Abstract
Free full text

Genome-wide identification of mouse congenital heart disease loci
Abstract
Empirical evidence supporting a genetic basis for the etiology of congenital heart disease (CHD) is limited and few disease-causing mutations have been identified. To identify novel CHD genes, we performed a forward genetic screen to identify mutant mouse lines with heritable CHD. Lines with recessive N-ethyl-N-nitrsourea-induced CHD-causing mutations were identified using a three-generation backcross. A hierarchical screening protocol was used to test the hypothesis that the fetal-to-neonatal circulatory transition unmasks the specific structural heart defects observed in CHD. Mice with heart defects were efficiently ascertained by selecting for pups exhibiting perinatal lethality and characterizing their cardiac pathology. A marked increase of perinatal lethality was observed in the mutagen-treated cohort compared with an untreated backcross population. Cardiac pathology on perinatal lethals revealed cardiovascular defects in 79 pups from 47 of 321 mutagenized lines. All identified structural abnormalities were analogous to previously described forms of human CHD. Furthermore, the phenotypic recurrence and variance patterns across all lines were similar to human CHD prevalence and recurrence patterns. We mapped the locus responsible for heritable atrioventricular septal defects in six lines (avc1-6). Our screen demonstrated that ‘sporadic’ CHD may have major genetic component and established a practical, efficient approach for identifying CHD candidate genes.
INTRODUCTION
Congenital heart disease (CHD) represents the most common type of human birth defect and the most common non-infectious cause of death in children. Predictions of the incidence of CHD range from 1–5% of human live births (1–3). Knowledge of the developmental etiology of cardiovascular malformations will influence the prevention, risk ascertainment and physiological treatment of CHD. Although CHD mortality has decreased with the implementation of modern diagnostic and therapeutic management of affected children, little is known about the etiology of CHD (4,5). It is largely accepted that CHD has a genetic contribution, informed by observations that the incidence of CHD among siblings and offspring of affected individuals is higher than that found in the general population (6,7). However, the identification of causative mutations has proven difficult, and the degree to which CHD is genetically determined remains elusive.
Several factors have made elucidating the genetic basis of human CHD difficult. Until recent advances in surgical repair and palliation, most significant cardiac malformations have been lethal or have produced significant morbidity (4,5,8). Large pedigrees with segregation of CHD are uncommon, presumably due to negative selection pressure of reduced fecundity on disease-causing alleles. Subsequently, human Mendelian genetics has been a limited source for the identification of CHD disease genes; although a few genes have been identified using Mendelian genetics in human non-syndromic CHD in the last decade, the total number remains very small (9–14). Not surprisingly, the cardiac defects observed in these families represent the mildest forms of CHD (10,13). Furthermore, a priori knowledge of a role for the candidate gene in cardiac development preceded the identification of disease-causing mutations in most cases (15,16). Evidence to date indicates that the genes currently implicated can explain only a small minority of human CHD (17,18).
The inadequacy of present efforts to discover the more comprehensive genetic basis of human CHD informed our decision to conduct a forward genetic screen in mice. Previous effective strategies used to detect heart mutations in mice employed N-ethyl-N-nitrsourea (ENU) mutagenesis in association with fetal ultrasound (19,20). In contrast, we focused on the transition of fetal-to-neonatal circulation during perinatal life, assessing pathologic anatomy as the initial screening strategy to ascertain heart defects in the offspring of ENU-mutagenized mice.
Although numerous types of structural heart defects have been described, we noted that the specific pathologic anatomy of human CHD abnormalities fit into one of three categories, each capable of supporting fetal circulation, but not of supporting neonatal circulation: (i) septation defects (e.g. atrial septal defects, ventricular septal defects, patent ductus arteriosus, atrioventricular canal or truncus arteriosus), (ii) unilateral blocks (e.g. valvar stenosis or atresia of the tricuspid, mitral, pulmonary or aortic valve, or Ebstein's anomaly) and (iii) routing abnormalities (e.g. transposition of the great arteries, double-inlet RV or LV, or double-outlet RV or LV). Given that structural alterations occur in the cardiovascular system during the transition from fetal to neonatal circulation within a defined perinatal temporal window (Fig. 1) (21), we hypothesized that the perinatal circulatory transition unmasked the defects observed in more severe forms of CHD. A corollary to this hypothesis suggests that perinatal lethality could be used as a potential predictor for CHD. Specifically, we hypothesized that by using failure-to-survive-the-perinatal-physiologic transition as an initial selection in mice, we would delineate mutant animals with a higher probability of having CHD.
