Abstract
Free full text

Discrete Roles of STAT4 and STAT6 transcription factors in Tuning Epigenetic Modifications and Transcription during Helper T Cell Differentiation
Abstract
Signal transducer and activator of transcription 4 (STAT4) and STAT6 are key factors in the specification of helper T cells; however, their direct roles in driving differentiation are not well understood. Using chromatin immunoprecipitation and massive parallel sequencing, we quantitated the full complement of STAT-bound genes, concurrently assessing global STAT-dependent epigenetic modifications and gene transcription using cells from cognate STAT-deficient mice. STAT4 and STAT6 each bound over 4000 genes with distinct binding motifs. Both played critical roles in maintaining chromatin configuration and transcription of a core subset of genes through the combination of different epigenetic patterns. Globally, STAT4 had a more dominant role in promoting active epigenetic marks, whereas STAT6 had a more prominent role in antagonizing repressive marks. Clusters of genes negatively regulated by STATs were also identified, highlighting previously unappreciated repressive roles. Therefore, STAT4 and STAT6 play wide regulatory roles in T helper specification.
INTRODUCTION
In order to mount effective immunity towards diverse microbial pathogens, naive CD4+ T helper cells adapt distinct fates, becoming T helper 1 (Th1), Th2 or Th17 cells, and thereby orchestrating effector responses (Abbas et al., 1996; Murphy and Reiner, 2002; Weaver et al., 2006). These T helper effector subtypes are defined by their signature cytokines, namely interferon-γ (IFN-γ) for Th1 cells, interleukin-4 (IL-4) and IL-13 for Th2 cells and IL-17A for Th17 cells (Zhou et al., 2009). CD4+ T cells can also differentiate to become regulatory T cells, which serve to constrain immune-mediated damage (Sakaguchi et al., 2008). Thus, the attainment of these cell fates is critical not only for proper elimination of pathogens, but also for avoidance of damage to the host (Reiner, 2007).
The microenvironment of the naïve CD4+ T cell greatly influences its specification, with a key factor being the cytokine milieu. A major means by which cytokines exert their effect is through activation of signal transducer and activator of transcription (STAT) family transcription factors, which translocate to the nucleus, bind to the regulatory region of genes and induce transcription (Levy and Darnell, 2002; Schindler and Darnell, 1995). Among the 7 STAT family members, STAT4, in conjunction with STAT1, plays pivotal role in Th1 cell differentiation by transmitting IL-12 signals to produce IFN-γ and enhancing expression of T-box 21 (T-bet) (Thieu et al., 2008). STAT6, activated by IL-4, is the key player in Th2 cell differentiation and serves to enhance expression of IL-4, IL-13 and GATA binding protein 3 (Gata3) (Zhu and Paul, 2008). Among the STATs, STAT4 and STAT6 stand out as having highly specific functions, because the phenotype of gene-targeted mice is largely confined to Th1 or Th2 cell defects respectively and the mice develop otherwise normally with normal numbers of T cells (Wurster et al., 2000).
Transcriptional profiling has helped to clarify genes whose expression is controlled by STAT4 and STAT6, and these data argue that these transcription factors are clearly important for regulating a wide array of genes (Chen et al., 2003; Hoey et al., 2003; Lund et al., 2004; Lund et al., 2007; Rogge et al., 2000). However, this technology does not permit the discrimination of genes that are directly versus indirectly regulated. Chromatin immunoprecipitation (ChIP) can identify genes bound by transcription factors, but relatively few genes have been confirmed to be occupied by STATs. In the case of STAT4, known targets include Ifng, Il18r1, Il12rb2, Il2ra and Furin (Letimier et al., 2007; O'Sullivan et al., 2004; Pesu et al., 2006; Thieu et al., 2008), whereas STAT6-bound genes include Il4, Gata3, and Ccl17 (Kubo et al., 1997; Wirnsberger et al., 2006). Therefore it was far from evident what proportion of genes are directly occupied by STAT4 or STAT6, and ultimately contribute to Th1 or Th2 cell phenotypes or whether STATs work predominantly in an indirect manner. ChIP coupled with hybridization to arrays (ChIP-on-Chip) or massive parallel sequencing (ChIP-seq) provides the opportunity to identify transcription factor binding sites on a broader scale (Good et al., 2009; Jothi et al., 2008). This has expanded our understanding of STAT-bound genes; however, relating transcription factor binding to functional consequence, i.e. transcriptional control, has been difficult (Beima et al., 2006; Good et al., 2009; Wunderlich and Mirny, 2009).
Although transcription factors like STATs play a critical role in regulating differentiation, gene expression is also dependent on the status of chromatin structure, which is regulated by DNA methlylation, ATP-dependent remodeling of nucleosomes, incorporation of histone variants, and posttranslational modification of histone tails (Bernstein et al., 2007). As STATs play a critical role in initiating a defined gene transcription program, it is of great interest to determine how the distribution of histone epigenetic marks and recruitment of STAT transcription factors coordinate to orchestrate gene regulation in differentiating T cells (Wei et al., 2009; Wilson et al., 2009). Whether histone modifications are prerequisite events for STAT recruitment or vice versa has not been resolved. In the case of the Ifng and Il4, Il13 and Il5 loci, there is evidence that epigenetic modifications and higher order structural remodeling are STAT-dependent (Bernstein et al., 2007; Lee et al., 2006; Wilson et al., 2009; Zhang and Boothby, 2006). However, other than the signature cytokine genes, there is a paucity of information on the epigenetic control of genes that participate in T helper cell differentiation and the involvement of STATs in the process (Thieu et al., 2008; Yu et al., 2007). Whether signature cytokine genes (Ifng and Il4) represent a very limited subset of STAT-dependent T helper cell genes with respect to epigenetic control or whether they are prototypical of a large number of similarly regulated genes has not been determined; only very limited segments of the genome have previously been interrogated. By contrast, recent genome-wide mapping of STAT1 binding sites in HeLa cells indicated that epigenetic modifications preceded IFNγ-triggered STAT1 binding (Robertson et al., 2008; Robertson et al., 2007). Thus, the exact interplay between transcription factors and epigenetic modifications is still an open question.
We set out to address these questions by simultaneously measuring genome-wide STAT recruitment, histone modifications and mRNA expression during differentiation of Th1 and Th2 cells in wild-type and STAT-deficient cells. By mapping the distribution of H3K4me3, H3K27me3 and H3K36me3 along with STAT4 and STAT6 binding, we determined the entire spectrum of specific STAT4- and STAT6-occupied genes and linked STAT binding to corresponding gene transcription and epigenetic modifications. For the majority of STAT-bound genes, the absence of the cognate STAT had little effect on epigenetic modifications and gene transcription. However, we identified a core subset of STAT-occupied genes whose gene expression and chromatin configuration was highly STAT-dependent. Furthermore, we found that the global effects of STAT4 and STAT6 are distinct in that STAT4 primarily promoted accessible epigenetic marks whereas STAT6 had a prominent role in antagonizing repressive marks. This analysis also allowed the identification of subset of genes for which STAT4 and STAT6 appeared to serve as repressors. Therefore, our global analysis identified wide regulatory roles for STAT4 and STAT6 in regulating epigenetic state and transcription in their cognate T helper lineages.
