Abstract
Free full text

Association between mitochondrial DNA variations and Alzheimer's Disease in the ADNI cohort
Abstract
Despite the central role of amyloid deposition in the development of Alzheimer's disease (AD), the pathogenesis of AD still remains elusive at the molecular level. Increasing evidence suggests that compromised mitochondrial function contributes to the aging process and thus may increase the risk of AD. Dysfunctional mitochondria contribute to reactive oxygen species (ROS) which can lead to extensive macromolecule oxidative damage and the progression of amyloid pathology. Oxidative stress and amyloid toxicity leave neurons chemically vulnerable. Because the brain relies on aerobic metabolism, it is apparent that mitochondria are critical for the cerebral function. Mitochondrial DNA sequence-changes could shift cell dynamics and facilitate neuronal vulnerability. Therefore we postulated that mitochondrial DNA sequence polymorphisms may increase the risk of AD. We evaluated the role of mitochondrial haplogroups derived from 138 mitochondrial polymorphisms in 358 Caucasian ADNI subjects. Our results indicate that the mitochondrial haplogroup UK may confer genetic susceptibility to AD independently of the APOE4 allele.
1. Introduction
Despite the remarkable effort and resources devoted to Alzheimer's disease (AD) this last decade, the pathophysiology of AD has not been well characterized. The current animal models of AD pathogenesis and the human genome-wide association studies (GWAS) have not resulted in a single common origin for the irregular but common incidences of AD, thus implying extensive heterogeneity in the underlying pathophysiology. One clear underlying pathophysiologic feature of AD has been identified and this centered on “the amyloid and tau pathology” that is believed to lead to neuronal loss, decreased synaptic density, brain atrophy, and a subsequent progressive cognitive decline associated with AD (Yankner et al., 2008). Nevertheless, recent research into AD provides compelling evidence for a variety of additional pathological events including oxidative stress and apoptosis (Mamelak, 2007).
It is has been postulated that the accumulation of reactive oxygen species (ROS) over a period time is negatively correlated with mitochondrial function and significantly contributes to the aging process (Wallace and Fan, 2009). As the primary source of ROS, mitochondria play pivotal roles in maintaining cellular energy balance and lie at the nexus of the signaling pathways controlling apoptosis. As such, it is conceivable that mitochondria may mediate the development and clinical outcome of AD. In fact, numerous research findings suggest a link between altered cerebral metabolic rate for glucose (CMRglc) in subjects with mitochondrial mutations (Lindroos et al., 2009). PET imaging studies have demonstrated a detectable decline in cerebral metabolism prior to any brain atrophy or abnormality found by neuropsychiatric testing in subjects who later developed AD (Reiman et al., 2004). Postmortem studies on AD brains have revealed a reduction in the mitochondrial respiratory chain proteins in the posterior cingulate (Liang et al., 2008). The projection neurons of the cerebral cortex which tend to die first in AD, and show increased vulnerability to decreased mitochondrial efficiency (Gotz et al., 2009).
The Down-syndrome (DS) brain, with accelerated amyloid deposition and high propensity for AD, implies a possible synergy between amyloid deposition and ROS production. The structural changes that caused DS harbor APP, a precursor molecule of amyloid β as well as gene SOD1 encoding an enzyme named superoxide dismutase 1. SOD1 marks the first important step in scavenging and neutralizing ROS in cells (Engidawork and Lubec, 2001). The amyloid β, a product of APP processing, has been shown to be transported into the mitochondria through the TOMM complex (Devi et al., 2006) and inhibit the oxidative phosphorylation system (OXPHOS) in line with APOE (Crouch et al., 2005). In addition, DNA sequence variations within both the APOE and TOMM40 genes have been found to be significantly associated with AD in genome-wide association studies (Potkin et al., 2009; Roses et al., 2009; Shen et al., 2010). Mitochondria efficiency may decrease in response to amyloid toxicity and interfering proteins like APOE and Drp1 (Cho et al., 2009) that initiate a “vicious cycle” in ROS production facilitating apoptosis and leading to cell death (Reddy, 2009).
Therefore, it is plausible link AD to the functional status of mitochondria that critically relies on the mitochondrially expressed OXPHOS proteins (Fosslien, 2001). Because the mitochondria genome differs from the nuclear genome in the rate of accumulation of mutations, it has been proposed that some common mitochondrial DNA (mtDNA) polymorphisms probably alter protein functions and compromise mitochondria efficiency (Tuppen et al., 2009). There is growing evidence that certain mtDNA clusters and polymorphisms as well as the somatically acquired mutations could predispose to psychiatric disorders (Coskun et al., 2004; Jou et al., 2009; McMahon et al., 2000; Zecavati and Spence, 2009).
To further elucidate the relation between mitochondrial DNA sequences polymorphisms and risk of AD, we calculated the association of mitochondrial haplogroups derived from 138 mitochondrial polymorphisms in 358 ADNI subjects. To the best of our knowledge this is the first report considering mitochondrial SNPs in the context of a longitudinal clinical study of AD.
2. Methods
2.1 Ethics
The ADNI data was previously collected across 50 research sites. Study subjects gave written informed consent at the time of enrollment for imaging and genetic sample collection and completed questionnaires approved by each participating sites' Institutional Review Board (IRB).
