Abstract
Free full text

Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia
Associated Data
Abstract
Although the role of Hedgehog (Hh) signalling in embryonic pattern formation is well established1, its functions in adult tissue renewal and maintenance remain unclear, and the relationship of these functions to cancer development has not been determined. Here we show that the lossof Smoothened (Smo), an essential component of the Hh pathway2, impairs haematopoietic stem cell renewal and decreases induction of chronic myelogenous leukaemia (CML) by the BCR–ABL1 oncoprotein3. Loss of Smo causes depletion of CML stem cells—the cells that propagate the leukaemia—whereas constitutively active Smo augments CML stem cell number and accelerates disease. As a possible mechanism for Smo action, we show that the cell fate determinant Numb, which depletes CML stem cells, is increased in the absence of Smo activity. Furthermore, pharmacological inhibition of Hh signalling impairs not only the propagation of CML driven by wild-type BCR–ABL1, but also the growth of imatinib-resistant mouse and human CML. These data indicate that Hh pathway activity is required for maintenance of normal and neoplastic stem cells of the haematopoietic system and raise the possibility that the drug resistance and disease recurrence associated with imatinib treatment of CML4,5 might be avoided by targeting this essential stem cell maintenance pathway.
CML arises owing to a translocation between the BCR serine/threonine kinase gene and the ABL1 tyrosine kinase3. Imatinib mesylate, which binds to the ABL1 kinase domain and inhibits phosphorylation of substrates, has been used to treat CML, but it is not curative because the cancer stem cells that propagate the leukaemia are resistant to therapy and are not eradicated5–7. Furthermore, imatinib resistance due to mutations in the drug-binding site can occur, especially in advanced stage disease, leading to disease relapse and progression4. The design of effective new therapies thus critically depends on the identification of signals that are required for CML propagation and in particular signals required for CML cancer stem cell maintenance.
Given the parallels in signalling between development and cancer8,9, we examined whether Hh signalling, an important regulator of development and oncogenesis in many tissues1,10, might also have a role in CML. To this end we conditionally deleted Smo—a protein that is essential for Hh signal transduction2. Smo is negatively regulated by the Hh receptor Patched (Ptch) in the absence of Hh protein signals. This inhibition is relieved when Ptch is bound by the Hh proteins Shh, Ihh or Dhh. Subsequently, Smo causes activation of Hh pathway targets via the Gli family of transcriptional effectors2.
Smo was deleted in the haematopoietic system by crossing mice carrying a loxP-flanked Smo allele11 to mice in which Cre is driven by Vav regulatory elements12. Smo was efficiently deleted in haematopoietic cells of Vav-Cre;SmoloxP/loxP mice as compared to control SmoloxP/loxP mice lacking Cre (Fig. 1a, b). Although the frequency of haematopoietic stem cells (HSCs) and differentiated cells was unchanged in the Smo-deficient (Smo−/−) mice (Supplementary Fig. 1), these animals had a clear defect in long-term HSC function in primary and secondary transplants (Fig. 1c and Supplementary Fig. 1). Decreased reconstitution was not due to decreased homing or to a preferential loss of any specific lineage (Supplementary Figs 1 and 2), and could be recapitulated with whole bone marrow transplants (Supplementary Fig. 3). Cumulatively, these data indicate that Smo is required for HSC renewal in vivo.
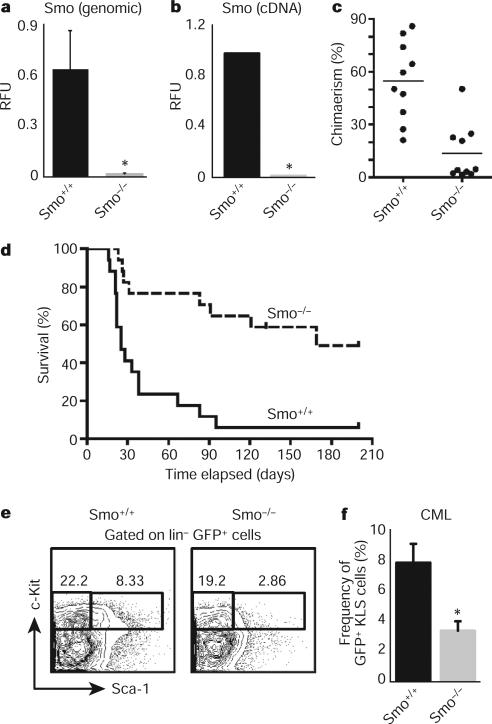
a, Real-time PCR (rtPCR) using primers specific for the loxP sites was carried out on Smo−/− or Smo+/+ bone marrow genomic DNA (n = 3, *P = 0.03). RFU, relative fluorescent units. b, rtPCR for Smo using complementary DNA from Smo−/− or Smo+/+ KLSF cells (n = 7, *P < 0.00001). c, KLSF cells from Smo−/− or Smo+/+ mice were transplanted together with competing bone marrow cells into congenic recipients and donor-derived chimaerism was monitored. The graph shows the average donor-derived chimaerism after long-term reconstitution (n = 2 with 20 mice, P = 0.0002). d, Survival curve of mice transplanted with BCR–ABL1-infected Smo−/− or Smo+/+ KLSF cells (n = 3 with 34 mice, P = 0.0002). e, Representative example of CML stem cells (GFP+ Lin−c-Kit+ Sca+) in the bone marrow of mice transplanted with BCR–ABL1-infected Smo−/− or Smo+/+ KLS cells. f, Graph of the average percentage of CML stem cells in mice receiving BCR–ABL1-infected Smo+/+ (8 mice) or Smo−/− KLS cells (9 mice). Frequency shows c-Kit+Sca+ cells as as a percentage of the GFP+Lin− population. *P = 0.006. Error bars in all bar graphs are s.e.m.
