upon heat shock, Pol III transcribed Alu RNA inhibits transcription by binding to Pol II at promoters of repressed genes. Additionally, Alu RNA contains functional modular domains that either bind Pol II or mediate transcriptional repression. This study established the importance of a human SINE ncRNA as a trans-regulator of mRNA transcription] [Abstract] [Google Scholar]
[Using a series of biochemical assays, this study revealed that B2 and Alu RNAs repress transcription by assembling with Pol II and GTFs into complexes at promoters and blocking Pol II from properly interacting with promoter DNA. This mechanism of transcriptional repression is unique and could poise genes for rapid reactivation upon removal of the repressor ncRNA] [Europe PMC free article] [Abstract] [Google Scholar]
[A new activity was found for TFIIF; it facilitates the dissociation of B1 RNA from Pol II. The repression domains of Alu RNA stabilize ncRNA interaction with Pol II in the presence of TFIIF. In cells TFIIF might utilize this newly discovered activity to remove non-repressor RNAs from Pol II, thereby allowing repressor ncRNAs to bind Pol II and control transcription] [Europe PMC free article] [Abstract] [Google Scholar]
[Bidirectional Pol II and Pol III transcription of a B2 SINE is necessary and sufficient to establish a chromatin boundary, which results in a permissive chromatin state in the mouse growth hormone locus and enables transcription at this locus in the developing pituitary gland. Given the vast number of SINEs in mammalian genomes, it is likely that transcription through many other SINEs establishes boundaries to control transcription of neighboring genes] [Abstract] [Google Scholar]
[The authors investigated Alu exonization in multiple human genes and tissues and found diverse Alu-derived splicing patterns and tissue specificity. Studies of human, chimpanzee and macaque tissues found increased transcript inclusion and muscle specificity of SEPN 1 Alu exonization after the divergence of human and chimpanzees. These observations suggest that Alu exonization may have contributed to primate evolution] [Europe PMC free article] [Abstract] [Google Scholar]
[The authors examined the effects of splice site sequence and U1 snRNA on Alu exonization. They found that the strength of the Alu 5′ splice sequence, the interplay between U1 snRNA and adjacent Alu splice sites, and exonic regulatory elements all contribute to Alu exonization. These results provide insight into the mechanism by which Alu sequences set patterns of alternative RNA splicing] [Europe PMC free article] [Abstract] [Google Scholar]
[This study demonstrates that the left arm of Alu is important for ensuring that Alu derived exons are alternatively rather than constitutively spliced. By preventing Alu exons from being constitutively spliced, the cell is able to sample new isoforms and minimize Alu derived deleterious effects] [Europe PMC free article] [Abstract] [Google Scholar]
[This study demonstrated that insertion of two inverted Alu elements into an intron resulted in alternative splicing of a downstream exon. This likely involved the formation of double stranded RNA structures that underwent A-to-I editing] [Europe PMC free article] [Abstract] [Google Scholar]
[By comparing the sequences of the gorilla, chimpanzee and human NARF DNA and RNA, the authors propose a model for the evolution of an Alu derive exon in the NARF gene] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.gde.2010.01.004
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2859989?pdf=render
Citations & impact
Impact metrics
Article citations
Ere, a Family of Short Interspersed Elements in the Genomes of Odd-Toed Ungulates (Perissodactyla).
Animals (Basel), 14(13):1982, 05 Jul 2024
Cited by: 0 articles | PMID: 38998094
SINEs as Potential Expression Cassettes: Impact of Deletions and Insertions on Polyadenylation and Lifetime of B2 and Ves SINE Transcripts Generated by RNA Polymerase III.
Int J Mol Sci, 24(19):14600, 27 Sep 2023
Cited by: 1 article | PMID: 37834047 | PMCID: PMC10572872
The origin of genetic and metabolic systems: Evolutionary structuralinsights.
Heliyon, 9(3):e14466, 11 Mar 2023
Cited by: 0 articles | PMID: 36967965 | PMCID: PMC10036676
Differential regulation of transposable elements (TEs) during the murine submandibular gland development.
Mob DNA, 12(1):23, 22 Oct 2021
Cited by: 7 articles | PMID: 34686213 | PMCID: PMC8540199
The transcriptional trajectories of pluripotency and differentiation comprise genes with antithetical architecture and repetitive-element content.
BMC Biol, 19(1):60, 25 Mar 2021
Cited by: 2 articles | PMID: 33765992 | PMCID: PMC7995781
Go to all (80) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
InvAluable junk: the cellular impact and function of Alu and B2 RNAs.
IUBMB Life, 61(8):831-837, 01 Aug 2009
Cited by: 33 articles | PMID: 19621349 | PMCID: PMC4049031
Review Free full text in Europe PMC
Transcriptome-wide effects of inverted SINEs on gene expression and their impact on RNA polymerase II activity.
Genome Biol, 17(1):220, 25 Oct 2016
Cited by: 13 articles | PMID: 27782844 | PMCID: PMC5080714
Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock.
Mol Cell, 29(4):499-509, 01 Feb 2008
Cited by: 312 articles | PMID: 18313387
Alu mobile elements: from junk DNA to genomic gems.
Scientifica (Cairo), 2012:545328, 16 Dec 2012
Cited by: 9 articles | PMID: 24278713 | PMCID: PMC3820591
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: R01 GM068414
Grant ID: R01 GM068414-07





