Abstract
Free full text

mRNA-seq with agnostic splice site discovery for nervous system transcriptomics tested in chronic pain
Abstract
mRNA-seq is a paradigm-shifting technology because of its superior sensitivity and dynamic range and its potential to capture transcriptomes in an agnostic fashion, i.e., independently of existing genome annotations. Implementation of the agnostic approach, however, has not yet been fully achieved. In particular, agnostic mapping of pre-mRNA splice sites has not been demonstrated. The present study pursued dual goals: (1) to advance mRNA-seq bioinformatics toward unbiased transcriptome capture and (2) to demonstrate its potential for discovery in neuroscience by applying the approach to an in vivo model of neurological disease. We have performed mRNA-seq on the L4 dorsal root ganglion (DRG) of rats with chronic neuropathic pain induced by spinal nerve ligation (SNL) of the neighboring (L5) spinal nerve. We found that 12.4% of known genes were induced and 7% were suppressed in the dysfunctional (but anatomically intact) L4 DRG 2 wk after SNL. These alterations persisted chronically (2 mo). Using a read cluster classifier with strong test characteristics (ROC area 97%), we discovered 10,464 novel exons. A new algorithm for agnostic mapping of pre-mRNA splice junctions (SJs) achieved a precision of 97%. Integration of information from all mRNA-seq read classes including SJs led to genome reannotations specifically relevant for the species used (rat), the anatomical site studied (DRG), and the neurological disease considered (pain); for example, a 64-exon coreceptor for the nociceptive transmitter substance P was identified, and 21.9% of newly discovered exons were shown to be dysregulated. Thus, mRNA-seq with agnostic analysis methods appears to provide a highly productive approach for in vivo transcriptomics in the nervous system.
Microarray-based transcriptome studies have provided a productive approach to the discovery of therapeutic targets in neurological disorders. For example, gene expression profiling of the dorsal root ganglion (DRG) in chronic pain (Griffin et al. 2003) identified several altered genes (Costigan et al. 2002), some of which were subsequently shown to be key modulators of pain (Tegeder et al. 2006). Gene expression analysis using microarray technology suffers from well-known limitations including poor sensitivity and dynamic range, a requirement for substantial requisite amounts of RNA, and a limited capacity to identify new transcripts or RNA splice sites. Given these limitations, it is unlikely that microarray analysis can reveal the full extent of transcriptome reprogramming underlying neurological disorders, such as chronic pain.
Ultra-high-throughput RNA sequencing has emerged as a revolutionary technology with superior dynamic range and reproducibility compared with microarrays (Marioni et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Wilhelm et al. 2008; Wold and Myers 2008; Wang et al. 2009). Conceptually, ultra-high-throughput mRNA sequencing (mRNA-seq) should be capable of overcoming virtually all limitations of microarray technology by permitting de novo capture of the full transcriptome of any experimental tissue. In practice, however, progress toward unbiased, comprehensive transcriptome analysis by short mRNA reads has been incremental. In fact, the development of these novel methods of bioinformatic analysis has proved to be less straightforward than might have been anticipated. In particular, pre-mRNA splice site mapping has been challenging.
De novo (unbiased) genome-wide mapping of pre-mRNA splice sites, termed “agnostic” here to emphasize the independence from pre-existing data sets (such as annotations found in databases), is not the norm. Published reports have relied on mapping mRNA-seq reads to reference databases of already known splice sites and/or to a limited number of hypothesized sequences (based on alternative exon joining) (Pan et al. 2008; Wang et al. 2008). While these approaches were pragmatically geared toward first-generation Illumina data consisting of very short reads (25–36 bp), they had a limited capacity for discovery of new splice sites; i.e., they were “biased” toward known sites. Trapnell et al. (2009) reported the TopHat software, which improved on previous strategies by mapping splice junction reads to an extended set of exon boundaries derived from empirical mRNA-seq data. TopHat thereby implemented a partially agnostic strategy, as splice junctions could be found independently of pre-existing annotations, albeit only nearby exon candidate regions established by other experimental data. TopHat also used other constraints like the GT-AG motif, which makes identification of canonical splice sites faster at the price of missing noncanonical events. The original report validated the approach using very short reads (25 bp) emphasizing strategies to reduce run-time (Trapnell et al. 2009). A report using a splice site model for prediction was published after submission of the present study (Filichkin et al. 2010).
The present study applies mRNA-seq to an animal model of neurological disease with the intent of determining if the extent of transcriptome reprogramming in the nervous system exceeds the breadth of previously documented alterations. Chronic pain was chosen because it is a common neurological disorder, which is incompletely understood at the functional genomic level. An important goal of our study was to proceed in an agnostic fashion relying only on the mRNA-seq data obtained and the nonannotated reference genome. Available transcript annotation data (exon–intron boundaries and gene classes) were used only to assess the precision of our methods for mapping exons and splice sites. Although the emphasis on bioinformatics was originally motivated by our interest in nervous system transcriptomes and by our interest in investigating novel transcripts and mRNA splice forms in that context, the resultant computational methods may be valuable for a variety of mRNA-seq studies in mammalian transcriptomes with a read length of at least 50 bp. Taken together, this study demonstrates how the discovery potential of mRNA-seq can be strengthened by newly developed bioinformatics with a strong statistical foundation. Equally importantly, mRNA-seq demonstrates that transcriptome reprogramming in the nervous system may be more extensive than recognized by microarray studies, raising the possibility that some neurological disorders such as pain may be recast in the future as diseases of altered gene expression amenable to transcription therapy.
Results
Single-DRG (L4) mRNA-seq
The L5 spinal nerve ligation (SNL) rat model of chronic neuropathic pain was chosen because it induces stable allodynia for months and separates unsevered and surgically compromised portions of the peripheral nerves clearly by spinal nerve level, i.e., L4 (anatomically intact) versus L5 (ligated and then cut). Experimental and sham control rats were prepared, and mechanical allodynia was confirmed (Fig. 1A) by behavior testing as detailed in our previous studies (Storek et al. 2008). L4 DRG were harvested from animals sacrificed 2 wk and 2 mo after SNL and flash-frozen. As illustrated in Figure 1A, total RNA was extracted (typical yield 1–1.5 μg per DRG); poly(A)-purified (twice); chemically fragmented (Mortazavi et al. 2008); and reverse-transcribed using hexamer priming. Eight sequencing libraries were constructed using cDNA from independent animals (two SNL and two control for each time point, 2 wk and 2 mo) and sequenced on an Illumina Genome Analyzer II (GAII) as described (Bentley et al. 2008).