RESULTS
Genomic distribution of STAT binding sites and identification of consensus binding motifs
To identify genes directly bound by STAT4 or STAT6, we polarized naïve CD4+ T cells under Th1 or Th2 cell conditions, and re-activated the cells with anti-CD3 and anti-CD28 antibodies and the respective cytokines for 2 hours prior to ChIP-seq analysis. The appropriate polarization of Th1 and Th2 cells was confirmed by intracellular staining (data not shown) and mRNA expression, as determined by microarray analysis. A total of 7 to 8 million non-redundant reads were uniquely aligned onto mouse genome (mm9 Build 37 assembly by NCBI). We used CisGenome to identify peaks (Ji et al., 2008), and preimmune serum as a negative control to define nonspecific binding. Genome-wide, we identified 10,831 STAT4-occupied peaks and 8,434 STAT6-bound peaks in wild-type cells cultured under Th1 and Th2 cell conditions, respectively (Table S1).
To examine the comparative distribution of STAT4 and STAT6 binding sites, we divided the mouse genome into four regions: promoter (Transcriptional start site (TSS) to upstream 10 kb), exonic, intronic, and intergenic regions. As shown in Figures 1A, 31% of STAT4-occupied sites and 36% of STAT6-occupied sites localized to promoters. This represents a statistically significant (p<10−6) enrichment, as promoter regions constitute only 2% of the total mouse genome (Figure S1). In contrast, only 5% of STAT4 and STAT6 binding sites localized to introns and less than 1% of the binding sites localized to exons, (Figures 1A), with intronic and exonic sequences accounting for 37% and 0.2% of the total mouse genome (Figure S1) respectively. Notably, 63% of STAT4 binding sites and 59% of STAT6 binding sites localized to intergenic regions, where they could potentially function as distal enhancers or promoters of unannotated transcripts (Figures 1A).
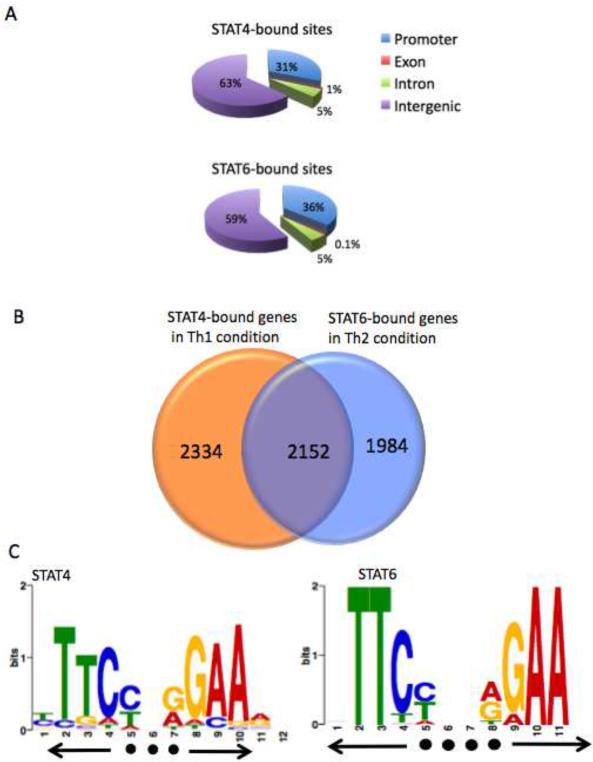
(A, B) ChIP-seq analysis was used to map STAT4 and STAT6 binding from polarized Th1 and Th2 cells. The distribution of (A) STAT4 and STAT6 binding sites was analyzed based on location: promoter (within 10 kb upstream from the transcription start site), exon, intron, and intergenic regions. (B) Venn diagram showing the number of genes uniquely bound by STAT4, STAT6 or genes bound by both STATs. STAT-bound genes were identified if at least one peak of binding was present within the gene region defined as 10 kb upstream of TSS to the transcription end site. (C) A parallel version of MEME (Bailey and Elkan, 1994) was used to perform a de novo search of consensus binding motifs for STAT4 and STAT6 and resulted in distinctive GAA palindromes for STAT4 with 3 bp spacer and STAT6 with 4 bp spacer.
To enumerate STAT-bound genes, we defined a gene locus as a region encompassing potential promoter elements (10 kb upstream from TSS), as well as the exonic and intronic regions of the gene itself (TSS to transcription termination site). Using this definition, we identified 2,334 genes uniquely bound by STAT4 in Th1 cells and 1,984 genes selectively bound by STAT6 in Th2 cells. Interestingly, 2,152 genes were bound by both factors (Figure 1B).
Previous in vitro studies have indicated that the STAT family proteins bind to a GAA palindromic motif (Decker et al., 1997). To determine if such an element was identifiable in our genome-wide analysis, we carried out de novo searches of consensus motifs of STAT4- and STAT6-bound regions using MEME (Bailey and Elkan, 1994). We identified the consensus motif for STAT4 as a GAA palindrome with a 3-nucleotide spacer. In contrast, STAT6 preferentially bound to a palindrome with a 4 base spacer (Figures 1C). Of the total of 4486 STAT4-bound genes, 100% exhibited at least one copy of the motif that was depicted in Figure 1C. However, only 30% of 4136 STAT6-bound genes contained at least one copy of the 4-space motif that was identified in Figure 1C. Among 2152 genes that were bound by both STAT4 and STAT6, 800 genes (37%) contained both 3-space and 4-space consensus motifs.
Colocalization of STAT binding with histone modifications
Other genome-wide surveys of transcription factor binding have suggested that in many cases, binding does not necessarily correlate with regulation of gene expression, particularly in eukaryotes (Wunderlich and Mirny, 2009). The interplay between transcription factor binding and epigenetic events has been equally unclear (Robertson et al., 2008; Zheng et al., 2007). To begin to relate epigenetic changes and occupancy of STAT transcription factors, we also mapped the distribution of H3K4me3, H3K27me3 and H3K36me3 using ChIP-seq (Table S1). We found ~60% of STAT4 and STAT6 binding sites colocalized with H3K4me3 islands (Figures 2A and 2B). Only 0.3% of STAT4 binding sites colocalized with H3K27me3 islands, whereas 5% of STAT6 binding sites colocalized with this modification. Of note, under the culture conditions used, roughly 1.7 fold more total H3K27me3 islands were present in Th2 cells than in Th1 cells, whereas the numbers of total H3K4me3 islands were similar (Figures 2A and 2B). We next compiled the localization of H3K4me3, H3K27me3 and H3K36me3 marks for all genes, relating this to their transcriptional start site (TSS) and transcriptional end site (TES) to visualize tag density profile for each mark (Figures 2C and 2D). As previously reported, H3K4me3 (in red) was highly enriched around the TSS (Roh et al., 2006). STAT4 and STAT6 binding sites (in blue) were also enriched around TSS; however, the precise peak of STAT binding coincided with a valley of H3K4me3 binding (Figures 2C and 2D lower panels). This suggests that maximal STAT binding likely occurs in a relatively nucleosome-poor region in the midst of abundant H3K4me3 modification encompassing several nucleosomes. Of note, H3K36me3 modifications (in purple) increased along gene bodies and peaked at the transcription end sites (Figures 2C and 2D upper panels), whereas H3K27me3 marks (in green) were relatively depleted in regions in which STATs were bound (Figures 2C and 2D lower panels).