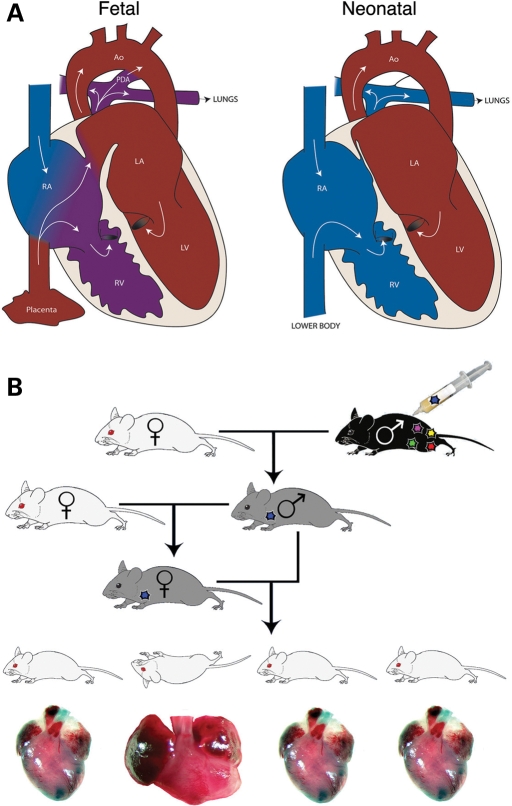
Forward genetic screen is based on the mammalian circulatory transition which occurs at birth. (A) Schematic depictions of fetal (L) and postnatal (R) cardiac structure and circulation. During fetal life, shunts normally exist within the cardiovascular system between the atria (the foramen ovale) and the great vessels (the ductus arteriosus). Shortly after birth, the foramen ovale and ductus arteriosus close, separating venous and systemic circulation. (B) ENU mutagenesis scheme; G0 males are ENU-treated, and G2 females are backcrossed to G1 males, allowing the effect of recessive mutations to be screened in G3 litters. Recessive mutants displaying perinatal lethality were then screened for congenital heart defects.
We carried out an ENU mutagenesis screen in mice to identify mutants with recessive CHD utilizing this rationale. We report that the incidence of recessive perinatal lethality in ENU mutagenesis screens is five times greater than background. This approach enabled us to ascertain a small fraction of all animals screened as candidates for detailed anatomic analysis and facilitated the use of serial section cardiac histology for definitive phenotypic evaluation of the heart during primary screening. Our results demonstrated that screening for perinatal lethality effectively selected for animals with a high probability of CHD: 15% of the lines with perinatal lethality demonstrated structural heart defects. Using this screening strategy, we identified 12 lines of inherited cardiovascular defects, including 6 with atrioventricular septal defects (AVSDs). We mapped the six loci that cause AVSDs to distinct genomic locations. These lines will provide a valuable resource for understanding the mechanisms of atrioventricular septation and may serve as candidate genes for human AVSDs.
RESULTS
ENU mutagenesis-induced recessive perinatal lethality
A three-generation breeding scheme was used to allow efficient identification of recessive phenotypes (Fig. 1) (22). A two-generation outcross between mutagenized C57BL/6J and wild-type FVB/N was used in the breeding scheme to facilitate the mapping of ENU-induced mutant alleles (Fig. 1) (22). Therefore, variable genetic background was a possible confounding factor in our screen. We addressed this issue empirically by performing a control genetic screen to determine underlying rates of perinatal lethality and CHD in a mixed C57BL/6J:FVB/N population. We screened three-generation outcross litters from control untreated and test mutagen-treated lines and compared the rates of recovery of perinatal lethal lines between them to assess the true ENU induction rate of parental lethality. We subsequently evaluated 32 mutagenized lines derived from six independently ENU-treated founder males for heritable perinatal lethality.
No heritable perinatal lethality was observed in control matings from independent pedigrees. In contrast, we identified six lines demonstrating heritable perinatal lethality in the mutagenized cohort. We concluded that ENU treatment effectively induced heritable perinatal lethality. Control matings generated four perinatal deaths that were evaluated for CHD. Normal heart structure was observed in each of the four by serial histology (data not shown). To determine whether physiologically silent CHD might exist within controls, serial histology of the hearts of 242 control pups (at least 10 per line) was performed; all were normal (data not shown). Based on these data, the estimated background CHD rate in the control outcrosses was 0%.
Identification of lines with CHD
Mice from three distinct ENU mutagenesis screen cohorts were evaluated for structural heart defects: (i) Brigham and Women's Hospital (BWH); (ii) The Jackson Laboratory; and (iii) Baylor College of Medicine. From each cohort, animals that died in the perinatal period (defined as the first 5 days of life) were evaluated by cardiac pathology to ascertain CHD. From the BWH cohort, serial cardiac histology from 68 deceased animals revealed 12 with structural heart defects, or 18% of the total number that died perinatally (Table 1). Structural abnormalities observed included septal defects [atrial septal defect (ASD) (3), ventricular septal defects (VSD) (2), AVSDs (5) and routing abnormalities (double-outlet right ventricle: 1)], all analogues of human congenital heart defects (Table 1). Four of the 32 lines had multiple affected progeny. One such line demonstrated inherited AVSDs. A second line demonstrated multiple affected pups with atrial septal defects, ventricular septal defects, or both. Additional affected animals from the other two lines with affected mice were sought from four sequential litters each; however, none was identified. These results demonstrated that perinatal lethality was an effective pre-screen for ENU-induced CHD.