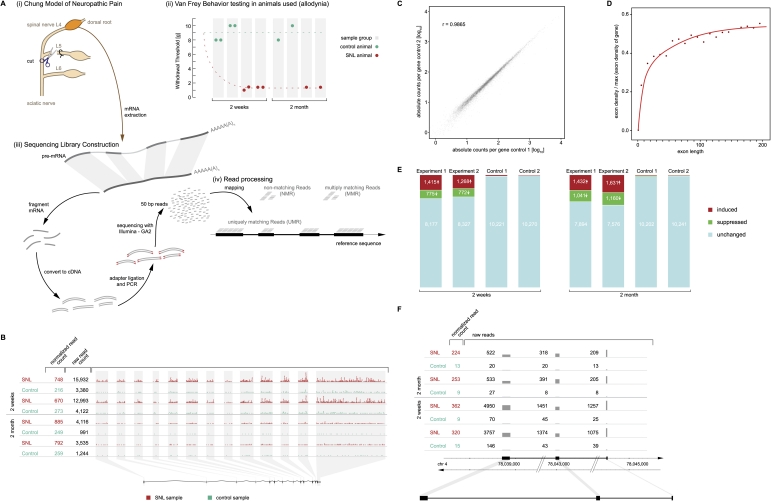
Expression of known genes altered in pain. (A) Experimental paradigm. (i) Neuropathic pain was modeled in rats by L5 SNL (“Chung model”), in which the L4 spinal nerve remains anatomically intact but the L4 DRG becomes dysfunctional, rendering the affected animal allodynic, i.e., the animal responds to light touch as if it were a noxious stimulus. (ii) Mechanical allodynia was confirmed prior to euthanasia by von Frey testing of the paw withdrawal threshold, which was abnormal (1.4–2 g) after SNL and normal (8–10 g) in sham-operated controls. (iii) Sequencing libraries were constructed from pools of two L4 DRG from different animals for the 2-wk time point (hence two behavior measurements per sample) and from a single DRG for the later time point (2 mo). RNA was isolated from the L4 DRG, poly(A)-purified, chemically fragmented, converted to a cDNA library, and sequenced. (iv) Resulting sequencing reads (50 bp long) were mapped to the entire rat genome and categorized as uniquely matching reads (UMR), multi-matching reads (MMR), and nonmatching reads (NMR) as detailed in Methods. (B) Gene-level analysis. Expression of each annotated gene was quantified by the total number of UMR mapping to its exons. The gene Gas7 (ENSRNOG00000003492), shown as an example, has 13 annotated exons. Each dot in the graph symbolizes 10 UMR observed. Sequencing depth at 2 wk was several-fold greater than at 2 mo, i.e., more reads were observed for each sample and gene, requiring correction in the comparative analysis. Normalized UMR counts (i.e., number of reads of the given gene per 106 reads obtained) are given in the left column, showing that the gene was induced 3.1-fold after SNL. (Each line marked as “SNL” or “Control” is an independent biological replicate.) (C) Reproducibility between biological replicates across 10,367 known protein-coding genes. Quantification of expression was highly reproducible across a wide range of gene expression levels because of the digital nature of the data. A tight correlation of read counts more than four orders of magnitude is shown. Correlation among biological replicates was high as indicated by a Pearson correlation coefficient of r = 0.99. (Correlation coefficients between all possible pairs in the study are shown in Supplemental Table 2). (D) Exon length versus average read coverage. A minimal exon length of 50 bp is required for a read to “fit” fully into an exon. Efficient quantification of expression by UMR is achieved for exons of ~100 bp length or greater. Depicted is the relationship between exon length and average read density (normalized to expression levels of individual genes) for the entire data set (all annotated exons). (E) Expression changes in known genes. Approximately 20% of genes were found to be dysregulated in the L4 DRG (after L5 SNL). Results are shown here for 10,367 known protein-coding genes. The absolute number of genes found to be induced or suppressed by more than 1.7-fold is shown. The analysis was designed with a predicted FDR of 0.5% (i.e., 52 genes predicted to be “induced” and “suppressed”). The empirically determined FDR supported the assumption (“Control”) demonstrating that more than 2000 genes were significantly altered in expression (as discussed in detail in Results and Methods). The bar graphs shown are independent biological replicates for both time points studied (2 wk and 2 mo). (F) External validation by published reports. A gene previously known to be strongly induced in the DRG in rat pain models (Bonilla et al. 2002; Costigan et al. 2002; Sun et al. 2002; Valder et al. 2003; Davis-Taber and Scott 2006), Neuropeptide Y, was found by mRNA-seq to be among the most strongly induced genes at both time points studied. A comprehensive list comparing mRNA-seq results with published reports is provided in Supplemental Table 5.
Genome-wide read mapping
Of 260 million high-quality mRNA-seq reads obtained, 142 million (53%) were matched to a unique site in the rat genome; these were termed uniquely matching reads (UMR) and were used to quantify the expression of known genes (Fig. 1) and to discover new exons (Fig. 2). There were 104 million (39%) nonmatching reads (NMR), which were used for splice unction (SJ) discovery (Fig. 3), and there were 20 million (8%) multiply matching reads (MMR) (i.e., aligning to more than one site in the reference genome), which were useful in completing the annotation of complex genes (Fig. 4).
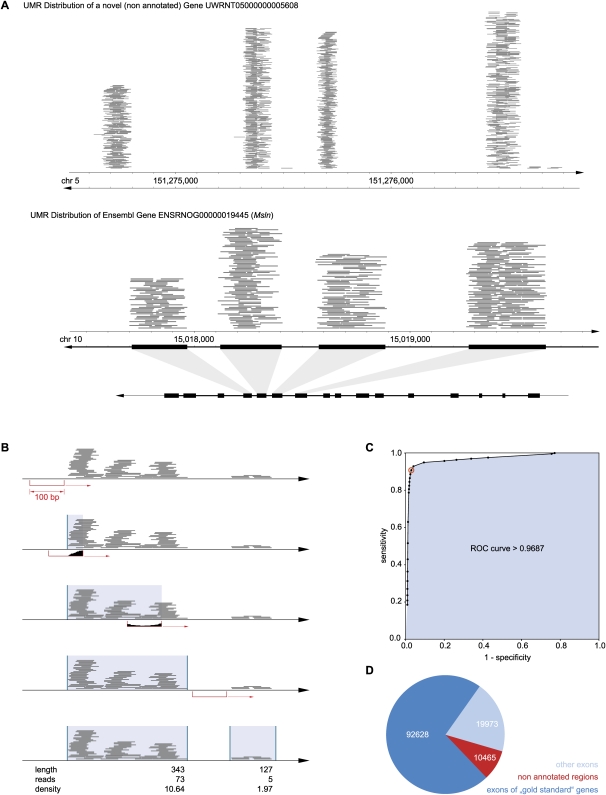
Mapping novel exons in nonannotated regions of the rat genome. (A) Typical UMR distribution. UMR mapped across nonannotated regions of the genome in nonrandom patterns, often resembling exon–intron structure. Depicted are UMR (each gray line = one read) mapped across a nonannotated region of chromosome 5. For comparison, UMR across an annotated region of chromosome 10 are shown along with the annotation of known exons illustrating correspondence. (B) Aggregation of UMR into read “clusters” resembling exons. A 100-bp “sliding window” was moved across the genome demarcating the beginning (UMR present, i.e., “filled” window) and end (UMR absent, i.e., “empty” window) of “UMR clusters.” Resulting UMR clusters differed in read density. Clusters consisting of high piles of hundreds of reads provide strong evidence for an exon, while clusters with few reads appear indeterminate. (C) Classifier for UMR clusters: exon versus no-exon. The newly defined UMR clusters were dichotomized into “exons” and “no exons” using average read density (read coverage) as a classifier. The ROC curve shown here was obtained by applying the classifier to an exploratory subset of 10,367 (annotated) genes, varying read density from 0.25 to 100. Sensitivity and specificity are plotted for 26 different read densities. The classifier was found to be a very precise test with an area under the curve of 0.97. At a read density of 4 (inflection point of the curve indicated as red circle), the sensitivity of the test was 92% and the specificity was 97%. The favorable test characteristics were confirmed in the validation set (sensitivity 91%, specificity of 97%). Applying the procedure to the full data set, 123,066 UMR exon clusters were found. (D) Location of UMR exon clusters. 10,464 new exons were found (i.e., UMR clusters with density >4 in a nonannotated region); the remaining clusters overlapped known exons belonging either to the 10,367 genes quantified above or to other annotated genes.