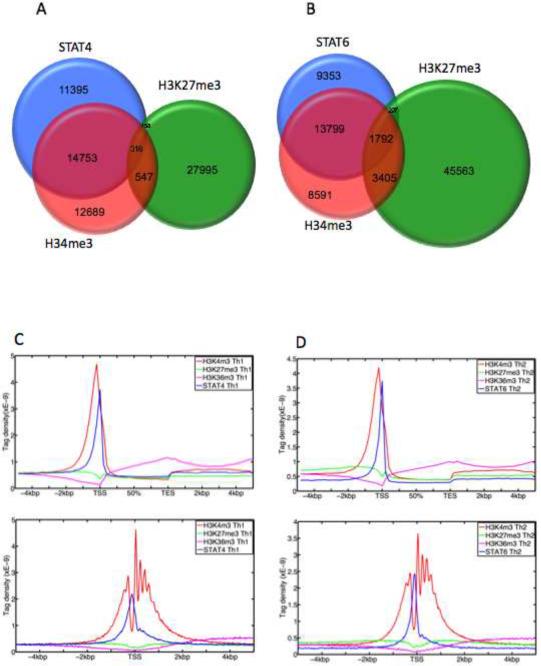
(A, B) ChIP-seq was performed to map histone epigenetic modification in Th1 cells and Th2 cells. The concordance between H3K4me3, H3K27me3 modifications and (A) STAT4 binding sites in Th1 cells and (B) STAT6 binding sites in Th2 cells is shown. (C, D) Compiled tag density profiles for H3K4me3, H3K27me3, H3K36me3 and (C) STAT4 in Th1 cells and (D) STAT6 in Th2 cells are shown. The diagrams represent all genes that showed positive signals for the respective marks. Upper panel: tag density profile across gene body ± 5 kb flanking regions with 200 bp resolution. Lower panel: tag density profile across promoter ± 5 kb flanking regions with 25 bp resolution.
ChIP-seq provides a comprehensive view of STAT-occupied genes that can participate in helper cell function
The ChIP-seq data set generated allowed us to assess global quantitative views of transcription factor binding and STAT-dependent epigenetic modifications, but its precision and resolution also provided a refined view at the gene level. Therefore, we next evaluated localized STAT-dependent epigenetic modification of STAT-occupied genes that contribute to the Th phenotype. Figure 3 depicts a compilation of browser views of selected genes showing relevant marks (STAT binding, H3K4me3 and H3K36me3) in wild type and STAT-deficient cells. The complete list of genes occupied by STAT4 and STAT6 are provided (Table S2) and genome-wide STAT binding and STAT-dependent epigenetic modifications can be visualized via the genome browser; http://www.niams.nih.gov/Research/Ongoing_Research/Branch_Lab/Molecular_Immunology_and_Inflammation/Supplemental/OShea/KannoImmunity10/
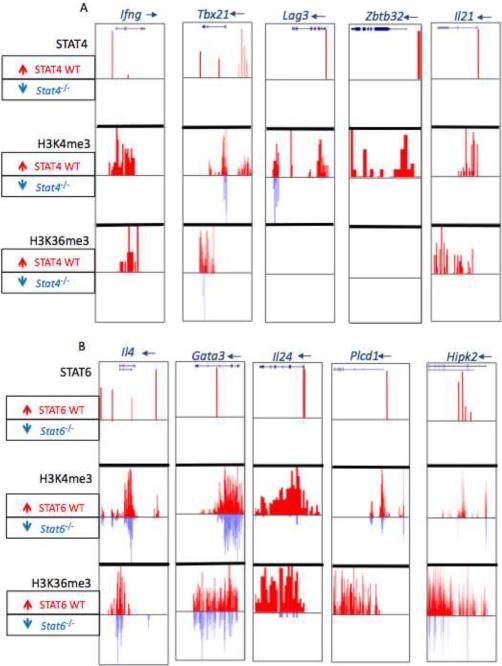
ChIP-seq signal profile maps are shown as a genome browser view. (A) Th1 cell genes, Ifng (chr10:117,875,074-117,885,977), Tbx21 (chr11:96,947,000-96,996,774), Lag3 (chr6:124,848,219-124,863,009), Zbtb32 (chr7:31,374,500-31,385,000), Il21 (chr3:37,119,708-37,136,092), (B) Th2 cell genes, Il4 (chr11:53,418,425-53,444,775), Gata3 (chr2:9,770,098-9,802,379), Il24 (chr1:132,778,117-132,786,123), Plcd1 (chr9:118,980,999-119,011,401) and Hipk2 (chr6:38,641,845-38,850,017). STAT4 and STAT6 binding in wild-type is shown in red upward peaks. Epigenetic marks (H3K4me3, H3K36me3) in wild-type cells are depicted as red upward peaks whereas the accompanying blue colored downward peaks depict the binding in STAT4-deficient cells. Where no signal was detected, the corresponding columns are blank.
Among selected Th1 cell genes shown in Figure 3A, the Ifng is well-known to be regulated by STAT4 (Xu et al., 1996). In addition to STAT4 binding in the promoter, we identified multiple peaks across the extended Ifng locus (Figure S2). Consistent with previous work, (Wilson et al., 2009), permissive H3K4me3 and H3K36me3 modifications were identified throughout the entire Ifng locus and nearly all permissive marks were STAT4-dependent (Figure S2). The Tbx21 gene encodes the key transcription factor T-bet, which is regulated by both IFN-γ-STAT1 and IL-12-STAT4 (Afkarian et al., 2002; Lighvani et al., 2001; Thieu et al., 2008). STAT4 binding localized to an upstream region, which included a previously described enhancer at −13 kb (Yang et al., 2007). Of note, H3K4me3 and H3K36me3 marks throughout the Tbx21 gene were highly STAT4-dependent. We found that STAT4 bound many other Th1 cell-expressed genes and influence their permissive epigenetic marks (see below).
A key feature of lineage commitment is the repression of alternative fates in concert with promotion of the desired lineage. Zbtb32, which encodes repressor of GATA3, is a factor that attenuates Th2 cell responses (Hirahara et al., 2008; Miaw et al., 2000). It was also a STAT4-occupied gene that exhibited STAT4-dependent epigenetic modifications. Thus, STAT4 not only directly regulates Ifng itself, but also regulates transcription factors that promote Th1 cell differentiation and repressors that inhibit Th2 cell differentiation (Figure 3A). Specific STAT4 binding of selected genes was confirmed by ChIP-qPCR (Figure S3, Table S3). By contrast, a number of other genes preferentially expressed in Th1 cells were not directly bound by STAT4 (e.g. Cdh3, Cxcr3, Etv5, and Cebpb) (Rogge et al., 2000).
IL-21-producing follicular helper T cells have recently been recognized as a new subset of T cells; however, their origins are complex (Fazilleau et al., 2009; King et al., 2008). They can arise separately as a lineage, but can also be generated from other polarized subsets. Recently, it has been shown that IL-12 can promote the generation of human Tfh cell (Schmitt et al., 2009). We found that the Il21 gene was a very prominent STAT4-bound gene whose epigenetic regulation was strongly STAT4-dependent (Figure 3A). These data argue that STAT4 is a factor that can regulate Th1 cell differentiation but can also contribute to Tfh cell differentiation.