Table 1.
ENU mutagenesis increased the rate of perinatal lethality and generated 12 lines of heritable cardiovascular defects
| Analysis of CHD by screena | Control screen | Mutagenized screen |
|---|---|---|
| Total lethals evaluated for CHD | 4 | 441 |
| Total evaluated for CHD | 246 | 441 |
| Number with CHD | 0 | 79 |
| CHD rate per lethal | 0% | 17.9% |
| CHD rate per liveborn | 0% | 1.3% |
| Total lines evaluated | 321 | |
| Lines with CHD | 47 | |
| CHD rate per line | 14.6% | |
| Analysis of CHD by classb | Per line | |
| Total affected | 47 | |
| Septation defects | 41 | |
 Atrial septal defect, secundum Atrial septal defect, secundum | 17 | |
 Ventricular septal defects Ventricular septal defects | 22 | |
  Membranous Membranous | 7 | |
  Muscular Muscular | 9 | |
  Conus Conus | 2 | |
 AVSDs AVSDs | 6 | |
  Atrial septal defect, primum Atrial septal defect, primum | 2 | |
  Complete common AV canal Complete common AV canal | 4 | |
| Unilateral blocks | 4 | |
 Pulmonary stenosis Pulmonary stenosis | 2 | |
 Tricuspid stenosis Tricuspid stenosis | 2 | |
| Routing abnormalities | 6 | |
 Interrupted aortic arch Interrupted aortic arch | 3 | |
 Truncus ateriosus Truncus ateriosus | 2 | |
 Double-outlet right ventricle Double-outlet right ventricle | 1 | |
aA control screen was performed simultaneously to determine the baseline rates of perinatal lethality and CHD (left column). No defects were observed in the control screen. Simultaneously, a mutagenesis screen was performed to ascertain the rates of induction of perinatal lethality and CHD caused by ENU treatment (right column). CHD was observed in animals demonstrating perinatal lethality in 47 of 321 mutagenized lines (14.6%).
bDistribution of CHD by class from mutagenized lines. Three classes of congenital heart defects (septation defects, unilateral blocks and routing abnormalities) were observed, corresponding to the types of defects found in human CHD. From 47 affected lines, 41 demonstrated defects in septation, 4 demonstrated unilateral blocks (stenotic lesions) and 6 demonstrated routing abnormalities of the great arteries.
Our screening for CHD was expanded via participation in two ongoing large-scale ENU screens, The Jackson Laboratory National Heart Lung and Blood Institute ENU Mutagenesis Program (23) and the Justice Laboratory at Baylor College of Medicine (24,25). Each utilized a three-generation backcross strategy to identify recessive alleles. From the Jackson Laboratory–National Heart Lung and Blood Institute ENU Mutagenesis Program, we studied 366 perinatal lethals from 282 independent lines. Based upon serial section histology, 66 had CHD structural heart abnormalities (18%). The Justice Laboratory screen utilized a chromosome 11 balancer, a strategy that permitted mutation mapping concomitant with phenotypic analysis (24,25). We evaluated the cardiac anatomy of mutant mice with perinatal lethality from seven independent lines. A heart defect was identified in mutant animals from one line, L11Jus16. The hearts of mutant mice from this line demonstrated abnormal (L) looping and AVSDs.
Each line that generated perinatal lethal mice with CHD (n = 47) was tested for heritability. G2 female parents of affected pups were mated to G1 males, and 50 or more liveborn G3 pups were screened. Ten lines with greater than one mouse affected with CHD were identified. We suggest that these 10 lines carry CHD mutations.
Taken together, examination of three independent cohorts of ENU-mutagenized mice yielded comparable rates of perinatal lethality (~18%) and produced 79 CHD lesions (Table 1). Affected mice were ascertained in 47 of 321 (14.6%) mutagenized lines (CHD lines). Septal defects (n = 67) were the most common CHD class ascertained, accounting for 85% of malformations and observed in 41 of 47 CHD lines (87%) (Table 1). Ventricular septal defects (n = 22), atrial septal defects (secundum type, n = 17) and AVSDs (n = 6) accounted for these prevalent CHD. Routing abnormalities, including interrupted aortic arch type B, truncus ateriosus and double-outlet right ventricle (n = 8), occurred in six lines (13%). Unilateral blocks including pulmonary stenosis and tricuspid stenosis occurred in four lines (9%). We estimate an overall CHD rate of 1.3% of liveborn mice from the mutagenized cohorts (7.5% perinatal lethality rate × 17.9% of lethals affected with CHD).