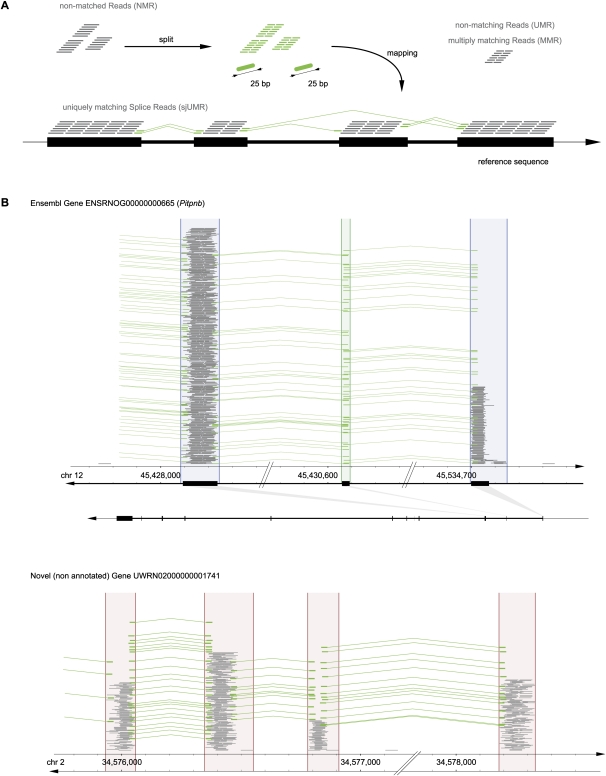
Agnostic splice junction (SJ) mapping. (A) Agnostic, i.e., de novo genome-wide SJ discovery from split reads (25 bp + 25 bp). NMR were split into 25-mer halves, which were matched independently to the genome reference sequence. Reads whose halves each matched uniquely to the genome defined SJs de novo; these were termed uniquely mapping splice junction reads (sjUMR). (B) SJ validation on known genes and their application to define connections of novel exons and exon borders. (Green) SJ-spanning uniquely mapping reads (sjUMR; mapped as described in the Methods section). Most SJs were supported by multiple sjUMR (28.6 on average), as depicted here by piles of green lines. (Top panel) A known gene (Pitpnb ENSRNOG00000000665) is shown here, demonstrating how sjUMR mapped precisely to the border of exons as annotated in the reference genome. (Bottom panel) sjUMR connecting novel UMR clusters to groups, thereby defining new genes. In some instances, UMR cluster borders fall into introns (as a result of the inclusiveness of the sliding-window algorithm), as depicted here (right sides of second and third cluster). In those cases, exon borders were defined precisely by sjUMR. Note (top panel, second exon) that very short (<50 bp) exons that were undetectable by UMR could be defined through mapping of SJ reads.
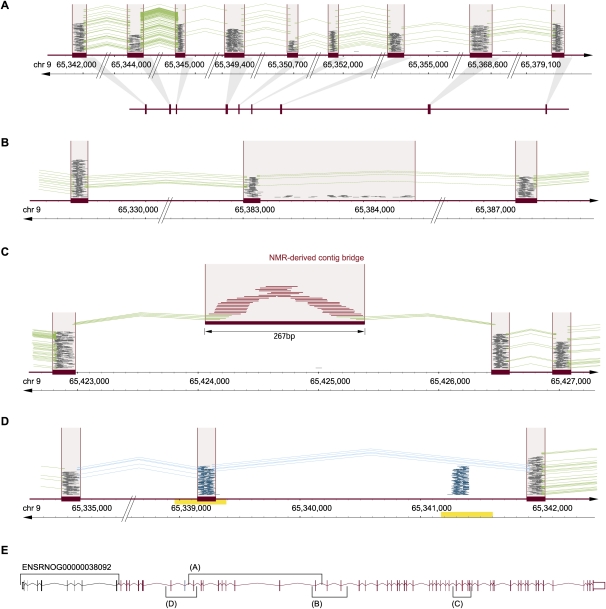
Integrating information from UMR, sjUMR, MMR, and NMR read classes for complex genome (re-)annotation. (A) UMR- and sjUMR-based definition of new exon groups. A novel gene candidate was identified on chromosome 9 through several UMR clusters (satisfying the classifier criterion of a read density > 4), which were connected through splice junctions defined by sjUMR. Expression of exons was coregulated (suppressed by 70% after SNL). An open reading frame encoding 409 amino acids was noted lacking a start and stop codon, suggesting that an incomplete gene fragment was identified consisting of nine exons. (B) sjUMR validation of a low-read density UMR cluster as a novel exon. Consistent with the limited sensitivity of the UMR cluster classifier (91%), a UMR read cluster with a read density of 3.9 was initially not classified as an exon (because its density was <4); but sjUMR-based SJs defined it unambiguously as a novel exon connecting the above gene fragment to a series of 18 3′-located UMR clusters. Note that the cluster density was low because a region of scattered intron reads was added to the read cluster by the sliding window, an imprecision that was rectified by agnostic splice site mapping, which defined precise exon borders. Through the step depicted in this panel, the candidate gene was extended to 28 exons. (C) NMR-derived contig bridge. A faulty or missing section in the reference genome can be indicated by runs of N (ambiguous bases) as encountered 3′ of the 28 exons assembled above (50 scattered N). Such missing sections in the reference are also an important reason why some mRNA-seq reads do not match. Accordingly, we found that in such cases, contigs of NMR can be assembled bridging faulty sequences, as shown here. As a result of the NMR contig-bridge, the gene candidate could be expanded to 49 exons. (D) MMR candidate exons discriminated by sjUMR. Exon copy numbers >1 in the reference sequence result in clusters of MMR (blue) matching collectively to more than one site. A case of two such MMR clusters in close proximity (within <2 kb; note duplicated region indicated by yellow bar) was observed 5′ of the above gene candidate. As shown here, the ambiguity was resolved by sjUMR; this led to a 5′ extension of the gene candidate by another 15 exons, which resulted in a complete gene with start and stop codon consisting of 64 exons. (E) A summary of the novel 64-exon gene is shown encoding a 3313-amino-acid-long protein (14,084-bp mRNA). While the study was under way, a homologous mouse protein with important CNS relevance was reported (“large previously unknown protein”) (Lu et al. 2009), termed UNC-80, which serves as a substance-P and neurotensin coreceptor. Down-regulation of the rat UNC-80 rat homolog after SNL may contribute to allodynia through alterations in peptide activity in the DRG.
We obtained 2.6 × 108 (274,622,530) 50-bp-long cDNA sequence reads equaling 13 Gb, or ~4.7 whole-genome equivalents, of sequence data. Of these 7,083,741 (2.58%) were discarded because of poor base call quality scores or unknown bases. The remaining 267,538,789 reads were aligned to the full rat reference genome (RGSC 3.4; mitochondrial genome included) using the ELAND aligner software (Illumina) and allowing for ≤2 mismatches per 32 consecutive bases. We took advantage of the extended read length of 50 bp by requiring that the first and the last 32 bp both mapped to the same site (offset by 18 bp) in order to declare a match. We found that 142,572,750 (53.3%) of all processed reads matched a unique genomic site (termed UMR), and were used to quantify the expression of known genes (Fig. 1) and to discover new exons (Fig. 2). Additionally, 20,408,306 (7.6%) reads matched to multiple (i.e., >1) sites in the genome (termed MMR). Finally, 104,557,733 (39%) were NMR. Nonmatching mRNA-seq reads may occur for several reasons, including pre-mRNA splicing, which creates sequences that are not present in the genome, i.e., spanning splice junctions (alignment of NMR to splice sites is discussed in a separate section below); exclusion of the Y chromosome and omission of other sequences from the current rat reference genome assembly RGSC 3.4; sequencing errors; and genetic differences (single nucleotide polymorphisms, insertions, deletions) between the rat strain employed (Sprague Dawley) and the reference genome strain (Brown Norway). Accordingly, the set of NMR was used for SJ discovery (Fig. 3), as described in detail below.
The average read density across known mRNA transcripts was 1.25-fold higher for 3′ exons than 5′ exons. A 3′ bias is to be expected with any mRNA detection method that employs a poly(A) purification step. In our case, the median difference was small, only 25%, because the library construction protocol employed (chemical mRNA fragmentation followed by random priming for cDNA synthesis) was designed to achieve even coverage.