Previous studies have argued that STAT6 directly contributes to the regulation of the Il4 and Gata3 genes; although limited regions of the genes were analyzed (Ansel et al., 2006; Pai et al., 2003; Zheng and Flavell, 1997). As shown in Figure 3B, STAT6 bound to both the Il4 and Gata3 loci, which were associated with strong permissive epigenetic modifications. Other STAT6-bound genes whose permissive epigenetic modifications showed dependency on STAT6 included: Il24 (Schaefer et al., 2001; Wang and Liang, 2005), Plcd1 and Hipk2 (Chen et al., 2003) (Figure 3B).
Differential patterns of epigenetic regulation by STAT4 and STAT6
Having established that the epigenetic modifications of many Th cell-defining genes are highly STAT-dependent, we sought to evaluate the impact of the cognate STAT on the global epigenetic patterns of occupied genes. To this end, we calculated the total tag counts of epigenetic modifications (K4me3, K27me3 and K36me3) encompassing each occupied gene. Using the K-means algorithm, we clustered STAT-bound genes based on the ratio of tag counts in wild-type and STAT-deficient cells for each of three epigenetic marks (Figure 4, Table 4). Based on this analysis, six epigenetic patterns for STAT4-occupied genes emerged (Figure 4A). For the largest cluster (75% of the total STAT4 bound genes), the absence of STAT4 had minimal effects on epigenetic modifications, suggesting that other factors were the major regulators (denoted as “indeterminate pattern”). Five other clusters of genes were also apparent. We identified clusters with high amounts of H3K4me3 (denoted “K4 high”, 5.7%) and H3K36me3 (“K36 high”, 4.3%) that were clearly STAT4-dependent (Figure 4A). In addition, the third cluster showed STAT-dependent inhibition of H3K27me3 marks (“K27 low”, 7.0%). Analysis of STAT6-occupied genes revealed similar but not identical clusters (Figure 4B). Like STAT4, roughly 25% of STAT-occupied genes showed strong dependency on STAT6 for their distinct epigenetic patterns. However, a discrete cluster of genes in which STAT6 induced high levels of H3K4me3 was not apparent. Although analysis of selected genes showed that STAT6 can positively regulate H3K4me3 (Figure 3B), these genes did not cluster as a discrete group. Rather, the proportion of genes for which STAT6 reduced H3K27me3 was greatly expanded compared to the corresponding cluster of STAT4-occupied genes (11.8% vs. 7.0%).
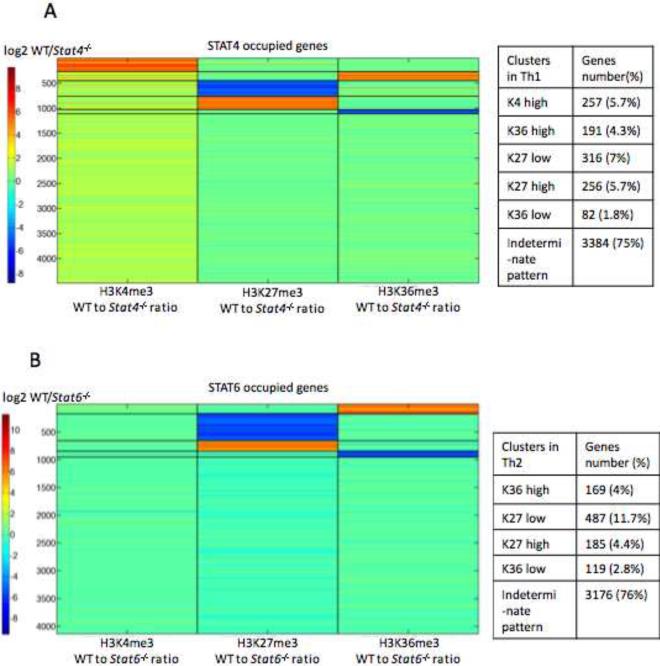
(A, B) The total tag count of each epigenetic modification (H3K4me3, H3K27me3, and H3K36me3) was computed across the body of each STAT-occupied gene in wild-type versus STAT-deficient cells. The ratio of tag counts for three epigenetic modifications was used to cluster STAT-occupied genes by applying the K-means clustering method with squared Euclidean distance with 1000 iterations. (A) STAT4-bound genes in Th1 cells clustered in 6 patterns based on the ratio of wild type vs. STAT4-deficient cells: H3K4me3(K4)-high, H3K36me3 (K36)-high, H3K27me3 (K27)-low, H3K27me3 (K27)-high, H3K36me3 (K36)-low, and an indeterminate pattern. (B) STAT6-bound genes in Th2 cells showed 5 distinct epigenetic patterns similar to A without the K4-high cluster. The accompanying tables list the number and percentage of genes in each cluster.
To confirm the clustering analysis, we plotted the global distribution of epigenetic marks on STAT-occupied genes in wild type and STAT-deficient cells. In the absence of STAT4, H3K4me3 modifications on STAT-bound genes were markedly reduced (Figure 5A upper panels), but as expected from the previous analysis, the absence of STAT6 did not affect the global pattern of H3K4me3 marks on the bound genes. The global distribution of H3K36me3 modifications (Figures 5A and B middle panels) showed minimum changes in the absence of STAT4 or STAT6. In contrast, STAT4 and STAT6 showed notably different influences on H3K27me3 marks. Although deficiency of STAT4 had little effect on the global pattern of H3K27me3 marks, the absence of STAT6 resulted in a significant, global increase of H3K27me3 marks (p= 8E-42, Student T-test) (Figure 5B lower panels). Taken together, our results suggested that STAT4 and STAT6 differentially affected distinct epigenetic modifications of their cognate genes. Specifically, STAT4 primarily promoted H3K4me3 marks whereas STAT6 predominantly acted to antagonize H3K27me3 modifications of its bound genes.
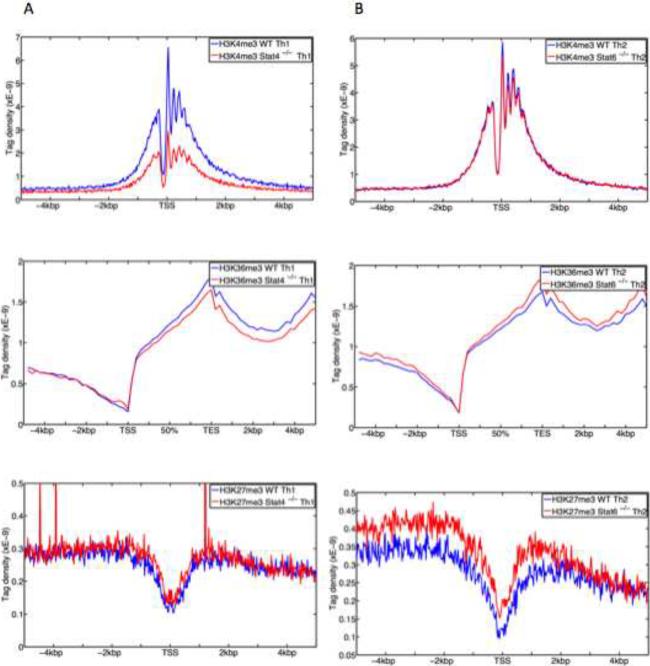
(A, B) Compiled tag density profiles for histone modification marks (H3K4me3, H3K36me3, H3K27me3) were calculated from all STAT4-bound genes in Th1 cells (A) and STAT6-bound genes in Th2 cells (B) in wild type cells (red) and in STAT-deficient cells (blue). The tag density profiles across promoter ± 5 kb flanking regions are shown for H3K4me3 (top panels) and H3K27me3 (bottom panels). The tag density profiles across gene body ± 5 kb flanking regions for H3K36me3 are shown in middle panels.