2.2 The Alzheimer's Disease Neuroimaging Initiative (ADNI)
ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations as a $60 million, 5-year public–private partnership. The primary goal of ADNI is to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians in the development of new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The principal investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California, San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. ADNI participants include approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years. Participants are evaluated at baseline, 6, 12, 18 (for MCI only), 24, and 36 months (although AD participants do not have a 36 month evaluation). For additional information see http://www.adni-info.org.
2.3 Identification of 138 mitochondrial SNPs
The genotyping procedure for the mtDNA and APOE variations was executed as described in (Potkin et al., 2009). Genotyping was performed on the Illumina Human610-Quad Infinium HD platform. The Illumina Human610-Quad BeadChip has of 550,000 polymorphic sites (SNP), plus an additional 60,000 genetic markers including 138 mitochondrial DNA sequence polymorphic sites. The 138 mitochondrial SNPs are based on the AF347015.1 mtDNA reference sequences, one of 53 African sequences deposited in Genbank by Ingman et al in 2001 (Ingman and Gyllensten, 2001). As the first step, the 138 mitochondrial AF347015.1 DNA SNPs were mapped to revised Cambridge Reference Sequence (rCRS) and manually annotated. The 138 SNPs consist of 21 noncoding, 91 protein coding, 4 rRNA, 11 tRNA and 1 termination sites. A subset of the 138 SNPs is shown in Table 1.
Table 1
| AF347015.1 mtDNA SNP | Corresponding rCRS SNP | Change | Type | Function | Haplogroup |
|---|---|---|---|---|---|
| MitoT9699C | 9698 | T>C | synonymous | Cytochtome C oxidase III (COIII) | U8 |
| MitoA11252G | 11251 | A>G | synonymous | NADH dehydrogenase 4 (ND4) | JT |
| MitoG12373A | 12372 | G>A | Synonymous | NADH dehydrogenase 5 (ND5) | UK |
The table illustrates a random subset of the 138 annotated mtDNA polymorphisms. The first column represents SNPs derived from the AF347015.1 mtDNA sequence. The second column represents the corresponding rCRS mtDNA sequence position. The last four columns describe the corresponding rCRS SNP characteristics.
2.4 Haplotyping and haplogroup assignment
Since variation in the mtDNA genome arose as result of the sequential accumulation of mutations over time (1 mutation/10,000 years), specific combinations of these ancient mutations can cluster as haplotypes, and subsequently define haplogroups. The hierarchical relationship amongst these haplogroups based on mutations can be represented by a phylogenetic tree via available programs (http://www.phylotree.org). The nucleotide changes of the 138 mitochondrial SNPs for each subject were compared to rCRS and used to identify motifs that characterize mitochondrial haplotype (van Oven and Kayser, 2009). Assignment of each individual mtDNA “sequence” to specific haplogroups was performed according to criteria as published (Torroni et al., 1996) (e.g. haplogroup J is define by mutations at locations 11251, 16126, and no mutation at 16294). Based on each subject's available genotyping, all 816 ADNI subjects were haplotyped and assigned to a haplogroup.
2.5 Population and stratification
Population heterogeneity has been cited as one of many difficulties in studying complex diseases (Schork et al., 2001). Although race and ethnicity were available as surrogates to evaluate genetic similarity amongst individuals, race does not always correctly reflect population of origin, particularly in heterogeneous admixed groups - as expected for the ADNI dataset. In order to avoid the confounding effect due to population stratification, we assessed evidence for genetic background heterogeneity and by leveraging publicly available, well-defined populations. Briefly, Phase 2 HapMap dataset (n=270 subjects) were merged with the entire genome-wide scan of 816 ADNI subjects (http://pngu.mgh.harvard.edu/~purcell/plink/res.shtml) (Enoch et al., 2006). For all individuals in the merged dataset, a pair-wise identity-by-state (IBS) distance matrix was created by a linkage agglomerative algorithm implemented in PLINK (Purcell et al., 2007). Subsequently, Multidimensional Distance Scaling (MDS) analysis was carried out on the genome-wide IBS pair-wise-distance matrix of the merged dataset to display the structure of the distance between individuals as a geometrical picture (JMP Genomics 4.00). For the purpose of this study, the ADNI individuals that clustered with CEU founders (Utah resident with ancestry from northern and western Europe), and belonged to the diagnostic AD (n=170) or control (n=188) groups were selected for further statistical analyses. Analysis comparing nuclear DNA genetic background differences among individuals in the different mitochondrial haplogroups did not reveal any obvious associations among European sample of individuals chosen for study.
2.6 Statistical analyses
Student's t-tests were used to evaluate the differences in means between AD and control (CTRL) groups for continuous variables. Statistical significance was assesses on the basis of a 2-sided test with α=0.05. Frequency differences for categorical outcomes in AD and CTRL groups were assessed via Chi-square (χ2) test statistic or Fisher's exact test (Table 2). Mitochondrial haplogroups and allelic frequencies were compared between AD and CTRL using χ2 test or Fisher's test. Effect size for the association was measured as an odds ratio (OR) with a 95% confidence interval (CI). Cochran-Mantel-Haenszel (CMH) tests were performed to adjust for APOE4 allele genotype (i.e. E4 “dose”). The homogeneity of odds ratios across strata was tested by the Breslow-Day test. Logistic regression was performed to assess the contribution of mtDNA haplogroup, APOE4 allele, and their potential interaction effects to AD risk. Adaptive permutation tests were employed for accommodating multiple-testing.