Transcriptome reprogramming
As a first step in our analysis, the expression of 10,367 Ensembl known protein-coding genes (out of 17,738 annotated) was quantified. Raw UMR counts for each gene (provided in Supplemental Table 1) correlated highly when controls were compared (biological replicates). For these pairs Pearson correlation coefficients were always r = 0.99 (Supplemental Table 2). The correlation among SNL-control pairs was lower (average 0.82), as was to be expected if gene expression was altered after SNL. For subsequent analyses, read counts were normalized to the total number of reads obtained for each sample (multiples of 106). Results are shown in Figure 1B–F (the remaining 7371 Ensembl known protein-coding genes were expressed at low levels). As shown in Figure 1, when SNL animals were compared with controls at the 2-wk time point, 1268 to 1415 genes were induced, and 772 to 775 genes were suppressed. A false discovery rate (FDR) of 0.5% (52 of 10,367 genes) in each group (i.e., “induced” and “suppressed”), resulted in a true-positive estimate of 1289 (12.4%) induced and 721 (7.0%) suppressed genes (statistical procedures and computations are detailed further in Methods). In other words, nearly 20% of the L4 DRG transcriptome was reprogrammed to a new expression level in response to SNL of the neighboring (L5) nerve root.
Alterations persist long term
Examining the same set of 10,367 genes, we found that 1432 to 1631 genes were induced and 1041 to 1160 were suppressed 2 mo after SNL. Fewer total mRNA sequences were obtained for this time point (one lane compared with four for the early time point), resulting in a slight increase in the measurement variance and an observed FDR of 0.7% (51 to 87 false-positive genes vs. 52 expected). Accounting for the observed (i.e., higher) FDR, 1458 genes (14.1%) were induced and 1027 (7.5%) were suppressed at 2 mo.
Of all genes induced ≥2-fold at 2 mo, 96% were induced ≥1.4-fold at 2 wk. Similarly, of all genes suppressed by a factor of ≥2 (i.e., to less or equal than half of controls) at 2 mo, 97% were suppressed by at least 1.4-fold at 2 wk. No genes were found to be induced or suppressed by a factor of ≥2 at one of the two time points but regulated in the opposite direction at the alternate time point. While the study was not designed as a time-series experiment and not powered to detect single genes (or very few) displaying dynamic regulation, the results strongly suggest that the large majority of transcriptome alterations persisted stably from 2 wk to 2 mo. Overall, >20% of measured genes were altered at 2 mo, demonstrating that transcriptional reprogramming persisted in neuropathic pain matching the chronicity of the condition (Fig. 1E,F).
Extent of DRG gene induction and suppression detectable by mRNA-seq
Genes were induced up to >100-fold and were suppressed as much as 10-fold in SNL animals (compared to sham controls). Ten genes were induced 32- to 121-fold, 42 genes were induced ≥10-fold, and 212 were induced ≥4-fold; 165 genes were suppressed ≥4-fold (at 2 wk and/or 2 mo). The magnitude and breadth of changes documented appear generally more extensive than alterations detectable in comparable microarray studies of the DRG in pain (Bonilla et al. 2002; Costigan et al. 2002; Valder et al. 2003; Davis-Taber and Scott 2006; Rodriguez Parkitna et al. 2006). The principle of quantification by mRNA-seq is digital counting, which is free of background hybridization problems (or similar artifacts limiting sensitivity) and does not suffer from saturation of probes (known to diminish precision when quantifying high-expressing genes in microarrays) (Wold and Myers 2008). The resulting superior dynamic range of mRNA-seq established by the original studies (Cloonan et al. 2008; Marioni et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Sultan et al. 2008; Wilhelm et al. 2008; Wold and Myers 2008; Wang et al. 2009) was found here to be immensely informative when applied to the nervous system.
Mapping nonannotated portions of the DRG transcriptome
Previous “genome-wide” nervous system transcriptome studies have been based on microarrays and, therefore, have been restricted to known mRNA. mRNA-seq is not bound to such limitations. We found 6,224,094 RNA reads (4.7% of UMR) mapping to nonannotated regions of the rat genome. Most of these occurred in dense aggregates of many reads (dozens to thousands) that were separated by long nontranscribed regions (Fig. 2A). To define which read-aggregates resembled novel exons, we developed a classifier. First, mRNA-seq reads were joined into “clusters” by a 100-bp sliding window (Fig. 2B). Next, newly defined clusters were compared with exons of well-expressed protein-coding genes (a subset of the 10,367 genes described above) containing 97,646 exons. One-tenth of the exons were used for test development. Test conditions were systematically varied requiring different average read densities (from <1 to >100 UMR per 50 bp) to declare that a cluster was an exon. Cluster classification results were compared with the actual exon annotation, recording the frequency with which an annotated exon was detected (true-positives) and how often an exon was erroneously predicted in an intron region (false-positives). A receiver operating characteristic (ROC) curve (Fig. 2C) demonstrated that this was an effective classifier (area under the curve 0.97). Optimal precision was achieved at a density cut-off of >4 with a sensitivity of 92% and a specificity of 97% in the exploratory exon set (10% of all gold standard exons). The sensitivity was 91%, and the specificity was 97% when the classifier was applied to the validation set (the remaining 90% of exons). Clusters meeting these criteria were termed “exon clusters.” Applying this procedure to the entire genome, we found 92,628 exon clusters in the 10,367 previously considered known protein-coding genes, 19,974 exon clusters in other annotated genes (e.g., weakly expressed protein-coding genes, RNA genes, or genes predicted on the basis of orthologs), and 10,464 new exon clusters in nonannotated regions of the genome (Fig. 2D). The new exon clusters had a total length of 7,290,658 bp, i.e., taken together they covered 0.27% of the genomic sequence.
We evaluated the possibility that contrast could be used as an additional criterion for classifying read clusters. Within the gold-standard regions, true-positive clusters had on average a steeper flank than false-positives. But the overlap was considerable (as seen in Supplemental Fig. 1), suggesting that only about two out of three clusters would be classified correctly. Improving a strong classifier by combining it with a weak one usually fails to improve the overall result—as one of us previously found in another area of informatics research, speech recognition (P. Beyerlein, unpubl.).
“Classic” genes—i.e., regions with well-defined start, end, and exon boundaries—may not be the only source of RNA. “Dark matter” (Johnson et al. 2005) regions of the genome may be transcriptionally active; for example, it has been reported that up to 94% of the human genome can be transcribed (The ENCODE Project Consortium 2007). We found 968,726 UMR that mapped neither to annotated areas of the genome nor to newly defined clusters. These were labeled “dark matter reads.” Dark matter reads were rare, comprising only 0.71% of UMR, and were dispersed sparsely, covering 1.4% of the genome sequence at an average density of 1.2×. Given this sparsity, statistical inferences cannot be derived from this portion of the data set, leaving DRG dark matter transcription an uncertain possibility that will require even larger sequencing efforts before we can arrive at a critical assessment.
Sequencing depth in the present study was ample for discovery of new exons (resembling those of annotated genes) as demonstrated by the test characteristics provided above (i.e., sensitivity and specificity in the high-90s percentage range) and by the high average read density, 43×, for the newly discovered 10,464 exon clusters.
Agnostic splice junction discovery
Pre-mRNA (cis-)splicing is fundamental to defining transcriptomes. Splicing determines how many different proteins (or regulatory RNA) are derived from a set of neighboring exons, and which of them should be grouped together in a single semantic unit, i.e., a gene. A remaining stated goal of mRNA-seq, for which no published solution yet exists, is to determine pre-mRNA SJs de novo, i.e., solely from the experimental data (mRNA-seq reads) without use of existing annotations or information from other sources or experiments.