Correlations between STAT-dependent epigenetic regulation and gene expression
Having identified distinct clusters of STAT-dependent epigenetic modifications, we next assessed how these patterns correlated with regulation of gene expression. Transcriptional profiling was performed in Th1 and Th2 cells (Table S5) to identify STAT-dependent genes (≥2-fold difference in expression between wild type versus STAT-deficient cells). Among them, clusters of genes that exhibited strong STAT-dependent H3K4me3 or K36me3 marks were overwhelmingly positively regulated by STAT in terms of mRNA expression (Figure 6). Genes assigned to these clusters for STAT4 include Ifng, Tbx21, and Il21. Interestingly for STAT6, the gene cluster for which this factor inhibited H3K27me3 marks included the major lineage-specifying genes such as Il4, Gata3, Il24, and Il4ra. The entire lists of genes included in each gene cluster are presented in Table S4 (also see Figure S4 for summary). For both STAT4 and STAT6, STAT-dependent positive epigenetic changes (K4 high, K36 high, K27 low) correlated well with the STAT-dependent positive gene expression (Figure 6).
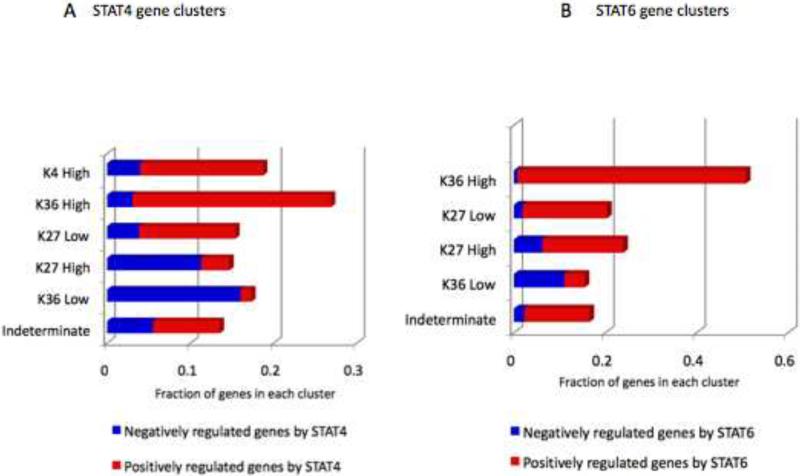
(A, B) STAT-dependent gene expression change was evaluated by microarray for each epigenetic cluster as described in Figure 4. Two-fold change was used as a cut off to sort genes into positively regulated (red bars) and those negatively regulated (blue bars) by STAT4 (A) and by STAT6 (B). Genes whose expression was not affected by STAT4 or STAT6 were not depicted in the graphs.
By comparing STAT4 and STAT6 regulated genes, we were particularly interested in a group of genes bound by both factors and how STATs regulated their function in their cognate lineages. For 5% of the genes bound by both STATs, both factors served to positively regulate these genes in the respective subsets. Examples of these genes include: Il10 (Saraiva et al., 2009), Il7r (Kallies, 2008), Socs3 (Dimitriou et al., 2008), and Id2 (Kusunoki et al., 2003). As shown in Figure 7A, permissive epigenetic marks on those genes, as well as gene expression, were dependent upon the cognate STAT.
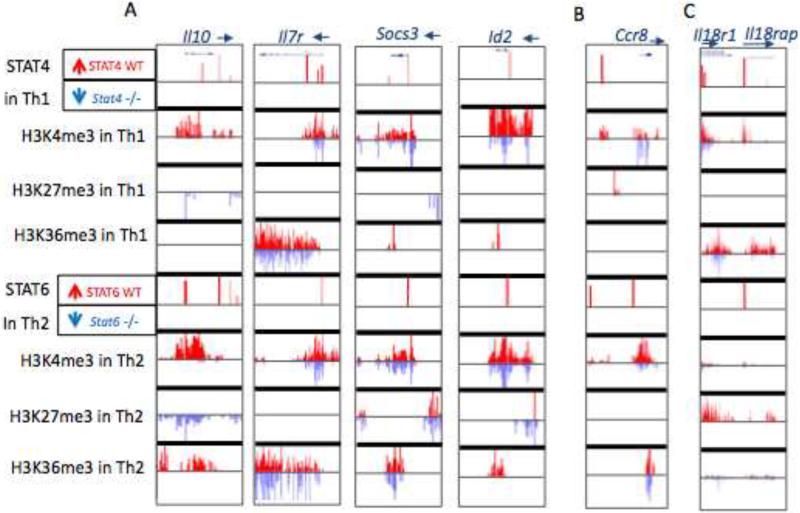
(A) ChIP-seq profiles for binding of STAT4 and STAT6, as well as epigenetic modifications in Th1 and Th2 cells. Illustrative genes include: Il10 (chr1:132,911,300-132,926,671), Il7r (chr15:9,430,427-9,470,839), Socs3 (chr11:117,821,153-117,836,725) and Id2 (chr12:25,769,886-25,787,125). (B, C) ChIP-seq data illustrating STAT binding and epigenetic modifications of (B) Ccr8 (chr 9:119,988,491-120,013,456) and (C) Il18r1-Il18rap (chr1:40,521,609-40,620,416).
Genes regulated by both STAT4 and STAT6: opposing function on key genes
Although ample evidence points to roles of STAT in driving gene expression, there are remarkably few examples in which STAT serve as direct functional repressors. In this regard, we were intrigued by clusters of genes in which STAT apparently inhibited H3K36me3 (K36 low) or increased H3K27me3 (K27 high), both indicative of repressive epigenetic changes (Figure 4). Indeed for the genes included in these clusters, STAT-induced gene inhibition was a more prominent feature than activation, particularly for STAT4 bound genes (Figure 6). One example of such a gene is Ccr8 (Figure 7B). STAT4 binding in the upstream region and adjacent H3K27me3 marks were present in wild type cells, but absent in STAT4-deficient cells. This was of interest as Ccr8 is a Th2 cell-preferentially expressed gene (Gonzalo et al., 2007), which was bound by STAT6. Thus, it appears that STAT4 actively promotes repression of this gene in Th1 conditions. Globally, we identified 265 genes bound and negatively regulated by STAT4, among which 150 (57%) were preferentially expressed in Th2 cell condition (Figure S5A). Therefore, a prominent portion of STAT4-repressed genes was preferentially expressed in Th2 cells. Conversely there were 213 STAT6-bound and negatively regulated genes of which 40% were Th1 cell preferentially expressed (Figure S5B). One such example though, is the region encompassing Il18r1-Il18rap, for which STAT6-dependent H3K27me3 marks were present in Th2 conditions, indicative of STAT6-induced epigenetic repression of the loci (Figure 7C). As such, Il18r1-l18rap is a provocative example for which STAT4 and STAT6 induce opposing epigenetic changes: STAT4 promotes permissive marks whereas STAT6 induce repressive marks (Figure 7C). Therefore, a critical subset of genes exists to which both STAT4 and STAT6 bind, and these factors act in an opposing manner to modulate epigenetic changes and associating gene expression. This divergent action presumably provides insurance of preferential, lineage-specific expression (Figure S4).