Table 2
| CTRL | AD | Statistical Significance (p-value) | |
|---|---|---|---|
| # Subjects | 188 | 170 | |
| Mean Age | 75.93±5.01 | 75.79±7.57 | p≥0.83 |
| Gender (Male/Female) | 98/90 | 101/91 | p≥0.706 |
| Smoker/non-smoker | 69/101 | 222/135 | p≥0.26 |
| Handedness (right/left) | 171/17 | 158/12 | p≥0.5 |
| Mean MMSE | 29.11±0.93 | 23.40±2.07 | p≤0.00001 |
| Mean ADAS-cog | 9.37±4.09 | 28.65±8.7 | p≤0.00001 |
| Mean years of Education | 16.18± 2.81 | 14.9± 3.0 | p≤5.4E-5 |
| APOE (ε2/ε3/ε4) | 27/297/52 | 8/189/143 | p≤4.7755E-17 |
Mean ± standard deviation (SD) and the frequency of each category are represented with test statistics.
2.6.1 Statistical Package
JMP Genomics Version 4.00. (SAS Institute Inc., Cary, NC, 1989-2007); PLINK (Purcell et al., 2007); STATA10 (StataCorp. 2007. Stata Statistical Software: Release 10. College Station, TX: StataCorp LP).
3. Results
3.1 Demographic
All subjects were part of the Alzheimer's Disease Neuroimaging Initiative (ADNI), a longitudinal multi-site observational study including. All the participants in this study were confined to group case (AD) or control (CTLR) defined at baseline diagnosis. Following MDS analysis (section 2.7), a total of 170 AD subjects and 188 healthy controls were included in this analysis. The two groups were compared at different variables and summarized in Table 2.
AD and cognitively normal participant (CTRL) groups do not differ in age, gender, smoking and handedness. However, AD subjects had a lower education level (p≤5.4E-5) and had a disproportionally higher APOE4 allele frequency (p≤4.77E-17) than the CTRL group. The two groups also significantly differed in ADAS-cog (p≤0.00001) and MMSE scores (p≤0.00001) reflecting the enrollment criteria for ADNI.
3.2 Association Analysis
3.2.1 Case-control differences in mitochondrial haplogroups
All subjects with Caucasian origin belonged to a major haplogroup N which was rooted approximately 70,000 years before present (YBP). Haplotype N consisted of 358 subjects who were distributed among the 9 designated haplogroups: H, I, J, K, T, U, V, W, and X. These 9 haplogroups were partitioned into 4 encompassing haplogroup clusters 1) HV, 2) JT, 3) UK, and 4) IWX based on their relation to ancestor lineage N (Finnila et al., 2001; Richards et al., 1996; Richards et al., 1998) as detailed in Figure 1.
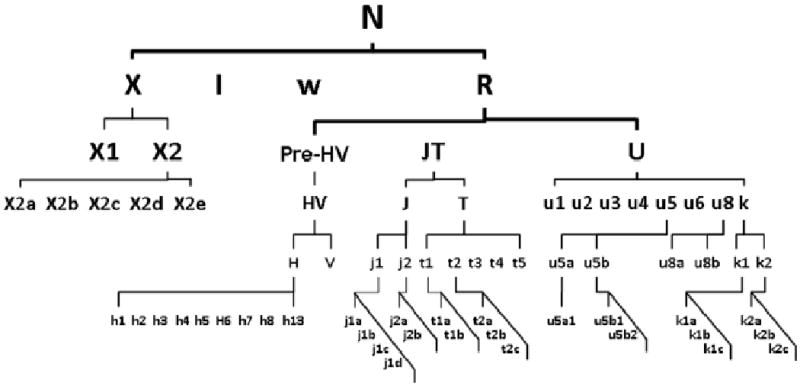
Schematic phylogeny of the basal European mtDNA haplogroups.
Legend: Simplified mtDNA phylogeny tree demonstrate the Caucasian lineages exclusively originated from haplogroup N approximately 50,000-70,000 YBR. The alphabetical symbols illustrate the descendant lineages of macrograph N and indicate the phylogenic relationship amongst haplogroups as a basis of the haplogroup designation to clusters. The global mtDNA variations are available at http://www.phylotree.org.
The frequency of the haplogroups for each of the 4 clusters is demonstrated in Figure 2. The frequency distribution of the clusters is consistent with the reported worldwide mitochondrial haplogroup distribution reported at the MITOMAP database (www.mitomap.org). From the 358 ADNI participants, 86 subjects (24%) belong to haplogroup UK, 67 subjects (19%) belong to haplogroup JT, 183 (51%) to haplogroup HV, and 22 subjects (6%) to haplogroup IWX.
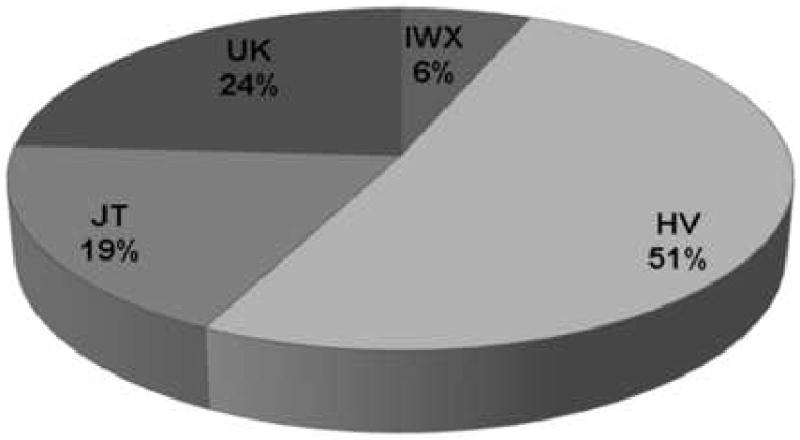
Overall distribution of mtDNA haplogroups of 358 subjects in the ADNI dataset.