In previous mRNA-seq studies, splicing was detected either by mapping reads to known SJs or to limited sets of hypothetical junctions derived by alternative (in silico) assembly of known exons (Pan et al. 2008; Wang et al. 2009). These approaches were the only practicable methods when mRNA-seq reads were very short, i.e., 25–36 bp. Such methods are termed biased because only the limited set of SJs that are predicted to exist can be experimentally detected. Accordingly, previous studies could not discover new SJs in nonannotated sections of the genome nor could they discover unsuspected SJs involving known genes (e.g., splicing of newly discovered exons to known genes). In our study, the length of sequences was longer (50 bp), thanks to improved technology, which allowed us to implement a computationally novel approach for unbiased SJ discovery that overcame the shortcomings of previous approaches. The new approach is termed “agnostic” because it operates free of preconceived notions about exon borders, mapping SJ de novo based solely on the information represented by the mRNA-seq data.
We reasoned that some reads would span SJ symmetrically, i.e., the splice site would be exactly in the middle of the read. In these instances the 25-bp sequence on each side of the junction would map to a different location on the same chromosome indicative of the positions and precise borders of two exons belonging to the same gene (Fig. 3A). Accordingly, the two 25-bp ends (each of the two halves) of every 50-bp read obtained in the study were mapped independently against the entire genomic reference sequence, allowing us to identify 4,539,891 reads containing an SJ exactly in the middle (1.7% of all sequences obtained in the study); such reads were termed sjUMR (algorithmic definition provided in Methods).
We found that 97% of 25-bp halves of all sjUMR mapped within the 123,066 UMR clusters defined above, providing a first validation of the approach (examples are shown in Fig. 3B,C). In other words, we confirmed that the overwhelming majority of sjUMR connected bona fide exons and not random positions. Next, sjUMR were grouped into clusters, termed SJ [read] clusters (SJC), each of which consisted of all the sjUMR spanning the same splice connection. From this analysis we identified 99,291 SJC connecting two of the previously defined exon clusters. To validate the predicted exon–exon splice connections, we determined if exons connected by SJC belonged to the same gene. We found that SJC that were located on one side in an exon cluster of an annotated gene connected to another exon cluster of the same gene in 94% of instances. In 3% of cases, they connected to a previously nonannotated exon cluster, and in another 3%, they connected to an exon cluster belonging to an apparently unrelated gene. While this indicates an error rate of 3%–6% “wrong” connections, at least 94% of the connections described by SJC were confirmed by the annotation as true connections, and up to 97% may, indeed, be correct considering that a frequent type of newly discovered exons are those belonging to (and thereby extending) a previously discovered gene.
The majority of spliced introns in mammals (as well as most lower eukaryotes) are flanked by a 5′-GT…AG-3′ consensus motif, i.e., 5′-GT at the beginning of the intron (directly following the last base of the splice donor exon) and AG-3′ at the end (directly preceding the first base of the splice acceptor exon). As an additional means of external validation of the agnostic SJ discovery method, we examined the frequency of the consensus motif. Among 65,108 unselected SJC consisting of 10 or more sjUMR, 97% flanked the 5′-GT…AG-3′ motif; this is virtually indistinguishable from the frequency observed in annotated known protein-coding genes (also 97%), further emphasizing the high degree of validity of our agnostic splice site discovery algorithm.
SJC can consist of sjUMR with different offsets (i.e., different start and end positions) appearing as “jitter” of sjUMR in a graphical representation. While the observation of a jitter may at first appear counterintuitive, it has two discernible causes, which were taken into account for the final splice site prediction, as follows: (1) Mismatches at the end of the sjUMR r1,…,r25. We allowed for up to two mismatches when aligning reads to the reference genome; these mismatches were permitted to occur at the end of r1,…,r25 or at the beginning of r26,…,r50. In other words, an sjUMR could be a perfect match if cut into two unequal fragments r1,…,r23 and r24,…,r50 (or r1,…,r24 and r25,…,r50; r1,…,r26 and r27,…,r50; r1,…,r27 and r28,…,r50); i.e., it is in fact an NMR aligning in a bona fide fashion to the identified SJ; yet, given the 25 bp + 25 bp cut method used, it aligns with mismatches. We resolved this by simply trimming mismatched bases of such sjUMR, which resulted in unambiguous SJs, which, in most cases were supported by additional reads with the correct offset (not requiring trimming). (2) A microhomology may exist between the 5′-end of an intron and the 5′-end of the subsequent exon. In this case, the position of an SJ based on any form of mRNA sequencing is ambiguous, albeit this has no consequence for the predicted mRNA, which is the same regardless of the SJ used. The 5′-GT…AG-3′ motif flanking 97% of introns could be used to resolve the overwhelming majority of such cases with single-base-pair precision.
Stable patterns of pre-mRNA splicing in chronic pain
Alteration in pre-mRNA splicing is a mechanism of cellular regulation. We considered the possibility that splicing might be altered in the DRG of animals with chronic pain as compared to controls. To test this possibility, we compared the usage of alternative splice sites with the same statistical approach used above for comparison of gene expression levels. Based on a false discovery rate analysis of the 573 best-supported alternative splice events, >99% of alternative splicing choices were changed <2.79-fold. When SNL and control samples were compared, only four to seven alternative splice events fell outside the 99% control range compared with five to six (1% of 573) expected false-positive events. Reads crossing the splice junctions at other locations (22 + 28, 23 + 27, 24 + 26, 26 + 24, 27 + 23, 28 + 22) were then added to the analysis to improve the degree of support by using all data. As a result the 99% confidence interval was tightened (−1.37 to 1.37 on a log2 scale), but the outcome remained principally unaltered—four to six alternative splice events now fell outside the 99% control range compared with an unchanged expected false-positive number of events (five to six). Thus, no differences were found to suggest that alterations in splicing were a common mechanism of gene regulation in pain.
De novo transcriptome annotation: Integration of all mRNA-seq read classes and agnostic SJ discovery
Applying the newly defined SJC to annotating previously nonannotated portions of the rat genome, we found that 3420 of the 10,464 nonannotated exon clusters (newly discovered as described above) were connected to annotated genes by SJC. We found 421 groups of exons consisting solely of newly discovered nonannotated exon clusters connected by SJC, i.e., transcriptional units resembling new genes. Of these, 90 were homologous to nonannotated regions of the mouse genome and 320 to known mouse genes. No homology was found for the remaining 11 (BLASTN E-value ≤ 0.01). Comparison with the human genome led to a smaller fraction of known homologs, 269, which was to be expected because of the greater phylogenetic distance. Integrating information from UMR, sjUMR, MMR, and NMR read classes provided complex genome annotation/reannotation including discovery of large novel genes. For example, we mapped a 64-exon-long homolog of UNC-80, which was previously not annotated in the rat genome. While our study was under way, a mouse form of UNC-80 was prominently reported as a novel substance-P and neurotensin coreceptor (Lu et al. 2009). Substance P has long been recognized as a critical peptide neurotransmitter at the spinal level in chronic pain. Down-regulation of the rat UNC-80 homolog after SNL may contribute to allodynia through alterations in peptide activity in the DRG. The case of UNC-80 serves as an example of how de novo transcriptome annotation may be highly informative in identification of novel candidate regulators that would not have been identifiable by previous technologies such as microarrays or qPCR.
Newly discovered transcripts are dysregulated in pain
Of the newly discovered exons, 9.0% were significantly induced and 12.9% were suppressed in SNL animals compared with sham controls, a rate comparable to the rate of altered regulation of known genes described above, further suggesting that at least some of the newly discovered genes will ultimately prove to be important regulators in chronic pain.
Discussion
mRNA-seq is a paradigm-shifting technology for transcriptomics. The present study developed novel bioinformatics analysis methods focusing on genome-wide exon discovery and agnostic pre-mRNA splice site mapping, both of which were supported by rigorous statistical validation. Taking advantage of this methodological progress, we have conducted an mRNA-seq study in a neurological disease model demonstrating an unprecedented breadth of nervous system transcriptome reprogramming extending to previously unmapped mRNA.