DISCUSSION
In this study, we examined the landscape of transcription factor binding of STAT4 and STAT6 through genome-wide approaches and identified the genes uniquely bound by each STAT, as well as those occupied by both factors. Our analysis revealed a core subset of STAT-occupied genes, whose expression and local epigenetic fingerprint are clearly dependent upon the presence of the given STAT. Using this strategy, a large number of genes were identified, many of which were not previously known to be directly regulated by STATs. Consequently, the direct functional roles of STAT4 and STAT6 are considerably broader than previously appreciated. Furthermore, we found that the two STATs differ in their ability to influence global epigenetic profiles. On the level of each bound gene, STATs induce a variety of localized positive or negative epigenetic patterns. However genome-wide, promoting H3K4me3 modifications is a more prominent global feature of STAT4, whereas antagonizing repressive marks (H3K27me3) is a major property of STAT6. Our analysis also uncovered a number of genes for which STAT4 and STAT6 appear to serve as repressors. Among them, a considerable proportion were bound by both factors, and regulated in an opposing manner.
With the ability to measure transcription factor binding on a genome-wide scale, a fundamental issue is the functional relevance of factor binding and how to evaluate it. In this study, we identified over 4000 genes bound by the cognate STAT in Th1 and Th2 conditions. Of these, 15% (684 genes for STAT4) to 29% (1200 genes for STAT6) showed STAT-induced expression changes. An emerging concept coming from genome-scale studies is that only a fraction of factor-bound genes showed clear functional dependence as evaluated by gene expression changes. For example, a recent study enumerating GATA-1 binding showed that ~40% of genes (790 genes out of 1800 bound genes) showed GATA-1 induced expression changes (Yu et al., 2009). Our study produced similar absolute numbers of functionally relevant transcription factor-bound genes when evaluated by microarray-based expression changes.
By adding analysis of STAT-induced epigenetic changes to gene expression changes, we were able to further separate highly relevant STAT binding that substantially impacted both epigenetic signature and gene transcription, from binding events that had minimal functional outcomes. Our analysis revealed subsets of gene clusters whose epigenetic signature was highly STAT dependent. These genes comprise roughly 1000 genes of total 4000 STAT-occupied genes. Our data firmly establish that there is a substantial subset of genes for which the recruitment of STAT during the course of T helper cell differentiation serves to maintain the distinctive epigenetic pattern of the genomic region. For these STAT-regulated genes, it will be of considerable interest to carefully dissect the kinetics of epigenetic pattern formation during the process of lineage specification.
Also of interest was the identity of the STAT-bound genes whose transcriptional and epigenetic regulation was highly dependent on the factor. The genes identified include phenotype-defining cytokines (Ifng, Il4 and Il24), receptors (Il18rap, Il18r1, Lag3, Il4ra), transcription factors (Gata3, Tbx21) and transcriptional repressor (Id2 and Zbtb32). In addition to these recognized regulators of T helper cell differentiation, we identified a number of previously unrecognized STAT-regulated genes with preferential expression in Th1 and Th2 cells. These include genes that encode transcriptional repressors (Ikzf3 [Aiolos] and Nfil3), and diverse signaling molecules that included kinases, phosphatases, G-proteins and adaptors (Hipk2, Plcd1, Gbp2 and Skap2). Clearly, it will be of interest to define the role of these molecules in T cells and assess how their actions relate to the specification of cognate lineages.
Tfh cells represent a recently recognized subset of helper T cells and we were intrigued to see that a major gene expressed by Tfh cells, Il21, was also a STAT4-bound and positively regulated gene. Although previous data has argued that IL-6 acting via STAT3 is the main driver of Tfh cell differentiation (Fazilleau et al., 2009; King et al., 2008), recent data indicates that in human T cells, IL-12 can also promote Tfh cell specification (Saraiva et al., 2009). Our data are entirely consistent with this result and suggest that this mechanism is not unique to human T cells. Interestingly, we also found that STAT4 bound the Bcl6 gene, but unlike IL-21, the expression of the former was modest in Th1 cell polarizing conditions. This would argue that IL-12-STAT4 signals alone are not sufficient to drive Bcl6 expression and presumably other signals are required. The data are also consistent with the evolving notions that Tfh cells represent a flexible subset. In related studies, we found that the Bcl6 and Il21 genes are the targets of STAT3 as well (Durant et al., 2010) and it will be important to differentiate the roles of different STATs in the transcriptional and epigenetic regulation of these important genes.
An unanticipated finding in our studies became apparent when we compared and contrasted the effects of STAT4 and STAT6 on epigenetic modification of their cognate genes. We found that STAT4 primarily promoted accessible epigenetic marks whereas STAT6 had a more prominent role in antagonizing a repressive mark on its cognate genes. One possibility was that the expression of chromatin modifying enzymes differed in Th1 and Th2 cells such that the balance is shifted toward promoting H3K27me3 modification in the absence of STAT6 in Th2 cell condition. The enhancement of H3K27me3 modifications might be explained by over-expression of the H3K27 methyltransferase, Ezh2, and down-regulation of the H3K27 demethylase, Jmjd3. However, this was not the case. We noted decreased expression of Ezh2 associated with a reciprocal increase of Jmjd3 in STAT6-deficient Th2 cells. Therefore, an alteration in the balance between the “writer” (Ezh2) and “eraser” (Jmjd3) (Wang et al., 2007) was evidently not the primary cause of increased H3K27me3 mark associated with the absence of STAT6. On the contrary, these enzymes appear to be compensating for STAT6-dependent alteration of H3K27me3 in Th2 cells. At present, we do not have an explanation for these findings, but clearly additional studies are warranted to dissect the molecular mechanisms underlying the differential regulation of H3K27me3 modifications in Th1 and Th2 cells.
The present study also revealed a large number of genes that are bound and negatively regulated by STAT4 and STAT6. Although the ability to concomitantly promote the expression of some genes and repress others is a common feature of transcription factors associated with lineage commitment (Cobaleda et al., 2007), there is a paucity of circumstances in which STATs have been documented to behave as functional repressors. This was a more prominent feature of STAT4 than STAT6. Clearly, it will be of considerable interest to carefully dissect the molecular basis of the repression of these genes. Particularly interesting were genes bound by both STAT4 and STAT6, for which these factors had opposing functions. In this case, the two STATs appear to function in concert to ensure differential expression of lineage-specifying genes (e.g. Il18r1-l18rap). These will be an interesting subset of genes to analyze in further detail.
In summary, our data provide a platform for understanding how STATs function to modulate epigenetic events to shape gene expression profile unique to T helper subtype. The emerging picture is that STATs bind to a broad, yet clearly defined set of genes and contribute in a very substantial manner to helper T cell differentiation. They do so in both a positive and negative manner. Investigating the function of newly identified genes as they relate to helper cell function and elucidating the mechanisms by which STATs regulate the expression, both positively and negatively, will clearly be important.