Genomic mapping of AVSD loci
We focused on AVSDs because these defects are a common form of human CHD, have a high morbidity and have an increased recurrence rate than other forms of human CHD (6). Six lines with inherited mutations causing AVSDs were mapped to distinct loci on five mouse chromosomes.
avc1
Gross evaluation of multiple deceased pups from line 16 of the initial mutagenized cohort revealed severe biatrial and biventricular chamber enlargement (6/6) (data not shown). Serial histology revealed complete AVSD in each mutant heart (Fig. 2). In addition, preaxial polydactyly was present bilaterally on both fore and hind limbs (Fig. 2). This mutation was mapped to proximal chromosome 5 and named atrioventricular canal1 (avc1) (Fig. 2). Identification of recombinant chromosomes in affected animals from an intercross between backcross carriers narrowed the interval containing avc1 to 0.9 megabases (Mb), between D5Mit229 and D5Mit420 (Fig. 2).
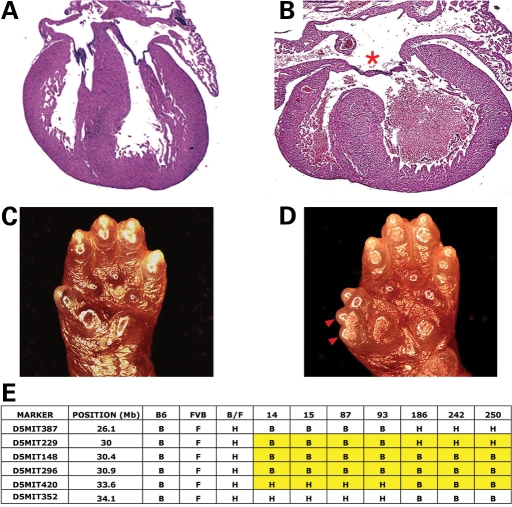
Avc1 caused AVSDs and polydactyly and mapped to chromosome 5. (A and B) Morphology of control litter mate (A) and avc1 (B) mutant newborn mouse hearts. Avc1 hearts demonstrated highly penetrant AVSDs (red asterisk). (C and D) Morphology of control litter mate (C) and avc1 mutant mouse (D) upper limbs. avc1 upper limbs demonstrated penetrant preaxial polydactyly (red arrowhead). (E) Avc1 maps to proximal chromosome 5. Recombinant chromosomes in affected animals refined the interval containing avc1 to 3.6 Mb between D5Mit229 and D5Mit420.
avc2
Perinatal lethal pups from The Jackson Laboratory line G2-02-158 each displayed an incomplete AVSD (an atrial septal defect of the primum type) and preaxial polydactyly of all limbs (Fig. 3). The causative mutation was mapped to the proximal 43.2 Mb region of mouse chromosome 15 and named atrioventricular canal 2 (avc2) (Fig. 3). The phenotype in this line was incompletely penetrant and lost after repeated outcrossing to FVB. However, the phenotype was re-established after a two-generation backcross to a C57BL/6-A/J chromosome 15 chromosome substitution strain (C57BL/6J-Chr15A/NaJ). The molecular region of interest containing avc2 was narrowed to a 24.2 Mb region at the proximal end of mouse chromosomes 15 using recombinant chromosomes identified in phenotypically affected animals (Fig. 3). The gene-poor region of interest contained less than 70 genes, none previously implicated in cardiovascular development.
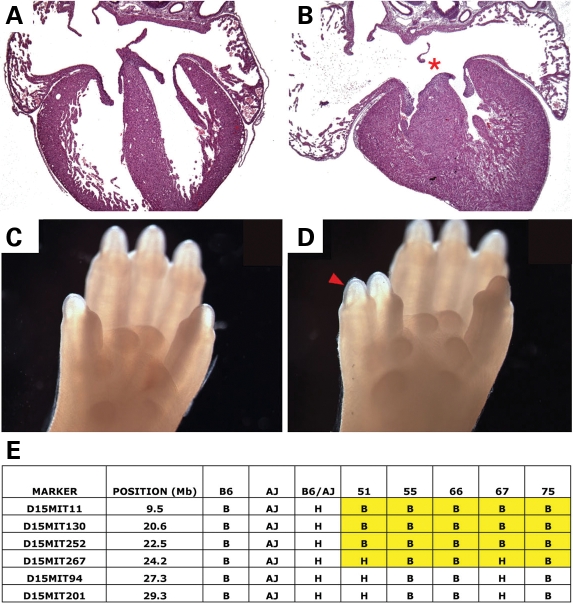
Avc2 caused AVSDs and polydactyly and mapped to chromosome 15. (A and B) Morphology of control litter mate (A) and avc2 (B) mutant newborn mouse hearts. Avc2 hearts demonstrated highly penetrant AVSDs (B, red asterisk). (C and D) Morphology of control litter mate (C) and avc2 mutant mouse (D) upper limbs. avc2 upper limbs demonstrated penetrant preaxial polydactyly (red arrowhead). (E) Avc2 maps to proximal chromosome 15. Recombinant chromosomes in affected animals refined the interval containing avc2 to 24.2 Mb proximal to D15Mit267.