The acquisition of 13 Gb of raw transcriptome sequence (267 million 50-bp reads) permitted precise quantification of more than 10,000 genes in an important site of the nervous system, the DRG, for two conditions and two time points (Fig. 1). The results included discovery of novel genes (Fig. 2), allowed agnostic mapping of pre-mRNA splice sites (Fig. 3), guided discovery of large genes (Fig. 4), and demonstrated that one in five genes, including previously unmapped genes with novel candidate neural regulator roles, were significantly altered, some by as much as 100-fold, in animals with neuropathic pain (Fig. 1; Supplemental Table 3).
Using mRNA-seq, we discovered an order of magnitude more gene expression changes than were expected based on previous microarray studies performed on DRG in similar rat pain models (Bonilla et al. 2002; Costigan et al. 2002; Valder et al. 2003; Davis-Taber and Scott 2006; Rodriguez Parkitna et al. 2006). At the same time, the results obtained using mRNA-seq cross-validated results obtained in microarray experiments by others.
A χ2 test performed on 330 genes, which were reported in the microarray literature (as up- or down-regulated) and quantified in our experiments, demonstrated a highly significant positive correlation, P < 10−10 (Supplemental Table 4). Of those genes, 67 were up-regulated and 61 down-regulated in both data sets. There was no instance of regulation in the opposite direction, further supporting our results. A number of genes, which were found in published studies to be regulated, were found unchanged in our data set. This was not unexpected. The published microarray studies have all used a model of nerve injury at the level of the sciatic nerve, which contains sensory fibers from three anatomical levels (L4–L6). In published studies, DRG from the L4 and L5 levels were pooled. In the process two qualitatively different types of DRG alterations were sampled and pooled—direct injury (dendrites anatomically damaged) and indirect effects, i.e., pure neural dysfunction (dendrite anatomically intact). We performed a spinal nerve ligation (SNL) at L5 and harvested the L4 DRG selectively for mRNA-seq analysis. Therefore, our analysis focused selectively on the dysfunctional but anatomically intact L4 DRG. Accordingly, it could be expected that we would not detect expression changes related to direct nerve injury, which may account for the majority of the 202 genes found to be unaffected in our model. Cases of concordance included well-known examples like neuropeptide Y and activating transcription factor-3 (ATF3). ATF3 was found in our study to be induced 11-fold in the L4 DRG 2 wk after L5 SNL, consistent with the ~10-fold induction of ATF3 that had been described in DRG microarray studies (Costigan et al. 2002; Valder et al. 2003; Davis-Taber and Scott 2006). An overview of the relationship with the microarray literature is presented in Supplemental Table 4. A gene-by-gene comparison is provided in Supplemental Table 5. Both are based on all positive findings available from all published DRG microarray studies (Bonilla et al. 2002; Costigan et al. 2002; Valder et al. 2003; Davis-Taber and Scott 2006; Rodriguez Parkitna et al. 2006). While the referenced microarray studies relied on less selective injury models (a mixture of direct axonal injury and indirect dysfunction of unsevered neurons) and required pools of many DRG (from different anatomical levels and multiple animals), mRNA-seq provided single-DRG sensitivity, allowing us to perform the present study in the surgically selective L5 SNL model (Chung et al. 2004). The neural structure investigated, the L4 DRG, was anatomically intact (unsevered L4) in all animals but was dysfunctional in the experimental group driving abnormal behavior, i.e., allodynia, which was experimentally validated (Fig. 1A). Thus, mRNA-seq demonstrated that transcriptome reprogramming in an intact site of the nervous system (L4 DRG) is a genomic correlate of altered behavior, i.e., hind paw withdrawal in response to soft von Frey hairs (allodynia).
The mRNA-seq library construction methods used (Mortazavi et al. 2008) did not preserve strand information. Detected expression changes may therefore in some cases be due (at least in part) to alterations in transcription of the opposite strand as observed in the FANTOM3 data set (Katayama et al. 2005).
New computational methods were developed for nearly every aspect of our analysis, especially novel exon discovery and agnostic splice-site mapping, which proved critical for the discovery aspect of the study. New programs were developed (in C, Java, and Perl; available at http://www.th-wildau.de/bioinformatics/wios/ or http://mayoresearch.mayo.edu/mayo/research/beutler_lab/wios.cfm), which should facilitate implementation of the analysis paradigms presented here into other mRNA-seq studies.
Rigorous validation of the analysis tools through comparison of mRNA-seq results with “gold standard” reference annotations and the computation of comprehensive statistics supporting the approach was a major focus of our study. We demonstrated that gene quantification was reproducible, confirming original reports on mRNA-seq technology (Mortazavi et al. 2008). We demonstrated that new exon discovery could be achieved with high sensitivity and specificity (91% and 97%, respectively; ROC curve > 0.968 shown in Fig. 2C). We developed an approach to agnostic SJ discovery and showed that splice junctions could be mapped genome-wide with great precision (94%–97% with one validation approach and 97% with another).
We previously reported the use of mRNA-seq in a study of cerebellar RNA from humans with schizophrenia (Mudge et al. 2008). While this constituted a first foray into neurobiology, conclusions from the study were limited because of the absence of an experimental intervention or effective controls, as well as by the lack of bioinformatics innovation toward new transcript and pre-mRNA splice site discovery, and the very short read length of first-generation mRNA-seq technology employed. To overcome these limitations, we designed the present study with newer technology, i.e., library construction requiring <1 μg of mRNA and sequencing to a read length of 50 bp. Using a widely accepted, well-controlled animal model of an important neurological disorder, the rat SNL model of chronic neuropathic pain, the present study could trace all transcriptome changes observed to a specific experimental manipulation in the nervous system.
A limited pathway analysis is presented in Supplemental Figure 2. Annotations of represented genes are provided in Supplemental Table 12. The pathway links extracellular neuroimmunological alterations (supported by our data) through multiple cytosolic intermediaries with a large number of transcriptional events. Suppression of neuroimmunological activation by DRG-directed gene therapy with immunosuppressive cytokines such as interleukin 10 is known to suppress neuropathic pain (Beutler et al. 2005), as would be predicted from the pathway shown in Supplemental Figure 2. The decision to stay otherwise clear of network analyses was deliberate. Most known “pathways” are cancer-centric, reflecting the area where most primary data of molecular interaction have been accumulated. Regarding nervous system pathways, sources from different anatomical sites are often combined to construct a pathway. Pathway depictions usually imply linearity, which may not reflect the complexity of regulatory feedback loops and interdependence of events. Furthermore, the true initiating events for complex transcriptional reprogramming may be chromatin remodeling events, whose study in the nervous system has just begun. Comprehensive pathway recognition in neuroscience experiments may be enhanced as it becomes easier to capture genome-scale data sets for defined sites and conditions.
In the future, neurological diseases such as pain might be investigated as problems of transcriptional reprogramming by integrating complete capture and quantification of the various classes of RNA with a genome-wide characterization of the chromatin state, for example, through chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) (Johnson et al. 2007) or through determining DNA methylation (Brunner et al. 2009). Such approaches may ultimately lead to the identification of targets for transcription therapy that might employ molecules exerting an analgesic effect by inducing or suppressing specific gene expression programs. Investigation of these various conceptual layers are currently merging at the technological level through the adoption of short read sequencing techniques such as mRNA-seq.