METHODS
Mice, isolation of cells and cell culture
C57BL/6J and STAT6-deficient mice were purchased from Jackson Laboratory. STAT4-deficient mice were provided by Dr. Mark Kaplan at Indiana University. Animals were handled and housed in accordance with the guidelines of the NIH Animal Care and User Committee. Splenic and lymph node T cells were obtained by disrupting organs of 8- to 10-week-old mice. All cell cultures were performed in RPMI supplemented with 10% fetal calf serum, 2 mM glutamine, 100 IU/mL penicillin, 0.1 mg/mL streptomycin, and 2 mM β-mercaptoethanol. T cells were enriched using a CD4+ T Cell Kit and AutoMacs isolator (Miltenyi Biotec, Auburn CA). Naive CD4+ T cells were further isolated by flow cytometry, staining with anti-CD4, anti-CD62L, anti-CD44, and anti-CD25 antibodies. Naïve CD4+ T cells were first cultured in the presence of plate-bound anti-CD3 and anti-CD28 (10 μg/mL each), IL-12 (20 ng/mL) and anti-IL-4 (10 μg/mL) for 3 days, followed by IL-2 (50 U/mL) and IL-12 (10 ng/mL) for 4 days (Th1 conditions) or plate bound anti-CD3 and anti-CD28, plus IL-4 (10 ng/mL) and anti-IFN-γ (10 μg/mL) for 3 days, followed by IL-2 (50 U/mL) and IL-4 (10 ng/mL) for 4 days (Th2 conditions). Before harvesting, cells were restimulated with plate-bound anti-CD3 and anti-CD28 and respective cytokines for 2 hours. Cytokines were from R&D Systems (Minneapolis, MN) and antibodies were from BD Phamingen (San Jose, CA).
ChIP-Seq analysis
ChIP-Seq experiments and data processing were performed as described previously (Wei et al., 2009; Zang et al., 2009). Briefly, T cells (2 × 107) were treated with MNase to generate mononucleosomes fraction to analyze histone modifications using following antibodies (anti-H3K4me3, ab8580; anti-H3K36me3, ab9050, Abcam, Cambridge, MA; and anti-H3K27me3, 07-449, Millipore, Billerica MA). For STAT-ChIP, cells were chemically cross-linked and sonicated to generate fractionated genomic DNA and immunoprecipitated with anti-STAT4 (sc486) or anti-STAT6 (sc981, Santa Cruz Biotechnology Inc., Santa Cruz, CA). The DNA fragments were blunt-end ligated to the Illumina adaptors, amplified and sequenced using the Illumina Genome Analyzer II (illumina, San Diego, CA). Sequenced reads of 25 bp were obtained using the Illumina Analysis Pipeline. All reads were mapped to the mouse genome (mm9) and only uniquely matching reads were retained. Unique read numbers for each library are listed in Table S1. Significant peaks (islands) were identified based on window tag-count threshold determined from a p-value of 0.05 (defined by Poisson background model) for histone modifications (Zang et al., 2009). For STAT bindings, CisGenome (Ji et al., 2008) was used to determine significant peaks compared to the negative control of normal rabbit serum IP with the FDR of 1%. A parallel version of MEME (Bailey and Elkan, 1994) was used to perform a de novo search of consensus binding motifs for STAT4 and STAT6.
Quantitative calculation of STAT binding, H3K4me3, H3K27me3 and H3K36me3 levels for all genes
A list of 24,946 unique Refseq genes was obtained from UCSC Genome Table Browser (mm9). A gene is considered to be bound by STAT if at least one peak is found with the region between 10 kb upstream of the transcription start site and the end of the transcript by the two-sample CisGenome analysis (Ji et al., 2008). Epigenetics data were analyzed based on the island approach as described before (Zang et al., 2009). The tag count for each island was first normalized to get a tag count per 1 million of total tag counts in that library.
Profiles of tag density across genes
For each gene, uniquely mapped tags were summarized in 200 bp windows for the regions from 5 kb upstream of the transcription start site (TSS) to the TSS and from the transcription end site (TES) to 5 kb downstream of TES. Within the gene bodies, tags were summarized in windows equal to 4% of the gene length. All window tag counts were normalized by the total number of bases in the windows and the total read number of the given sample.
Microarray data collection and analysis
Total cellular RNA from cells cultured under Th1 and Th2 conditions was extracted with mirVana kit (Applied Biosystems/Ambion, Austin, TX) according to the manufacturer's instructions. Approximately 10 μg of RNA was labeled and hybridized to GeneChip Mouse Genome 430 2.0 arrays (Affymetrix) according to the manufacturer's protocols. Expression values were determined using GeneChip Operating Software (GCOS) v1.1.1 software. All data analysis was performed using Affymetrix Power Tools (APT) software package 1.10.2 (Affymetix, Santa Clara, CA).
ChIP-qPCR
In order to confirm STAT-binding peaks detected by ChIP-seq, PCR primers were designed (Table S3) and qPCR was performed from the chromatin that was prepared similarly as ChIP-seq.