avc3
Perinatal lethal pups from The Jackson Laboratory line G2-02-419 demonstrated AVSDs of the complete common atrioventricular canal (CCAVC) type and diminished compact myocardium (Fig. 4). Using DNA from affected neonatal lethal pups, the mutation was mapped to mouse chromosome 9 and named atrioventricular canal 3 (avc3). Evaluation of embryonic specimens revealed complete phenotypic penetrance at E14.0 with 30% embryonic lethality (6/30). Recombinant chromosomes in phenotypically affected mutants from an intercross of backcross carriers narrowed the region of interest containing avc3 to 5.2 Mb, between D9Mit106 and D9Mit260 (Fig. 4). This interval contains the gene Smad6, and Smad6 null-mutant mice have cardiac valve abnormalities (26). We sequenced Smad6 exons from mutant embryos and found no non-synonymous or splice-site polymorphisms.
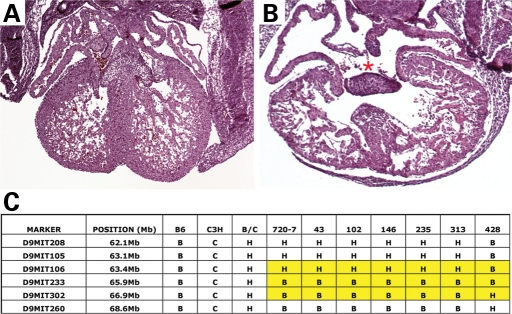
Avc3 caused AVSDs and mapped to chromosome 9. (A and B) Morphology of control litter mate (A) and avc3 mutant E17.5 mouse hearts. Avc3 hearts demonstrated highly penetrant AVSDs (B, red asterisk). (C) Avc3 maps to proximal chromosome 9. Recombinant chromosomes in affected animals refined the interval containing avc3 to 6.5 Mb between D9Mit106 and D9Mit302.
avc4
Perinatal lethal pups from The Jackson Laboratory line G2-03-133 demonstrated AVSD, CCAVC type (Fig. 5). The causative mutation was mapped to the distal end of mouse chromosome 12 and named atrioventricular canal 4 (avc4). Further investigation of animals homozygous for this chromosomal region revealed complete penetrance with variable expressivity. All mutants demonstrated AVSDs, varying from primum atrial septal defect to complete AVSD. In approximately half of affected mutants, the AVSD demonstrated an unbalanced hypoplastic right ventricle (Fig. 5). In addition, rare defects of the great vessels, including interrupted aortic arch (3/25) and hypoplastic pulmonary artery (2/25), were observed (Fig. 5). Recombinant chromosomes were identified in 14 affected animals from an intercross between backcross carriers and narrowed region containing avc4 to 11.5 Mb distal to D12Mit181 (Fig. 5).
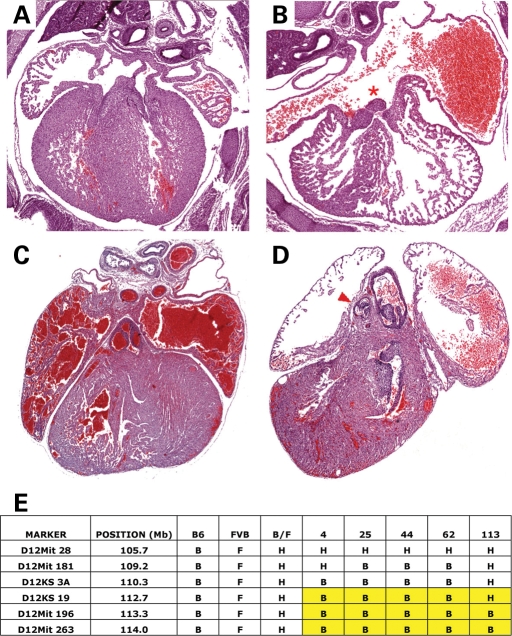
Avc4 caused AVSDs and mapped to chromosome 12. (A–D) Morphology of control litter mate (A and C) and avc4 (B and D) mutant newborn mouse hearts. Avc4 hearts demonstrated highly penetrant AVSDs (B, red asterisk) and pulmonary stenosis (D, red arrowhead). (E) Avc4 maps to distal chromosome 12. Recombinant chromosomes in affected animals refined the interval containing avc4 distal to D12KS19.
avc5
Mice with perinatal lethality from Line l11Jus16 of the Baylor College of Medicine/Justice Laboratory chromosome 11 mutagenesis screen demonstrated AVSDs as well as cardiac looping abnormalities. The causative mutation was named avc5 (Fig. 6). Analysis of genetically affected mutants demonstrated complete penetrance with variable expressivity. Observed cardiac situs abnormalities included L-loop ventricles and situs inversus. Cardiac structural defects included AVSD with levocardia (3/17), AVSD with dextrocardia (2/17), double-outlet right ventricle (3/17), transposition of the great arteries (2/17) and ventricular septal defect, muscular (1/17). The constellation of observed defects suggested that L11Jus16 represents a mouse model for heterotaxy syndrome. Initially mapped to the gene-rich region of chromosome 11 under the balancer region, the region containing avc5 has been narrowed using 32 affected animals with recombinant chromosomes to a 6.1 Mb region between D11Mit320 and D11Mit245.