Methods
Base calling and alignment of sequencing reads to the reference genome
The Illumina analysis software pipeline program Bustard was used for base calling. ELAND v1.3 was used to map reads to the Rattus norvegicus reference genome assembly RGSC 3.4 (release 50; ftp://ftp.ensembl.org/pub/release-50/). The maximum read length directly processed by ELAND (i.e., without additional computing steps) is 32 bp. Our reads were 50 bp long, r1,…,r50. We extracted two 32-bp substrings—r1,…,r32 and r19,…,r50—and aligned those individually allowing for up to two substitutions, i.e., base pair mismatches (MM). We then combined the alignment result for the two substrings classifying r1,…,r50 as a UMR if r1,…,r32 and r19,…,r50 matched once with an offset of 18 bp (50 − 32 bp) between r1 and r19; as a MMR if the event occurred more than once; and as a NMR if it did not occur. We compared our approach with the GERALD pipeline for processing of 50-bp reads (“ELAND extended”), which aligns r1,…,r32 to establish the uniqueness of the match followed by counting the number of MM in the remaining fragment to exclude that r1,…,r50 is nonmatching. While equally specific, this approach was less sensitive, overcalling MMR at the expense of UMR. Direct comparison showed that our modified approach increased the fraction of UMR by 1.8% (4.7 million) of all reads. Furthermore, we validated a subset of the results with another alignment tool, Bowtie (Langmead et al. 2009), which detected UMR with a sensitivity of 96%, an expected result (Supplemental Table 6).
Induction and suppression of 10,367 known protein-coding genes
For a given sample of mRNA-seq reads S (e.g., all reads obtained for a single DRG), the read count for an individual gene c(g, s) is the sum of read counts for the set E of all exons e1…en annotated by Ensembl (RGSC 3.4, release 50) as belonging to the gene g, i.e.,
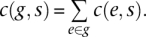
A read was counted as belonging to an exon if its start position fell within annotated exon boundaries. “Known protein-coding” (kpc) Ensembl genes were considered in this part of the analysis, requiring a c(g, s) ≥ 100 for at least two samples at the early time point (2 wk), thereby excluding low-expressing genes that contribute only noise. As a result 10,367 genes were included in the analysis. c(g, s) were normalized to the total number of UMR in each respective sample s:
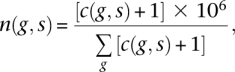
i.e., the count of reads for a gene per 1 million reads in the respective sample. A count of 1 was added to c(g, s) as a discounting step precluding the possibility of division by 0 in subsequent operations.
To validate mRNA-seq for the quantification of gene expression changes, a comparison with quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed. Fold-changes of gene expression measured by Taqman qRT-PCR were highly correlated (r = 0.956, slope 0.984) with mRNA-seq, as shown in Supplemental Figure 3, indicating the validity of mRNA-seq as a method for quantifying relative changes of transcript levels.
To estimate the number of induced and suppressed genes in SNL animals compared to controls, the FDR concept was applied. The relative change r of the normalized expression of a given gene n(g, s) between two samples r(g, s1, s2) was calculated as:
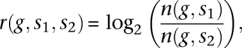
i.e., the fold-change on a log2 scale, where +1 corresponds to doubling of gene expression and −1 to a reduction by half.
Next, the ratio r(g, s1, s2) of each of the 10,367 genes was determined, with s1 and s2 corresponding to two biological replicates, i.e., either two control or two SNL animals. Under ideal conditions, all ratios on the log2 scale should be 0 (i.e., a change of 1×, which is “no change”). Examining the actual distribution of these control ratio distributions, we found that the respective medians were very close to 0, their shape was symmetric, and the average 99% confidence interval was ±0.8096, i.e., only 0.5% of control expression ratios r(g, s1, s2) were higher or lower than the confidence interval cut-off, corresponding to a FDR of 52 genes induced and suppressed among the total of 10,367. Next, we determined the number of genes induced and suppressed after SNL by calculating r(g, s1, s2) with s1 and s2 corresponding to SNL and control samples, repeating the procedure for independent biological replicates. Results are shown in Figure 1E.
Validation of mRNA-seq with qPCR
mRNA-seq was performed using two quality-control standards, the universal human reference RNA (Stratagene) and human brain reference RNA (Ambion). Fold-change differences among 755 transcripts quantified by mRNA-seq were calculated as described above and compared with established Taqman qRT-PCR data (Shi et al. 2006) as shown in Supplemental Figure 3.
Pathway analysis
Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity Systems; http://www.ingenuity.com). A network is a graphical representation of the molecular relationships between genes and gene products. Genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base. Human, mouse, and rat orthologs of a gene are stored as separate objects in the Ingenuity Pathways Knowledge Base, but are represented as a single node in the network. The intensity of the node color (Supplemental Fig. 2) indicates the degree of up- (red) or down- (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product. Annotation of genes is provided in Supplemental Table 12.
Novel gene discovery
A read histogram was constructed for each chromosome c by counting the number of reads covering a given base position x, denoted as hc(x), resulting in a group of chromosome-spanning histograms hc(·)(c = 1, … ,21) with single base position resolution covering the genome. For this analysis, data from all samples were pooled:
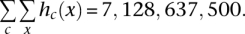
A sliding window with length l was shifted over the genome integrating the area under the curve of the read histogram hc(x) in the interval [p, p + l], i.e.,
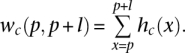
When wc(p, p + l) is zero, the consecutive interval of length l has no reads. An interval [a, b] on the chromosome c, for which all values wc(a, a + l), wc(a + 1, a + 1 + l), wc(a + 2, a + 2 + l), …, wc(b − l, b) are non-zero, is called a read cluster. We denote this read cluster with Ka,b,c. Setting the window length l = 100, we defined 882,913 Ka,b,c. In other words, we identified 882,913 read clusters consisting of one or several neighboring reads; clusters were separated by sections devoid of reads (no read within 100 bp).
Because an individual read cluster by this definition could consist of as little as a single read and as many as thousands, we reasoned that only some, i.e., those with many reads, would be strong predictors of exons. Therefore, we introduced read density as a measure to characterize clusters further. For each read cluster Ka,b,c, we calculated the read density as follows:
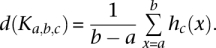
Next, we developed a classifier dichotomizing clusters into a set of exon-like clusters F and its complement based on read density, whereby
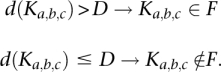
To establish the test characteristics of the procedure, we tested it for D = 0, …, 100 on a training set as described in the Results section, constructing the ROC curve shown in Figure 2C, which suggested D = 4 as a cut-off for optimal precision. Sensitivity and 1-minus-specificity values for each tested read density are provided in Supplemental Table 7.
De novo mapping of mRNA splice junctions
Pre-mRNA is processed to mRNA such that intron sequences are removed and neighboring exon sequences joined together creating novel SJ sequences that are not found in the genome reference sequence. mRNA-seq reads derived from splice junctions typically do not match to the genome sequence, and thus were classified as NMR in the alignment described above. Previous mRNA-seq studies have mapped SJs using a limited sequence database consisting of annotated splice sites and predicted sites obtained by in silico alternative exon joining. These approaches were the only practicable way of using very short sequence reads (25–32 bp) for this goal, but they were unable to predict SJs de novo genome-wide. We sought to overcome this limitation.
We based SJ discovery on reads classified as NMR. We focused on reads traversing the SJ exactly in the middle of the read, because we reasoned that those reads would on average have the best balance of sequence information available for mapping each end independently. Thus, every NMR r1,…,r50 was split into the two fragments r1,…,r25 and r26,…,r50, which were independently aligned with ELAND to the entire genome (as described above for 32-bp sequences). Alignment results for such sequence pairs r1,…,r25 and r26,…,r50 were combined by applying known constraints of splicing biology (specifically, the upper 99th percentile of intron lengths and an assumption of a single-molecule pre-mRNA structure) and selecting only those cases that consisted of pairs matched to a single site. We found 4,539,891 such reads; 124,403 of these consisted of pairs mapping with a negative distance, i.e., deletions. After removing deletions, 4,415,488 reads remained. In other words, we identified reads that as a 50-mer r1,…,r50 did not match (i.e., NMR) but whose 25-mer halves (r1,…,r25 and r26,…,r50) each matched uniquely to the genome, thereby defining an SJ. These reads were termed splice junction UMR (sjUMR). We identified 4,415,488 sjUMR (1.7% of all reads) by the 25 bp + 25 bp cut method.