Legend: Labels designate the name of the haplogroup clusters and the relative frequencies of the haplogroup expressed in percentages.
The chi-square (χ2) test statistics revealed significant association between disease status and mitochondrial haplogroups (χ2=7.99, df=3, p-value≤0.046). The strongest association among the 4 haplogroups involved in the UK haplogroup with a contrast between CTRL (χ2=2.75) versus AD (χ2=3.05) and associated odds ratio of (1.92, 95% CI: 1.13, 3.26; p-value < 0.013). A formal test for homogeneity confirmed that UK haplogroup appeared to have a stronger association with AD than the other haplogroups (χ2=7.97 df=3 p-value < 0.045). A score test for trend of odds supported this (χ2= 4.76, p-value < 0.03). The magnitude of association remained significant (χ2=7.63, df=3,p-value≤0.0057 CI 1.12, 195) after adjusting for APOE4 allele dose.
3.2.2 Case-control differences in mitochondrial single nucleotide polymorphism
An allelic association analysis was conducted for each of the polymorphic mtDNA sites. In the analysis, all the 138 SNPs were calculated for association regardless of haplogroup specificity or minor allele frequency. Adaptive permutation analysis for each polymorphism was performed to determine an empirical p-value. All the mtSNPs with an empirical p-value < 0.05 were considered statistically significant. The SNP DNA position, minor allele frequency in AD and CTRL, asymptotic p-value, odds ratio, confidence interval, functional consequence and haplogroup specificity are reported in Table 4.
Table 4
| SNP | Position in mtDNA (bp) | Allele frequency | P-value | OR | CI | Function | Haplogroup | ||
|---|---|---|---|---|---|---|---|---|---|
| AD | CTRL | L95 | U95 | ||||||
| MitoA11467G | 11467 | 0.28 | 0.14 | 0.003 | 2.22 | 1.3 | 3.78 | NADH dehydroenase (ND4) | U and Uk |
| MitoA12308G | 12308 | 0.3 | 0.17 | 0.006 | 2.03 | 1.23 | 3.34 | tRNA for leucine (CUN) | U and Uk |
| MitoG12372A | 12372 | 0.3 | 0.18 | 0.006 | 1.99 | 1.21 | 3.27 | NADH dehydroenase (ND5) | U and Uk |
| MitoC9698T | 9698 | 0.17 | 0.08 | 0.021 | 2.26 | 1.16 | 4.41 | Cytochrome C oxidase (COIII) | U8 (Uk) |
| MitoC16270T | 16270 | 0.09 | 0.03 | 0.048 | 2.52 | 1 | 6.36 | non-coding region | Not specific |
Analysis of individual SNP revealed that risk of AD was increased in subjects who carried minor allele MitoA11467G (OR=2.22; 95% CI, 1.30 3.78; p value≤0.003). The MitoA11467G is a synonymous polymorphism located in gene NADH dehydroenase 4 (ND4). The encoded protein of this gene is a subunit of a large enzyme complex known as complex I. Complex I is responsible for the first step in the oxidative phosphorylation process by transferring electrons from NADH to ubiquinone. The second most significant SNP was MitoA12308G (OR=2.03; 95% CI, 1.23 3.34; p value≤0.006) located in a tRNA which transfers the amino acid leucine for protein synthesis. This polymorphism was found highly significant in an interaction with 10398G (empirical P value = 0.0028) suggesting some women are at increased risk to develop breast cancer (Kinoshita et al., 1996). MitoG12372A (OR=1.996; 95% CI, 1.21 3.27., p value≤0.006) located in gene NADH dehydrogenase 5 (ND5) that encodes a subunit for complex I. MitoC9698T (OR=2.265; 95% CI, 1.16 4.41; p value ≤0.003) is synonymous variant encoding Cytochrome C oxidase III (COIII) enzyme found in complex IV. Defects in mitochondrial COIII gene implicated in Leber hereditary optic neuropathy (LHON) (Brown et al., 1992; Eichhorn-Mulligan and Cestari, 2008) and age-dependent accumulation of mutations in mitochondrial DNA (mtDNA) in cytochrome c oxidase has been implicated in the onset of sporadic AD (Davis et al., 1997; Lin and Beal, 2006) MitoC16270T (OR=2.527; 95% CI, 1.16 4.41; p value=≤0.048) is located in the hypervariable segment assumed to represent a mutational hotspot (see summary in Table 4)
Besides age as a major risk for AD, APOE 4 is the most consistently replicated genetic risk factor for late-onset AD. Since there is a significant APOE allele frequency difference in our cohort, a subsequent analysis was performed to assess the potential for an APOE4 cofounding or interactive effect. Logistic regression revealed no evidence for an interaction effect. However, APOE revealed strong association with AD at level of OR=4.11-5.35 independently of mtSNPs.
The location and potential effect on gene function of the significant mutations in relation to mtDNA are depicted in Figure 3.
