Abstract
Free full text
A role for SUMO modification in transcriptional repression and activation
Abstract
Since the discovery of the SUMO (small ubiquitin-like modifier) family of proteins just over a decade ago, a plethora of substrates have been uncovered including many regulators of transcription. Conjugation of SUMO to target proteins has generally been considered as a repressive modification. However, there are now a growing number of examples where sumoylation has been shown to activate transcription. Here we discuss whether there is something intrinsically repressive about sumoylation, or if the outcome of this modification in the context of transcription will prove to be largely substrate-dependent. We highlight some of the technical challenges that will be faced by attempting to answer this question.
How does SUMO regulate transcription?
There is now substantial evidence for the involvement of SUMO modification in the regulation of gene expression [1, 2]. From first principles, a number of mechanisms by which this regulation might occur are apparent and these are illustrated in Figure 1. Firstly, it could be that sumoylation of a transcription factor could affect its binding to DNA promoting either its association or dissociation from specific promoters (Figure 1A). This hypothesis is tempting because SUMO carries a negative charge (pI = 5.2), and so it is easy to imagine how this could interfere with DNA binding by a basic transcription factor. However, a clear example of this type of regulation shows that SUMO modification can also promote DNA binding. In the case with stress induced sumoylation of heat shock transcription factor 1 (HSF1), conjugation of SUMO in fact stimulates the ability of HSF1 to bind DNA and therefore presumably its capacity to regulate genes that respond to heat shock [3]. Alternatively, sumoylation could promote or inhibit protein-protein interactions leading to differential recruitment of activities that might activate or repress gene transcription (Figure 1B). Such regulation of protein interactions could also impinge on transcription indirectly, perhaps by antagonizing ubiquitin-mediated degradation or influencing the subcellular localisation of partner proteins required either for activation or repression [4, 5]. Finally, it is possible that SUMO modification could affect enzymatic activities of proteins that regulate gene expression (Figure 1C). For example the de novo DNA methyltransferase Dnmt3a [6] is reported to be a substrate for SUMO modification as are a number of histone deacetylases [7, 8] as well as a histone acetyltransferase Gcn5 [9] and a histone methyltransferase Clr4 [10]. It remains to be determined to what extent SUMO modification directly regulates the catalytic activities of these proteins and whether other enzymes that modulate histone modifications and gene expression, such as JmjC domain containing histone demethylases [11], can be subject to sumoylation. The regulatory mechanisms discussed here are not mutually exclusive and the challenge is to work out how each contributes to the regulation of various sumoylated proteins. None of these mechanisms immediately suggest that SUMO should preferentially either promote or antagonize transcription.
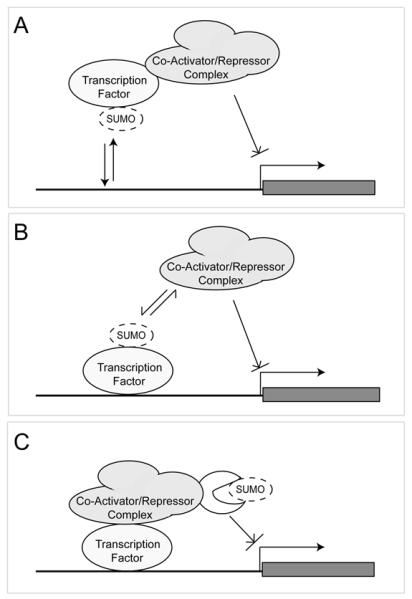
(A) SUMO may affect DNA binding of transcription factors. (B) SUMO could facilitate or inhibit protein-protein interactions between transcription factor and co-repressor or co-activator proteins. (C) SUMO could promote or inhibit enzymatic activities associated with co-repressor or co-activator complexes.
Technical difficulties in studying the role of SUMO
Before being able to investigate any of these putative mechanisms in detail, it is first necessary to identify proteins that are targets for sumoylation. Determining bona fide sumoylation targets in vivo and elucidating the physiological outcome of their modification is not a straightforward task. SUMO proteases can be much more promiscuous in cellular extracts made under native conditions than they are in vivo making biochemical analysis of many native sumoylated proteins extremely difficult [12]. Furthermore analyses of the role of SUMO in mammalian systems has depended heavily on the use of protein overexpression and transiently transfected reporter genes, which may not give a true reflection of the physiological roles of the proteins being studied in all cases. Even using RNAi to test the effect of depleting the endogenous pool of SUMO represents an extremely crude manipulation due to the number of different substrates of this modification and so there is a great danger of observing indirect effects. The most surgical experiments where lysines targeted for sumoylation are mutated carry several caveats: In mammalian systems these almost invariably involve overexpression of the sumoylation substrate and there is always the formal possibility that the observed effects are due to disruption of another post-translational modification of lysine such as ubiquitination, acetylation and methylation. However, mutations in glutamate within the conserved sumoylation motif (ΨKxE), which are less likely to affect other modifications of the neighbouring lysine, could be more informative. Also, in contrast to many kinases and phosphatases, specific small molecule inhibitors of SUMO ligases and proteases that can be administered to cells and used to dissect their function in vivo are not yet available.