Novel sjUMR were then combined into SJ clusters (SJC) each covering the same SJ; 154,577 SJC were found; i.e., in our data set, each SJC was supported by an average of 28.6 sjUMR. SJC were validated and used to establish connectivity between known and/or novel exons as detailed in the Results section and illustrated in the figures. We defined 421 new genes consisting solely of newly discovered exons and SJC. The full sequence and genome position of the entire data set are provided in Supplemental Tables 8–11. The relative frequency of noncanonical splice sites observed was provided in Supplemental Table 13.
Acknowledgments
Support was provided by the National Institute of Neurological Disorders and Stroke (NINDS) of the NIH (R01NS063022 and R21NS062271) (A.S.B.) and by the Schulze Family Foundation (A.S.B.).
Footnotes
[Supplemental material is available online at http://www.genome.org. Sequence reads from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE20895. Software is available for download from http://www.th-wildau.de/bioinformatics/wios/ and http://mayoresearch.mayo.edu/mayo/research/beutler_lab/wios.cfm.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101204.109.
References
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [Europe PMC free article] [Abstract] [Google Scholar]
- Beutler AS, Banck MS, Walsh CE, Milligan ED 2005. Intrathecal gene transfer by adeno-associated virus for pain. Curr Opin Mol Ther 7: 431–439 [Abstract] [Google Scholar]
- Bonilla IE, Tanabe K, Strittmatter SM 2002. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci 22: 1303–1315 [Abstract] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, et al. 2009. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res 19: 1044–1056 [Europe PMC free article] [Abstract] [Google Scholar]
- Chung JM, Kim HK, Chung K 2004. Segmental spinal nerve ligation model of neuropathic pain. Methods Mol Med 99: 35–45 [Abstract] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [Abstract] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D'Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ 2002. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci 3: 16 10.1186/1471-2202-3-16 [Europe PMC free article] [Abstract] [Google Scholar]
- Davis-Taber R, Scott VES 2006. Transcriptional profiling of dorsal root ganglia in a neuropathic pain model using microarray and laser capture microdissection. Drug Dev Res 67: 308–330 [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [Europe PMC free article] [Abstract] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC 2010. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [Europe PMC free article] [Abstract] [Google Scholar]
- Griffin RS, Mills CD, Costigan M, Woolf CJ 2003. Exploiting microarrays to reveal differential gene expression in the nervous system. Genome Biol 4: 105 10.1186/gb-2003-4-2-105 [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE 2005. Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet 21: 93–102 [Abstract] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein–DNA interactions. Science 316: 1497–1502 [Abstract] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [Abstract] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [Europe PMC free article] [Abstract] [Google Scholar]
- Lu B, Su Y, Das S, Wang H, Wang Y, Liu J, Ren D 2009. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature 457: 741–744 [Europe PMC free article] [Abstract] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y 2008. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [Europe PMC free article] [Abstract] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [Abstract] [Google Scholar]
- Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, Luo S, Zhang L, van Velkinburgh JC, Farmer AD, et al. 2008. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS One 3: e3625 10.1371/journal.pone.0003625 [Europe PMC free article] [Abstract] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [Europe PMC free article] [Abstract] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [Abstract] [Google Scholar]
- Rodriguez Parkitna J, Korostynski M, Kaminska-Chowaniec D, Obara I, Mika J, Przewlocka B, Przewlocki R 2006. Comparison of gene expression profiles in neuropathic and inflammatory pain. J Physiol Pharmacol 57: 401–414 [Abstract] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. 2006. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 24: 1151–1161 [Abstract] [Google Scholar]
- Storek B, Reinhardt M, Wang C, Janssen WG, Harder NM, Banck MS, Morrison JH, Beutler AS 2008. Sensory neuron targeting by self-complementary AAV8 via lumbar puncture for chronic pain. Proc Natl Acad Sci 105: 1055–1060 [Europe PMC free article] [Abstract] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [Abstract] [Google Scholar]
- Sun H, Xu J, Della Penna KB, Benz RJ, Kinose F, Holder DJ, Koblan KS, Gerhold DL, Wang H 2002. Dorsal horn-enriched genes identified by DNA microarray, in situ hybridization and immunohistochemistry. BMC Neurosci 3: 11 10.1186/1471-2202-3-11 [Europe PMC free article] [Abstract] [Google Scholar]
- Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, et al. 2006. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med 12: 1269–1277 [Abstract] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [Europe PMC free article] [Abstract] [Google Scholar]
- Valder CR, Liu JJ, Song YH, Luo ZD 2003. Coupling gene chip analyses and rat genetic variances in identifying potential target genes that may contribute to neuropathic allodynia development. J Neurochem 87: 560–573 [Abstract] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [Europe PMC free article] [Abstract] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [Abstract] [Google Scholar]
- Wold B, Myers RM 2008. Sequence census methods for functional genomics. Nat Methods 5: 19–21 [Abstract] [Google Scholar]
Articles from Genome Research are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gr.101204.109
Read article for free, from open access legal sources, via Unpaywall:
https://genome.cshlp.org/content/20/6/847.full.pdf
Free to read at www.genome.org
http://www.genome.org/cgi/content/abstract/20/6/847
Free after 6 months at www.genome.org
http://www.genome.org/cgi/content/full/20/6/847
Free after 6 months at www.genome.org
http://www.genome.org/cgi/reprint/20/6/847
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/gr.101204.109
Article citations
Integrated analyses reveal evolutionarily conserved and specific injury response genes in dorsal root ganglion.
Sci Data, 9(1):666, 02 Nov 2022
Cited by: 3 articles | PMID: 36323676 | PMCID: PMC9630366
NBBt-test: a versatile method for differential analysis of multiple types of RNA-seq data.
Sci Rep, 12(1):12833, 27 Jul 2022
Cited by: 1 article | PMID: 35896555 | PMCID: PMC9329447
Global gene expression and chromatin accessibility of the peripheral nervous system in animal models of persistent pain.
J Neuroinflammation, 18(1):185, 26 Aug 2021
Cited by: 9 articles | PMID: 34446036 | PMCID: PMC8390277
Combination of autophagy and NFE2L2/NRF2 activation as a treatment approach for neuropathic pain.
Autophagy, 17(12):4062-4082, 09 Apr 2021
Cited by: 36 articles | PMID: 33834930 | PMCID: PMC8726676
Circadian regulation of chemotherapy-induced peripheral neuropathic pain and the underlying transcriptomic landscape.
Sci Rep, 10(1):13844, 14 Aug 2020
Cited by: 20 articles | PMID: 32796949 | PMCID: PMC7427990
Go to all (53) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE20895
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Brain-derived neurotrophic factor increases in the uninjured dorsal root ganglion neurons in selective spinal nerve ligation model.
J Neurosci, 21(13):4891-4900, 01 Jul 2001
Cited by: 200 articles | PMID: 11425916 | PMCID: PMC6762362
Change in mRNAs for neuropeptides and the GABA(A) receptor in dorsal root ganglion neurons in a rat experimental neuropathic pain model.
Pain, 78(1):13-26, 01 Oct 1998
Cited by: 130 articles | PMID: 9822208
Upregulation of Cav3.2 T-type calcium channels in adjacent intact L4 dorsal root ganglion neurons in neuropathic pain rats with L5 spinal nerve ligation.
Neurosci Res, 142:30-37, 21 Apr 2018
Cited by: 18 articles | PMID: 29684385
A comparison of RNA-seq and exon arrays for whole genome transcription profiling of the L5 spinal nerve transection model of neuropathic pain in the rat.
Mol Pain, 10:7, 28 Jan 2014
Cited by: 59 articles | PMID: 24472155 | PMCID: PMC4021616
Funding
Funders who supported this work.
NINDS NIH HHS (4)
Grant ID: R01 NS063022
Grant ID: R01NS063022
Grant ID: R21 NS062271
Grant ID: R21NS062271