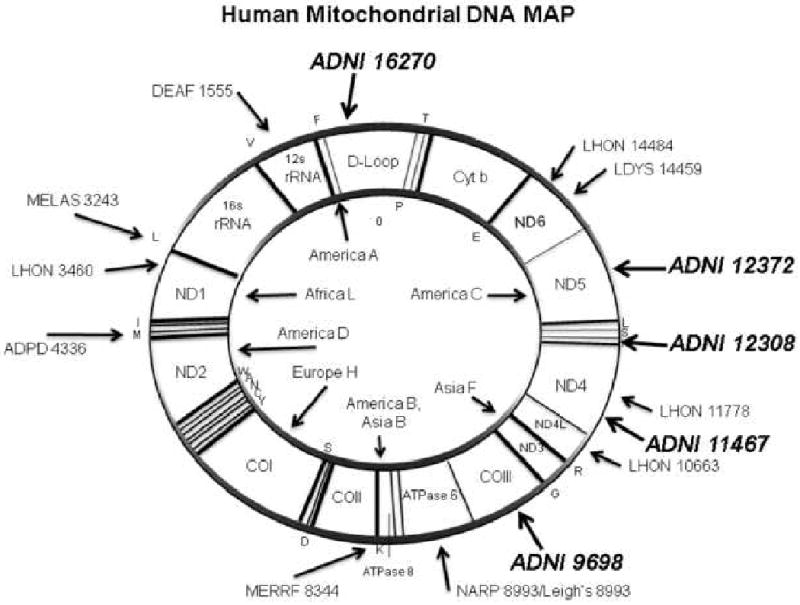
Semantic illustration of the Human Mitochondrial Genome
Legend: The 5 significant mitochondrial polymorphisms in italics (ADNI prefix followed by SNP number) are mapped to mtDNA. The most well-know pathogenic mutations are also outlined. The abbreviations are DEAF=deafness; MELAS=mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes; LHON=Leber's hereditary optic neuropathy; ADPD=Alzheimer's and Parkinson's disease; MERRF=myoclonic epilepsy and ragged red fiber disease; NARP=neurogenic muscle weakness, ataxia, retinitis pigmentosum; LDYS=LHON+dystonia modified from (Wallace, 2005).
Thus, based on χ2 tests and logistic regression analysis, the APOE4 allele, and the mitochondrial haplogroup UK, as well as specific mitochondrial polymorphisms, are significantly associated with AD in the ADNI cohort. Given the functional consequences of the significant mitochondrial polymorphisms, the possession of these polymorphisms may predispose to AD.
4. Discussion
Recent research studies have found links between mitochondrial dysfunction and common diseases of aging, such as Parkinson's (PD) and Alzheimer's disease (AD) (Wang et al., 2009; Wang et al., 2007). A growing body of evidence suggests a reasonable association between amyloid-β toxicity, mitochondrial dysfunction, oxidative stress and neuronal damage in AD pathophysiology (Mancuso et al., 2006) The impact of the dysfunctional mitochondria on the integrity of neuronal cells is not fully understood. Mitochondria are exclusively positioned to play a pivotal role in neuronal cell survival or death by controlling energy metabolism and apoptotic pathways. Mitochondrial malfunction can alter the delicate bioenergetics balance making neuronal cells vulnerable to challenge (Wallace et al., 2010). Mitochondrial haplogroups and polymorphisms have also received substantial consideration in psychiatric and neurodegenerative diseases (Tuppen et al., 2009). Mitochondrial associations were found between AD and haplogroup J with increased susceptibility, while haplogroup T was found to have protective effect (Chagnon et al., 1999). Increased risk of AD in males with haplogroup U (van der Walt et al., 2004) as well as the mitochondrial tRNA(Glu) 4336 SNP have been reported (Brown et al., 1996; Shoffner et al., 1993). In addition, we report 5 mitochondrial SNPs, 3 of which define haplogroup UK and 2 of them specific to certain UK haplotypes in association exhibit associations with AD in the ADNI cohort.
One potential explanation for the associations is that mtDNA haplogroups correlates with genetic background that are distinctive between geographically separated populations. However, our population stratification analysis did not find significant difference between the major haplogroups and nuclear DNA-based genetic background and hence argues against a population substructure confounding effect. Another potential explanation is that the underlying mechanism for the predisposition for Alzheimer disease is related to energy deficiency. Observations on longevity, neurodegenerative disease susceptibility (van der Walt et al., 2004; Wallace et al., 1998), sperm viability (Montiel-Sosa et al., 2006), and climate adaptation propose association between functional mtDNA variations and ATP production efficiency and correlated ROS and heat generation in different haplogroups (Arning et al., 2010). The mtDNA haplogroup most prone to energy deficiency in Europe are haplogroups U and Uk (Hendrickson et al., 2008). It is conceivable that additional genetic risk factors further compromise the mitochondrial ATP production causing it to fall below the threshold level needed for optimal neuronal functioning. It is important to note that mitochondria genome expresses 37 genes and other approximately 1500-2000 nuclear genes may play critical role in optimal mitochondria function (Wallace, 2008). This interaction between the two genomes may influence brain function (Roubertoux et al., 2003) and may contribute to the complexity of AD pathophysiology.
Despite few contradictory research findings in haplogroup association to AD (Chinnery et al., 2000; Coppede et al., 2007; Elson et al., 2006), the design of ADNI provides the opportunity to follow up our findings with neuroimaging and psychometric examination in addition to a nuclear DNA–based GWAS interrogation. This comprehensive approach has the potential to provide novel insight into the underlying pathomechanisms of Alzheimer's disease and possibly open up a new prospect for novel pharmacological targets and therapeutic strategies (Wallace, 2005).