SUMO modification in transcriptional repression
In spite of these difficulties, a growing number of substrates have been identified which are now allowing attempts to dissect mechanisms of transcriptional regulation by SUMO in detail. Many case studies have led to the suggestion that modification of transcription factors has a general role in transcriptional repression [1, 2]. Also the fact that targeting the SUMO E2 enzyme Ubc9 to a promoter was sufficient to repress transcription has led to this idea gaining further acceptance [13]. Another recent paper focussing on histone sumoylation in budding yeast revealed that mutation of sumoylation sites in histone H2B led to increased basal expression of several non-induced genes [14]. Also increased global histone acetylation was observed in a yeast strain with reduced histone sumoylation due to a temperature sensitive mutation in Ubc9 [14]. Given the association between histone acetylation and transcriptional activation, this evidence is consistent with a general role for SUMO modification in transcriptional repression. Furthermore, in fission yeast, SUMO is required for proper silencing of genes inserted into heterochromatic regions [10]. There is also evidence that SUMO is associated with repression in mammalian systems too. A series of elegant papers from the Sharrocks laboratory have uncovered a role for sumoylation of Elk-1 in the repression of genes activated by the MAP kinase signalling cascade [15-18]. In the basal state Elk-1 is SUMO modified leading to recruitment of HDAC2 and repression of target genes [15, 16]. However, when the pathway is activated by ERK, Elk-1 is phosphorylated concomitant with loss of SUMO and HDAC2 are lost leading to full activation of target genes [15]. Alternatively, if the pathway is activated by stress (p38) then sumoylation of Elk-1 is maintained in spite of its phosphorylation leading to only partial activation of target genes [18].
SUMO modification in transcriptional activation
Alongside this evidence for a role for SUMO in transcriptional repression, there are now a growing number of cases where sumoylation has been shown to activate transcription. For example, SUMO modification of p53 stimulates its ability to activate reporter genes possibly by competing against Mdm2-mediated ubiquitination, which targets p53 for degradation [19, 20]. Also, sumoylation of Tcf-4 by PIASγ increases its transcriptional activation in response to β-catenin although it is not currently known how this effect is brought about because SUMO modification does not inhibit the ability of Tcf-4 to associate with either β-catenin or DNA [21]. However, there are examples of SUMO modification contributing to gene activation where a mechanistic explanation is clearly emerging. In the case of de novo DNA cytosine methyltransferases Dnmt3a SUMO modification disrupts its interaction with HDAC1 and HDAC2 and inhibits its ability to repress in reporter assays [6]. Another nice illustration of a mechanism for SUMO mediated activation being unravelled is that repression by the transcription factor Ikaros was shown to be perturbed by double modification with SUMO at two lysines in the N-terminus of the protein [22]. In this case it was shown that sumoylation interferes with the interaction of Ikaros with the Sin3 and NuRD co-repressor complexes and so a mechanism for the derepression of transcription is clearly suggested [22]. Recently we and others have provided evidence that the methyl-CpG binding transcriptional repressor MBD1 is SUMO modified in a variety of cell lines [23, 24]. Uchimura et al found that modification of MBD1 by SUMO2/3 stimulates its interaction with the co-repressor MCAF in vitro [24]. They also showed that if SUMO-1 or SUMO-2 and 3 are depleted by RNAi, then MCAF is lost from MBD1-containing heterochromatic foci [24]. Given that MCAF is required for efficient reporter gene silencing by MBD1 [25], this evidence implies that sumoylation will have a positive role in MBD1-mediated transcriptional repression. In contrast, we found that conjugation of SUMO1 to MBD1 by PIAS1 and PIAS3 E3 SUMO ligases disrupts the interaction of MBD1 with the histone H3 lysine 9 (H3-K9) methyltransferase SETDB1. This leads to loss of the silencing chromatin modification (H3-K9 trimethylation) at the promoter of an endogenous MBD1 target gene, p53BP2, and derepression of transcription [23]. Interestingly, sumoylated MBD1 binds to SETDB1 in vitro but not in vivo suggesting that interaction of MBD1-SUMO1 with another partner protein may inhibit the assembly of the co-repressor complex with SETDB1 [23]. Thus conjugation of SUMO1 and SUMO2/3 has opposing effect on MBD1 function and may serve to recruit different partner proteins. In the case of MBD1, as with many other proteins, the balance between SUMO ligase and SUMO isopeptidase activities is probably crucial for the assembly of specific protein complexes. How SUMO conjugation/deconjugation machinery is regulated in response to physiological stimuli will be an intriguing question to answer.