ACKNOWLEDGEMENTS
We thank Drs. Keji Zhao, Jay Bream, and Vittorio Sartorelli for critical reading of the manuscript, Dr. W. Resch for help in data analysis, and J. Simone for technical help with cell sorting. This research was supported by the Intramural Research Programs of NIAMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. [Abstract] [Google Scholar]
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. [Abstract] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. [Abstract] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [Abstract] [Google Scholar]
- Beima KM, Miazgowicz MM, Lewis MD, Yan PS, Huang TH, Weinmann AS. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem. 2006;281:11992–12000. [Abstract] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. [Abstract] [Google Scholar]
- Chen Z, Lund R, Aittokallio T, Kosonen M, Nevalainen O, Lahesmaa R. Identification of novel IL-4/Stat6-regulated genes in T lymphocytes. J Immunol. 2003;171:3627–3635. [Abstract] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. [Abstract] [Google Scholar]
- Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. [Abstract] [Google Scholar]
- Dimitriou ID, Clemenza L, Scotter AJ, Chen G, Guerra FM, Rottapel R. Putting out the fire: coordinated suppression of the innate and adaptive immune systems by SOCS1 and SOCS3 proteins. Immunol Rev. 2008;224:265–283. [Abstract] [Google Scholar]
- Durant L. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010 2010. 10.1016/j.immuni.2010.05.003. [Europe PMC free article] [Abstract] [Google Scholar]
- Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. Follicular helper T cells: lineage and location. Immunity. 2009;30:324–335. [Europe PMC free article] [Abstract] [Google Scholar]
- Gonzalo JA, Qiu Y, Lora JM, Al-Garawi A, Villeval JL, Boyce JA, Martinez AC, Marquez G, Goya I, Hamid Q, et al. Coordinated involvement of mast cells and T cells in allergic mucosal inflammation: critical role of the CC chemokine ligand 1:CCR8 axis. J Immunol. 2007;179:1740–1750. [Abstract] [Google Scholar]
- Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, O'Malley JT, Perumal NB, Kaplan MH. Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming. J Immunol. 2009;183:3839–3847. [Europe PMC free article] [Abstract] [Google Scholar]
- Hirahara K, Yamashita M, Iwamura C, Shinoda K, Hasegawa A, Yoshizawa H, Koseki H, Gejyo F, Nakayama T. Repressor of GATA regulates TH2-driven allergic airway inflammation and airway hyperresponsiveness. J Allergy Clin Immunol. 2008;122:512–520. e511. [Abstract] [Google Scholar]
- Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, Sun YL, Kaplan MH. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. 2003;22:4237–4248. [Europe PMC free article] [Abstract] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. [Europe PMC free article] [Abstract] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. [Europe PMC free article] [Abstract] [Google Scholar]
- Kallies A. Distinct regulation of effector and memory T-cell differentiation. Immunol Cell Biol. 2008;86:325–332. [Abstract] [Google Scholar]
- King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu Rev Immunol. 2008;26:741–766. [Abstract] [Google Scholar]
- Kubo M, Ransom J, Webb D, Hashimoto Y, Tada T, Nakayama T. T-cell subset-specific expression of the IL-4 gene is regulated by a silencer element and STAT6. EMBO J. 1997;16:4007–4020. [Europe PMC free article] [Abstract] [Google Scholar]
- Kusunoki T, Sugai M, Katakai T, Omatsu Y, Iyoda T, Inaba K, Nakahata T, Shimizu A, Yokota Y. TH2 dominance and defective development of a CD8+ dendritic cell subset in Id2-deficient mice. J Allergy Clin Immunol. 2003;111:136–142. [Abstract] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. [Abstract] [Google Scholar]
- Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. EMBO J. 2007;26:1292–1302. [Europe PMC free article] [Abstract] [Google Scholar]
- Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. [Abstract] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O'Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–15142. [Europe PMC free article] [Abstract] [Google Scholar]
- Lund RJ, Chen Z, Scheinin J, Lahesmaa R. Early target genes of IL-12 and STAT4 signaling in th cells. J Immunol. 2004;172:6775–6782. [Abstract] [Google Scholar]
- Lund RJ, Loytomaki M, Naumanen T, Dixon C, Chen Z, Ahlfors H, Tuomela S, Tahvanainen J, Scheinin J, Henttinen T, et al. Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. J Immunol. 2007;178:3648–3660. [Abstract] [Google Scholar]
- Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–333. [Abstract] [Google Scholar]
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. [Abstract] [Google Scholar]
- O'Sullivan A, Chang HC, Yu Q, Kaplan MH. STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J Biol Chem. 2004;279:7339–7345. [Abstract] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. [Abstract] [Google Scholar]
- Pesu M, Muul L, Kanno Y, O'Shea JJ. Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood. 2006;108:983–985. [Europe PMC free article] [Abstract] [Google Scholar]
- Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33–36. [Abstract] [Google Scholar]
- Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–1917. [Europe PMC free article] [Abstract] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. [Abstract] [Google Scholar]
- Rogge L, Bianchi E, Biffi M, Bono E, Chang SY, Alexander H, Santini C, Ferrari G, Sinigaglia L, Seiler M, et al. Transcript imaging of the development of human T helper cells using oligonucleotide arrays. Nat Genet. 2000;25:96–101. [Abstract] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. [Abstract] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. [Abstract] [Google Scholar]
- Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O'Garra A. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. [Europe PMC free article] [Abstract] [Google Scholar]
- Schaefer G, Venkataraman C, Schindler U. Cutting edge: FISP (IL-4-induced secreted protein), a novel cytokine-like molecule secreted by Th2 cells. J Immunol. 2001;166:5859–5863. [Abstract] [Google Scholar]
- Schindler C, Darnell JE., Jr. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. [Abstract] [Google Scholar]
- Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, Ueno H. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31:158–169. [Europe PMC free article] [Abstract] [Google Scholar]
- Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–690. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. [Abstract] [Google Scholar]
- Wang M, Liang P. Interleukin-24 and its receptors. Immunology. 2005;114:166–170. [Abstract] [Google Scholar]
- Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. [Abstract] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. [Abstract] [Google Scholar]
- Wirnsberger G, Hebenstreit D, Posselt G, Horejs-Hoeck J, Duschl A. IL-4 induces expression of TARC/CCL17 via two STAT6 binding sites. Eur J Immunol. 2006;36:1882–1891. [Europe PMC free article] [Abstract] [Google Scholar]
- Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–440. [Europe PMC free article] [Abstract] [Google Scholar]
- Wurster AL, Tanaka T, Grusby MJ. The biology of Stat4 and Stat6. Oncogene. 2000;19:2577–2584. [Abstract] [Google Scholar]
- Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. [Abstract] [Google Scholar]
- Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood. 2007;110:2494–2500. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu Q, Thieu VT, Kaplan MH. Stat4 limits DNA methyltransferase recruitment and DNA methylation of the IL-18Ralpha gene during Th1 differentiation. EMBO J. 2007;26:2052–2060. [Europe PMC free article] [Abstract] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med. 2006;203:1493–1505. [Europe PMC free article] [Abstract] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. [Abstract] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. [Abstract] [Google Scholar]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. [Abstract] [Google Scholar]
- Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.immuni.2010.06.003
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1074761310002062/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.immuni.2010.06.003
Article citations
BACH2 regulates diversification of regulatory and proinflammatory chromatin states in TH17 cells.
Nat Immunol, 25(8):1395-1410, 15 Jul 2024
Cited by: 0 articles | PMID: 39009838
A distal enhancer of GATA3 regulates Th2 differentiation and allergic inflammation.
Proc Natl Acad Sci U S A, 121(27):e2320727121, 26 Jun 2024
Cited by: 0 articles | PMID: 38923989 | PMCID: PMC11228505
Serum level of interleukin-24 and its polymorphism in eczematic Iraqi patients.
Medicine (Baltimore), 103(25):e38635, 01 Jun 2024
Cited by: 0 articles | PMID: 38905384 | PMCID: PMC11191866
Regulation of T helper cell differentiation by the interplay between histone modification and chromatin interaction.
Immunity, 57(5):987-1004.e5, 12 Apr 2024
Cited by: 1 article | PMID: 38614090
Deciphering the developmental trajectory of tissue-resident Foxp3+ regulatory T cells.
Front Immunol, 15:1331846, 28 Mar 2024
Cited by: 0 articles | PMID: 38605970 | PMCID: PMC11007185
Review Free full text in Europe PMC
Go to all (210) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The biology of Stat4 and Stat6.
Oncogene, 19(21):2577-2584, 01 May 2000
Cited by: 191 articles | PMID: 10851056
Review
STAT4/6-dependent differential regulation of chemokine receptors.
Clin Immunol, 118(2-3):250-257, 18 Jan 2006
Cited by: 17 articles | PMID: 16413227
Expansion of T follicular helper-T helper 1 like cells through epigenetic regulation by signal transducer and activator of transcription factors.
Ann Rheum Dis, 77(9):1354-1361, 31 May 2018
Cited by: 35 articles | PMID: 29853448
STAT4 and STAT6, their role in cellular and humoral immunity and in diverse human diseases.
Int Rev Immunol, 43(6):394-418, 26 Aug 2024
Cited by: 0 articles | PMID: 39188021
Review
Funding
Funders who supported this work.
Intramural NIH HHS (1)
Grant ID: Z99 AR999999