Table 3
| Diagnostic category | Haplogroup frequency | HV | JT | IWX | UK |
|---|---|---|---|---|---|
| CTRL | Observed (%) | 28.50% | 3.35% | 11.10% | 9.50% |
| Expected (%) | 26.80% | 3.22% | 9.80% | 12.61% | |
| AD | Observed (%) | 22.63% | 2.80% | 7.60% | 14.53% |
| Expected (%) | 24.27% | 2.91% | 8.90% | 11.40% |
The contingency table shows the number of observed and expected subjects for each haplogroup by diagnostic categories.
Acknowledgments
The authors express their appreciation to Jordan Hiller (JMP Genomics) Liv McMillan, Divya Rajpoot, Jacklyn Walter and Jerod Rasmussen for their technical support. The genotyping was performed by Jennifer Webster and Drs. David Craig and Matt Huentelman of TGen. We want to acknowledge the support and numerous contributions by the ADNI investigators and the ADNI Industry Advisory Board (Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories), and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration and the Foundation for the National Institutes of Health. We gratefully acknowledge the participation of the ADNI subjects and their family members. Genotyping was performed at The Translational Genomics Institute, Phoenix AZ, by Jennifer Webster and Drs. David Craig and Matt Huentelman. Sample processing, storage and distribution were provided by the NIA-sponsored National Cell Repository for Alzheimer's Disease by Dr. Tatiana Foroud and Kelley Faber. Sample verification and quality control bioinformatics were provided by Drs. Li Shen, Sungeun Kim and Kwangsik Nho of the Indiana University Center for Neuroimaging, Nathan Pankratz of the IU Dept of Medical and Molecular Genetics, and Bryan DeChairo of Pfizer, Inc.
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu\ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principle Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada.
FUNDING: This work was supported by National Institutes of Health grants AG16573. NS211328, AG24373, DK73691, AG16573, and NS41850; a Doris Duke Clinical Interfaces Award, and CIRM Comprehensive Grant RC1-00353-1 awarded to D.C.W. Dr. Schork is funded in part by The National Institute on Aging Longevity Consortium [grant number U19 AG023122-01] and the Scripps Translational Sciences Institute Clinical Translational Science Award [NIH/NCRR Grant Number UL1 RR025774]. This analysis was supported by grants to The Transdisciplinary Imaging Genetics Center (TIGC-P20 RR020837-01), the Alzheimer's Disease Neuroimaging Initiative (ADNI U01 AG024904-01, and supplement 3U01AG024904-03S5), the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering (NIH), the Functional Imaging Biomedical Informatics Research Network (FBIRN U24-RR021992, National Center for Research Resources), commercial support from Vanda Pharmaceuticals, and private support from an anonymous Foundation and anonymous donations. Additional contributions made through the Foundation for the NIH from Merck & Co. Inc., Pfizer, Inc., and Gene Network Sciences, Inc. partially supported the genotyping results reported here. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH grant and supplement). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
#Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://www.loni.ucla.edu\ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. ADNI investigators include (complete listing available at http://www.loni.ucla.edu\ADNI\Collaboration\ADNI Manuscript Citations.pdf
Conflict of interest statement. None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arning L, Haghikia A, Taherzadeh-Fard E, Saft C, Andrich J, Pula B, Hoxtermann S, Wieczorek S, Akkad DA, Perrech M, Gold R, Epplen JT, Chan A. Mitochondrial haplogroup H correlates with ATP levels and age at onset in Huntington disease. J Mol Med. 2010;88:431–6. [Abstract] [Google Scholar]
- Brown MD, Shoffner JM, Kim YL, Jun AS, Graham BH, Cabell MF, Gurley DS, Wallace DC. Mitochondrial DNA sequence analysis of four Alzheimer's and Parkinson's disease patients. Am J Med Genet. 1996;61:283–9. [Abstract] [Google Scholar]
- Brown MD, Voljavec AS, Lott MT, MacDonald I, Wallace DC. Leber's hereditary optic neuropathy: a model for mitochondrial neurodegenerative diseases. Faseb J. 1992;6:2791–9. [Abstract] [Google Scholar]
- Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am J Med Genet. 1999;85:20–30. [Abstract] [Google Scholar]
- Chinnery PF, Taylor GA, Howell N, Andrews RM, Morris CM, Taylor RW, McKeith IG, Perry RH, Edwardson JA, Turnbull DM. Mitochondrial DNA haplogroups and susceptibility to AD and dementia with Lewy bodies. Neurology. 2000;55:302–4. [Abstract] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Coppede F, Mancuso M, Lo Gerfo A, Manca ML, Petrozzi L, Migliore L, Siciliano G, Murri L. A Ser326Cys polymorphism in the DNA repair gene hOGG1 is not associated with sporadic Alzheimer's disease. Neurosci Lett. 2007;414:282–5. [Abstract] [Google Scholar]
- Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–31. [Europe PMC free article] [Abstract] [Google Scholar]