Given that there are now a number of examples of sumoylation contributing to gene activation and light is being thrown on the general mechanisms by which this can occur, further examples of roles for SUMO in positive regulation of transcription are likely to emerge. The most challenging task, however, remains identification of proteins recognizing not only SUMO itself but interacting specifically with SUMO modified proteins.
Conclusions
Many cases are now documented where sumoylation either disrupts or stimulates protein-protein interactions, so it seems likely that this will prove to be far and away the most common mechanism by which SUMO-dependent regulation of transcription is achieved. Thus far, in many cases only relatively simple models have been produced. It will be of great interest to determine whether there are cases where sumoylation simultaneously inhibits the association of a substrate with one protein whilst promoting the formation of a complex with another. For example does sumoylation of MBD1 simply achieve derepression of a target gene by disrupting the interaction of MBD1 with SETDB1 leading to passive loss of histone H3 lysine K9 methylation at promoter nucleosomes during DNA replication? Or does MBD1 sumoylation also lead to recruitment of a demethylase activity making gene induction an active process? Similar questions can be asked in other cases where conjugation of SUMO has been shown to promote or inhibit a single protein-protein interaction. Answers to such questions might give us better understanding of how promoters can produce subtle and temporally regulated changes in transcription in response to different stimuli – a central problem in molecular biology.
Acknowledgements
The work in Stancheva lab is supported by Cancer Research UK, the Wellcome Trust, EMBO Young Investigators Programme and EU Network of excellence “The Epigenome”. M.J. Lyst is a BBSRC funded PhD student.
References
Full text links
Read article at publisher's site: https://doi.org/10.1042/bst0351389
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2871292?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Sumoylation regulates functional properties of the oocyte transcription factors SOHLH1 and NOBOX.
FASEB J, 37(2):e22747, 01 Feb 2023
Cited by: 4 articles | PMID: 36607631 | PMCID: PMC10129296
Sumoylation is Largely Dispensable for Normal Growth but Facilitates Heat Tolerance in Yeast.
Mol Cell Biol, 43(1):64-84, 01 Jan 2023
Cited by: 3 articles | PMID: 36720466 | PMCID: PMC9936996
ATF3 and CH25H regulate effector trogocytosis and anti-tumor activities of endogenous and immunotherapeutic cytotoxic T lymphocytes.
Cell Metab, 34(9):1342-1358.e7, 01 Sep 2022
Cited by: 20 articles | PMID: 36070682 | PMCID: PMC10496461
SUMO E3 ligase SIZ1 connects sumoylation and reactive oxygen species homeostasis processes in Arabidopsis.
Plant Physiol, 189(2):934-954, 01 Jun 2022
Cited by: 2 articles | PMID: 35238389
SUMOylation Is Essential for Dengue Virus Replication and Transmission in the Mosquito Aedes aegypti.
Front Microbiol, 13:801284, 27 Apr 2022
Cited by: 0 articles | PMID: 35572621 | PMCID: PMC9093690
Go to all (78) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Regulation of the dual-function transcription factor Sp3 by SUMO.
Biochem Soc Trans, 35(pt 6):1393-1396, 01 Dec 2007
Cited by: 21 articles | PMID: 18031229
Review
Modification of cardiac transcription factor Gata6 by SUMO.
Biochimie, 170:212-218, 01 Feb 2020
Cited by: 4 articles | PMID: 32017966
Regulation of the sumoylation system in gene expression.
Curr Opin Cell Biol, 20(3):288-293, 28 May 2008
Cited by: 38 articles | PMID: 18468876 | PMCID: PMC2495007
Review Free full text in Europe PMC
Modification with SUMO. A role in transcriptional regulation.
EMBO Rep, 4(2):137-142, 01 Feb 2003
Cited by: 281 articles | PMID: 12612601 | PMCID: PMC1315836
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Cancer Research UK (1)
Epigenetic gene silencing in normal and tumour cells
Dr Irina Stancheva, University of Edinburgh
Grant ID: A2702
Wellcome Trust (1)
Characterization of protein complexes associated with methyl-CpG binding protein MBD1 and their targeting to specific DNA sequences.
Dr Irina Stancheva, University of Edinburgh
Grant ID: 077691