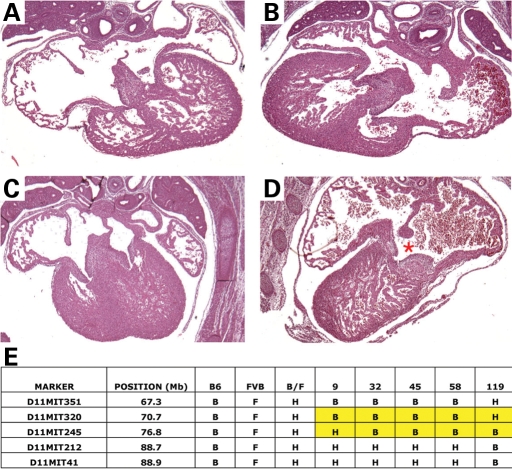
Avc5 caused AVSDs and mapped to chromosome 11. (A–D) Morphology of control litter mate (A and C) and avc5 (B and D) mutant newborn mouse hearts. Avc5 hearts demonstrated highly penetrant L-hand looping (B and D) and AVSDs (D, red asterisk). (E) Avc5 maps to chromosome 11. Recombinant chromosomes in affected animals refined the interval containing avc5 to 6.1 Mb between D11Mit320 and D11Mit245.
avc6
Deceased pups from line 44 of the initial mutagenized cohort revealed AVSD, CCAVC type (Fig. 7). The causative mutation was mapped to chromosome 11 and named avc6. Further investigation of genotyped animals revealed complete penetrance with variable expressivity. Isolated complete AVSD or AVSD with posterior malaligned ventricular septal defects consistent with Tetralogy of Fallot were identified (Fig. 7). Utilizing recombinant chromosomes from affected mutants, the region containing avc6 has been narrowed to a 32.5 Mb interval of mouse chromosome 11 between D11Mit320 and D11Mit360.

Avc6 caused AVSDs and mapped to chromosome 11. (A–D). Morphology of control litter mate (A and C) and avc6 mutant (B and D) newborn mouse hearts. Avc6 hearts demonstrated highly penetrant AVSDs (red asterisk). (E) Avc6 maps to proximal chromosome 11. Recombinant chromosomes in affected animals refined the interval containing avc6 to 26.5 Mb between D11Mit245 and D11Mit360.
The identification of the map position of these mutations allows us to identify obligate heterozygous mice by analysis of flanking markers. We performed gross cardiac pathology and detailed serial histology for three heterozygotes from each line discussed earlier. No defects were observed in any of the mice.
DISCUSSION
Using a forward genetic screen in mice, we ascertained heritable CHD malformations transmitted as Mendelian traits. Using perinatal lethality as a predictor of CHD, we performed efficient anatomical analysis by serial section cardiac histopathology. We conducted a parallel assessment of a comparable population of unmutagenized control mice, and demonstrated no underlying CHD. In contrast, 15% of lines from the mutagen-treated background demonstrated CHD, and 26% of affected lines demonstrated inherited CHD. To date, mutations from six lines with inherited AVSDs have been mapped. Given the paucity of genetic etiologies for AVSD in humans (27), we suggest that the identification of mutations at these loci will expand our understanding of the genetic etiology and developmental ontogeny of AVSDs.
Epidemiology of ENU-induced mouse CHD
Notably, most defects ascertained in our screens comprised septal defects. This finding is consistent with recent human prevalence data showing that the overwhelming majority of CHD in children and adults are septal defects (28). Although many human septal defects do not threaten survival, our selection of perinatal mortality may have defined mutations that alter a distinct subset of genes in cardiac septation. Alternatively, mouse cardiopulmonary physiology may be less tolerant of cyanotic challenges than human physiology. Regardless, the prevalence of different types of septal defects closely corresponded to human prevalence data. As observed in human CHD cohorts, in the mouse screens, VSDs were ascertained more commonly than ASDs, and both were more common than AVSDs. Furthermore, it has been noted that human AVSDs are more likely to be associated with a syndrome than other forms of CHD (reviewed in 29). Consistent with this observation in humans, three of six heritable mouse lines with AVSD defects (avc1, avc2, avc5) had malformations of other organ systems.