- Crouch PJ, Blake R, Duce JA, Ciccotosto GD, Li QX, Barnham KJ, Curtain CC, Cherny RA, Cappai R, Dyrks T, Masters CL, Trounce IA. Copper-dependent inhibition of human cytochrome c oxidase by a dimeric conformer of amyloid-beta1-42. J Neurosci. 2005;25:672–9. [Abstract] [Google Scholar]
- Davis RE, Miller S, Herrnstadt C, Ghosh SS, Fahy E, Shinobu LA, Galasko D, Thal LJ, Beal MF, Howell N, Parker WD., Jr Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1997;94:4526–31. [Europe PMC free article] [Abstract] [Google Scholar] Retracted
- Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J Neurosci. 2006;26:9057–68. [Abstract] [Google Scholar]
- Eichhorn-Mulligan K, Cestari DM. The genetics of leber hereditary optic neuropathy--prototype of an inherited optic neuropathy with mitochondrial dysfunction. Semin Ophthalmol. 2008;23:27–37. [Abstract] [Google Scholar]
- Elson JL, Herrnstadt C, Preston G, Thal L, Morris CM, Edwardson JA, Beal MF, Turnbull DM, Howell N. Does the mitochondrial genome play a role in the etiology of Alzheimer's disease? Hum Genet. 2006;119:241–54. [Abstract] [Google Scholar]
- Engidawork E, Lubec G. Protein expression in Down syndrome brain. Amino Acids. 2001;21:331–61. [Abstract] [Google Scholar]
- Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. [Abstract] [Google Scholar]
- Finnila S, Lehtonen MS, Majamaa K. Phylogenetic network for European mt. DNA Am J Hum Genet. 2001;68:1475–84. [Europe PMC free article] [Abstract] [Google Scholar]
- Fosslien E. Mitochondrial medicine--molecular pathology of defective oxidative phosphorylation. Ann Clin Lab Sci. 2001;31:25–67. [Abstract] [Google Scholar]
- Gotz J, Schonrock N, Vissel B, Ittner LM. Alzheimer's disease selective vulnerability and modeling in transgenic mice. J Alzheimers Dis. 2009;18:243–51. [Abstract] [Google Scholar]
- Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, Kingsley L, Goedert JJ, Vlahov D, Donfield S, Wallace DC, O'Brien SJ. Mitochondrial DNA haplogroups influence AIDS progression. Aids. 2008;22:2429–39. [Europe PMC free article] [Abstract] [Google Scholar]
- Ingman M, Gyllensten U. Analysis of the complete human mtDNA genome: methodology and inferences for human evolution. J Hered. 2001;92:454–61. [Abstract] [Google Scholar]
- Jou SH, Chiu NY, Liu CS. Mitochondrial dysfunction and psychiatric disorders. Chang Gung Med J. 2009;32:370–9. [Abstract] [Google Scholar]
- Kinoshita H, Imayama H, Sou H, Shibata J, Ogami N, Tamae T, Nakayama T. A case of obstructive icterus caused by incarceration of a pancreatic stone in the common channel of the pancreatobiliary ducts. Kurume Med J. 1996;43:79–85. [Abstract] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105:4441–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–95. [Abstract] [Google Scholar]
- Lindroos MM, Borra RJ, Parkkola R, Virtanen SM, Lepomaki V, Bucci M, Virta JR, Rinne JO, Nuutila P, Majamaa K. Cerebral oxygen and glucose metabolism in patients with mitochondrial m.3243A>G mutation. Brain. 2009;132:3274–84. [Abstract] [Google Scholar]
- Mamelak M. Alzheimer' s disease, oxidative stress and gammahydroxybutyrate. Neurobiol Aging. 2007;28:1340–60. [Abstract] [Google Scholar]
- Mancuso M, Coppede F, Migliore L, Siciliano G, Murri L. Mitochondrial dysfunction, oxidative stress and neurodegeneration. J Alzheimers Dis. 2006;10:59–73. [Abstract] [Google Scholar]
- McMahon FJ, Chen YS, Patel S, Kokoszka J, Brown MD, Torroni A, DePaulo JR, Wallace DC. Mitochondrial DNA sequence diversity in bipolar affective disorder. Am J Psychiatry. 2000;157:1058–64. [Abstract] [Google Scholar]
- Montiel-Sosa F, Ruiz-Pesini E, Enriquez JA, Marcuello A, Diez-Sanchez C, Montoya J, Wallace DC, Lopez-Perez MJ. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene. 2006;368:21–7. [Abstract] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One. 2009;4:e6501. [Europe PMC free article] [Abstract] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [Europe PMC free article] [Abstract] [Google Scholar]
- Reddy PH. Amyloid beta, mitochondrial structural and functional dynamics in Alzheimer's disease. Exp Neurol. 2009;218:286–92. [Europe PMC free article] [Abstract] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci U S A. 2004;101:284–9. [Europe PMC free article] [Abstract] [Google Scholar]
- Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt HJ, Sykes B. Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet. 1996;59:185–203. [Europe PMC free article] [Abstract] [Google Scholar]
- Richards MB, Macaulay VA, Bandelt HJ, Sykes BC. Phylogeography of mitochondrial DNA in western. Europe Ann Hum Genet. 1998;62:241–60. [Abstract] [Google Scholar]
- Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, Reiman EM. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J 2009 [Europe PMC free article] [Abstract] [Google Scholar]
- Roubertoux PL, Sluyter F, Carlier M, Marcet B, Maarouf-Veray F, Cherif C, Marican C, Arrechi P, Godin F, Jamon M, Verrier B, Cohen-Salmon C. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nat Genet. 2003;35:65–9. [Abstract] [Google Scholar]