The AVSDs identified in these mouse screens modeled patterns of AVSD presentation in humans. AVSDs are among the most common of the severe CHD lesions in children and adults (28). Patients with AVSDs have an increased recurrence rate of CHD among their offspring compared with patients with other types of CHD (6). Furthermore, AVSD is the most reproducible CHD phenotype in familial non-syndromic CHD; phenotypic concurrence between affected family members is much higher for AVSD than for all other types of CHD (7,30). We observed similar phenotypic expressivity patterns among mutant mouse lines. In avc1-6, AVSDs were uniformly expressed, facilitating the mapping of the disease-causing alleles. In contrast, heritable CHD observed in other lines consistently included more than one defect, suggesting that the mutations causing defects other than AVSDs were more variably expressed. Although AVSDs were present in only 13% of affected lines, AVSDs represented the dominant phenotype in 50% of lines with heritable CHD. We believe that the mapped mutations identified in our screen to date, avc1-6, serve as appropriate mouse models of human AVSD and provide important resources for our detailed investigation of potentially novel genes in atrioventricular septation.
None of the heritable mutations we have discovered has observable effects as heterozygotes. This is not surprising; many developmental loci that are disease causing in humans when haploinsufficient do not have comparable deleterious consequences when heterozygous in mouse models. For example, mutations in GATA4 result in dominant disease in humans (10) but are recessive in mice (31).
ENU-induced sporatic mouse CHD
Our results also have implications for the etiology of sporadic CHD. It has been assumed that human CHD is multifactorial in origin with an empirically undetermined genetic contribution. All of the structural defects ascertained in our screen, both heritable and non-heritable, appear to have a genetic etiology, including the 12 lines with multiple affected animals and the other lines with sporadic CHD. This conclusion is based on the marked differences in rates of CHD in lines derived from mutagenized founders versus non-mutagenized controls. Among screens of control mice, no sporadic CHD cases were observed, although screens revealed that 18% of perinatal lethal pups in mutagenized lines had CHD. The majority of mutagenized lines had sporadic CHD affecting only one affected mouse. Because no structural heart defects were observed in our control screen and because the only difference between these two screens was the presence or absence of ENU treatment of founder males three generations preceding anatomical cardiac screening, we conclude that these events reflect a genetic etiology.
There are a number of possible explanations for the sporadic appearance of many of the genetic lesions in these studies, most of which are relevant to human disease ascertainment. ENU treatment causes single-base changes, a high proportion of which are missense or splice-site mutations (32). These can generate hypomorphic alleles (rather than null mutations), which may have stochastic effects on developmental processes, and consequently demonstrate variable expressivity or incomplete penetrance. In addition, two of the three ENU screens we analyzed were done in mixed genetic backgrounds, in which modifying loci affecting cardiac development may be segregating. Finally, given the mutational load in ENU-treated mice (one sequence change per 1–2 Mb (33), it is possible that some defects are multigenic in origin.
Although the mouse models for CHD described here are complex, they very closely mimic the anatomy and occurrence patterns observed in human CHD. The majority of human CHD cases are also sporadic rather than heritable (3). Our results provide support for a genetic basis for sporadic CHD. These observations have important implications for how the causality of these congenital defects should be investigated. Specifically, the sporadic nature of many congenital defects has led to the proposition that somatically acquired copy-number variation is likely to be a common basis of disease, which can be assessed using methods such as comparative genome hybridization (34). Our results indicate that single-base changes are also likely to be causal, even in apparently sporadic disease, and suggest that whole-exome sequencing analysis should be considered for etiologic investigation.
MATERIALS AND METHODS
ENU mutagenesis
Three independent sources of ENU-mutagenized mice were screened for perinatal lethality and CHD: (i) a cohort of 32 lines from BWH/Harvard Medical School; (ii) a cohort of 282 independent lines from the Jackson Laboratory–National Heart Lung and Blood Institute ENU Mutagenesis Program; and (iii) a cohort of seven independent lines from the Justice Laboratory at Baylor University. The breeding protocol used for the identification of lines with recessive alleles in each screen is depicted in Figure 1.
Genetic background
In each cohort, male C57BL/6J mice (G0) were mutagenized by three weekly doses of 90 or 100 mg ENU/kg body weight. In the BWH/HMS cohort, the G0 male was outbred to two wild-type FVB/N females, chosen for their fecundity, large litter size and high degree of polymorphism with C57BL/6J. The first-generation (G1) males were also crossed with wild-type FVB/N females, and their second-generation (G2) daughters were backcrossed to G1 males, to enrich for recessive mutations in the third-generation (G3) offspring, which were screened at birth for perinatal lethality and CHD. This breeding strategy was designed to allow expedient recovery and genetic mapping of recessive mutations (22,35). To control for the mixed genetic background, a control screen was performed in non-mutagenized mice using the same breeding strategy. Both control and mutagenized lines were housed, maintained and bred in the facility, using identical protocols approved by the University of Chicago or BWH/Harvard Medical School. The rate of perinatal lethal line recovery was calculated from those lines that showed death of mice homozygous for the mutagen-treated chromosome from birth to postnatal day 5. The Jackson Laboratory–National Heart Lung and Blood Institute ENU Mutagenesis Program screen was performed in an inbred C57BL/6J background. The Justice Laboratory cohort utilized a balancer chromosome region on mouse chromosome 11, allowing phenotype analysis concomitant with initial mapping (24,36,37).