- Schork NJ, Fallin D, Thiel B, Xu X, Broeckel U, Jacob HJ, Cohen D. The future of genetic case-control studies. Adv Genet. 2001;42:191–212. [Abstract] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Jr, Weiner MW, Saykin AJ. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage 2010 [Europe PMC free article] [Abstract] [Google Scholar]
- Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MF, Mirra SS, Beal MF, Yang CC, Gearing M, Salvo R, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–84. [Abstract] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–50. [Europe PMC free article] [Abstract] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2009;1797:113–128. [Abstract] [Google Scholar]
- van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Murali Doraiswamy P, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. [Abstract] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–94. [Abstract] [Google Scholar]
- Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–80. [Abstract] [Google Scholar]
- Wallace DC. Mitochondria as chi. Genetics. 2008;179:727–35. [Europe PMC free article] [Abstract] [Google Scholar]
- Wallace DC, Brown MD, Melov S, Graham B, Lott M. Mitochondrial biology, degenerative diseases and aging. Biofactors. 1998;7:187–90. [Abstract] [Google Scholar]
- Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2009;10:12–31. [Europe PMC free article] [Abstract] [Google Scholar]
- Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J Neurosci. 2009;29:9090–103. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang X, Su B, Perry G, Smith MA, Zhu X. Insights into amyloid-beta-induced mitochondrial dysfunction in Alzheimer disease. Free Radic Biol Med. 2007;43:1569–73. [Abstract] [Google Scholar]
- Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. [Abstract] [Google Scholar]
- Zecavati N, Spence SJ. Neurometabolic disorders and dysfunction in autism spectrum disorders. Curr Neurol Neurosci Rep. 2009;9:129–36. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.neurobiolaging.2010.04.031
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2918801?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.neurobiolaging.2010.04.031
Article citations
MitoH3: Mitochondrial Haplogroup and Homoplasmic/Heteroplasmic Variant Calling Pipeline for Alzheimer's Disease Sequencing Project.
J Alzheimers Dis Rep, 8(1):575-587, 08 Apr 2024
Cited by: 0 articles | PMID: 38746629 | PMCID: PMC11091720
CoQ10 and Mitochondrial Dysfunction in Alzheimer's Disease.
Antioxidants (Basel), 13(2):191, 02 Feb 2024
Cited by: 3 articles | PMID: 38397789 | PMCID: PMC10885987
Review Free full text in Europe PMC
Mitochondrial DNA Haplogroup K Is Protective Against Autism Spectrum Disorder Risk in Populations of European Ancestry.
J Am Acad Child Adolesc Psychiatry, 63(8):835-844, 08 Dec 2023
Cited by: 1 article | PMID: 38072244
The contributions of mitochondrial and nuclear mitochondrial genetic variation to neuroticism.
Nat Commun, 14(1):3146, 30 May 2023
Cited by: 1 article | PMID: 37253732 | PMCID: PMC10229642
Role of Aβ in Alzheimer's-related synaptic dysfunction.
Front Cell Dev Biol, 10:964075, 26 Aug 2022
Cited by: 22 articles | PMID: 36092715 | PMCID: PMC9459380
Review Free full text in Europe PMC
Go to all (72) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Exploratory analysis of mtDNA haplogroups in two Alzheimer's longitudinal cohorts.
Alzheimers Dement, 16(8):1164-1172, 16 Jun 2020
Cited by: 18 articles | PMID: 32543785 | PMCID: PMC9847473
No consistent evidence for association between mtDNA variants and Alzheimer disease.
Neurology, 78(14):1038-1042, 21 Mar 2012
Cited by: 27 articles | PMID: 22442439 | PMCID: PMC3317529
Mitochondrial DNA variants in a Japanese population of patients with Alzheimer's disease.
Mitochondrion, 10(1):32-37, 22 Aug 2009
Cited by: 34 articles | PMID: 19703591
Mitochondrial alterations, oxidative stress and neuroinflammation in Alzheimer's disease.
Int J Immunopathol Pharmacol, 25(2):345-353, 01 Apr 2012
Cited by: 86 articles | PMID: 22697066
Review
Funding
Funders who supported this work.
NCRR NIH HHS (7)
Grant ID: P20 RR020837
Grant ID: P20 RR020837-01
Grant ID: U24 RR021992
Grant ID: U24 RR021992-02
Grant ID: UL1 RR025774
Grant ID: U24-RR021992
Grant ID: U24 RR021992-01A1
NHLBI NIH HHS (2)
Grant ID: P01 HL058120
Grant ID: P01 HL058120-06A19005
NIA NIH HHS (16)
Grant ID: U24 AG021886
Grant ID: AG24373
Grant ID: P50 AG016573-060005
Grant ID: P50 AG016573-070005
Grant ID: R01 AG024373-02
Grant ID: U19 AG023122
Grant ID: 3U01AG024904-03S5
Grant ID: R01 AG024373
Grant ID: R01 AG024373-01
Grant ID: U01 AG024904-01
Grant ID: U19 AG010483
Grant ID: U19 AG023122-01
Grant ID: AG16573
Grant ID: R01 AG024373-03
Grant ID: P50 AG016573
Grant ID: U01 AG024904
NIDDK NIH HHS (3)
Grant ID: DK73691
Grant ID: R01 DK073691
Grant ID: R01 DK073691-01
NINDS NIH HHS (6)
Grant ID: R01 NS041850-01
Grant ID: NS211328
Grant ID: R01 NS041850
Grant ID: R01 NS021328
Grant ID: NS41850
Grant ID: R01 NS021328-15