Screening for congenital heart defects
Cages for the control and mutagen-treated screens were checked morning and night for deceased neonatal mice. All mice that died during the first 5 days of life were examined for structural heart defects. By gross cardiac examination, direct inspection of the exterior of the heart was used to reveal outflow tract or situs abnormalities. Specifically, the great vessels were inspected for routing abnormalities including transposition or malposition of the great arteries, double-outlet right or left ventricle or interrupted or right aortic arch (38). They were also evaluated for septation abnormalities such as AP window or truncus arteriosus, which can often be identified by external examination (Van Praagh, 1975). Gross evaluation of the cardiac chambers was used to suggest the presence of an underlying structural abnormality within the heart (39), although all hearts subsequently underwent serial histological analysis.
Definitive anatomic evaluation of the mutant heart in our screen was by serial section histology. After gross examination of the hearts from mice that died perinatally, the hearts were then embedded in paraffin, cut in 5 μm sections and stained with hematoxylin and eosin. The evaluation of heart-looping abnormalities during serial section histology was aided by marking the left thorax with black ink prior to embedding. When multiple animals from an individual line were found to have cardiac structural abnormalities, the DNA from each affected pup was submitted for linkage analysis.
Genomic mapping
SNPs that were previously identified as polymorphic between inbred strains of mouse, including C57BL/6J, FVB/NJ and C3H/FeJ (40), were genotyped on the Illumina platform (40) using DNA from mice with selected CHD. Segregation of SNPs and CHD was assessed by χ2 analysis. Using three degrees of freedom for intercross data, a χ2 value of >16.3 indicated linkage with a P-value of <0.001 (41).
Conflict of Interest statement. None declared.
FUNDING
This work was funded by grants from the NIH [R01 HL092153; (I.P.M.), R01 HD036404; (D.R.B.) and U01 HD39372; and R01 CA115503 (M.J.J.)], the March of Dimes (I.P.M.), the AHA (I.P.M.) and the Schweppe Foundation (I.P.M.).
REFERENCES
Articles from Human Molecular Genetics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/hmg/ddq211
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/hmg/article-pdf/19/16/3105/1748514/ddq211.pdf
Free to read at hmg.oxfordjournals.org
http://hmg.oxfordjournals.org/cgi/content/abstract/19/16/3105
Free after 12 months at hmg.oxfordjournals.org
http://hmg.oxfordjournals.org/cgi/content/full/19/16/3105
Free after 12 months at hmg.oxfordjournals.org
http://hmg.oxfordjournals.org/cgi/reprint/19/16/3105.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
A phenotype-based forward genetic screen identifies Dnajb6 as a sick sinus syndrome gene.
Elife, 11:e77327, 18 Oct 2022
Cited by: 3 articles | PMID: 36255053 | PMCID: PMC9642998
The arginine methyltransferase Carm1 is necessary for heart development.
G3 (Bethesda), 12(8):jkac155, 01 Jul 2022
Cited by: 3 articles | PMID: 35736367 | PMCID: PMC9339313
In vivo identification and validation of novel potential predictors for human cardiovascular diseases.
PLoS One, 16(12):e0261572, 17 Dec 2021
Cited by: 4 articles | PMID: 34919578 | PMCID: PMC8682894
Live mechanistic assessment of localized cardiac pumping in mammalian tubular embryonic heart.
J Biomed Opt, 25(8):1-19, 01 Aug 2020
Cited by: 7 articles | PMID: 32762173 | PMCID: PMC7403774
Some Isolated Cardiac Malformations Can Be Related to Laterality Defects.
J Cardiovasc Dev Dis, 5(2):E24, 02 May 2018
Cited by: 15 articles | PMID: 29724030 | PMCID: PMC6023464
Review Free full text in Europe PMC
Go to all (13) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Interrogating congenital heart defects with noninvasive fetal echocardiography in a mouse forward genetic screen.
Circ Cardiovasc Imaging, 7(1):31-42, 06 Dec 2013
Cited by: 31 articles | PMID: 24319090 | PMCID: PMC3962690
Microcomputed tomography provides high accuracy congenital heart disease diagnosis in neonatal and fetal mice.
Circ Cardiovasc Imaging, 6(4):551-559, 12 Jun 2013
Cited by: 26 articles | PMID: 23759365 | PMCID: PMC3908688
Genome-wide association studies highlight novel risk loci for septal defects and left-sided congenital heart defects.
BMC Genomics, 25(1):256, 07 Mar 2024
Cited by: 0 articles | PMID: 38454350 | PMCID: PMC10918883
Toward a molecular understanding of congenital heart disease.
Circulation, 91(2):494-504, 01 Jan 1995
Cited by: 76 articles | PMID: 7805255
Review
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: R01 CA115503
NHLBI NIH HHS (2)
Grant ID: R01 HL092153
Grant ID: R01HL092153
NICHD NIH HHS (2)
Grant ID: R01 HD036404
Grant ID: U01HD39372