We next investigated the role of Hh signalling in CML initiation and propagation. CML can be modelled by transplanting BCR–ABL1-transduced haematopoietic progenitors into irradiated mice13. Whereas control cells transduced with BCR–ABL1 caused CML in 94% (16 out of 17) of recipients within 3 months, similarly transduced Smo−/− cells caused CML in only 47% (8 out of 17) of recipients (Fig. 1d), and these tumours showed an increased latency. The reduced CML incidence was not due to altered homing or engraftment (Supplementary Fig. 4). These data suggest that Hh activity is required for the initiation and propagation of CML. The propagation of several cancers has been shown to depend on cancer stem cells, a critical subset of cancer cells that are capable of transferring the disease to a new host8. Because deletion of Smo led to impaired CML growth, we postulated that the loss of Smo might affect CML stem cells. As shown in Fig. 1e, f, the frequency of c-Kit+ Lin− Sca-1+ (KLS) cells (which have previously been shown to be responsible for propagating CML6) was significantly reduced in the absence of Smo.
The fact that loss of Smo led to decreased numbers of CML stem cells led us to investigate whether activation of the Hh pathway might lead to an increased frequency of CML stem cells and acceleration of disease. To test this, we used a transgenic mouse expressing an activated form of Smo (SmoM2) fused to yellow fluorescent protein (YFP)14; expression of this gene from the Rosa26 locus promoter is blocked by a loxP-flanked stop sequence, and is thus inducible by Cre. When SmoM2–YFP mice were crossed with Vav-Cre mice, progeny expressed YFP in >70% of c-Kit+ Lin−/lo Sca-1+ Flk2− (KLSF) cells (Fig. 2a). Although the infection of both control and SmoM2 KLSF cells with BCR–ABL1 resulted in CML, the frequency of CML stem cells in animals receiving SmoM2 cells was fourfold greater than in mice receiving control cells (Fig. 2b, c). As expected, SmoM2 was expressed in transgenic but not in control CML stem cells (Fig. 2d). Most importantly, the increased CML stem cell frequency led to a significant acceleration of CML progression (Fig. 2e). SmoM2 expression also accelerated CML growth when activated post-embryonically (Supplementary Fig. 5). Thus genetic loss- and gain-of-function experiments indicate that Hh pathway activity controls the frequency and maintenance of CML stem cells, and consequently the incidence and latency of CML formation.
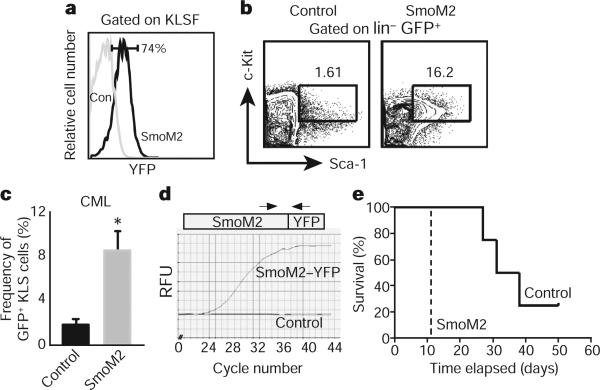
a, Analysis of YFP fluorescence, which reflects SmoM2 expression in control (con) and SmoM2 KLSF cells (n = 4). b, Analysis of CML stem cells (GFP+ KLS) in mice transplanted with BCR–ABL1-infected control (left) and constitutively activated Smo (SmoM2, right) KLSF cells. c, The average percentage of CML stem cells in mice receiving BCR–ABL1-transduced control (n = 4) and SmoM2 KLSF cells (n = 12). Error bars are s.e.m., *P = 0.048. d, rtPCR analysis of SmoM2 expression in CML stem cells. e, Survival curve of mice receiving BCR–ABL1 transduced control (solid line) or SmoM2 KLSF cells (dashed line; n = 2, 16 mice, P = 0.0082).
Our previous studies suggested that in normal haematopoietic stem cells, maintenance of the undifferentiated state depends on low levels of the cell fate determinant Numb15. We therefore tested whether Numb may act similarly in CML stem cells, and whether the effects of loss of Smo might be due to altered levels of Numb. We found that a greater frequency of Smo−/− CML KLS cells had high levels of Numb as compared to control CML KLS cells (Fig. 3a–c). Additionally, ectopically expressed Numb inhibited in vitro propagation of BCR–ABL1-infected haematopoietic cells (Fig. 3d) and of CML stem cells from established leukaemias (Fig. 3e). These data suggest that increased Numb expression may contribute to the loss of CML stem cells in the absence of Smo.
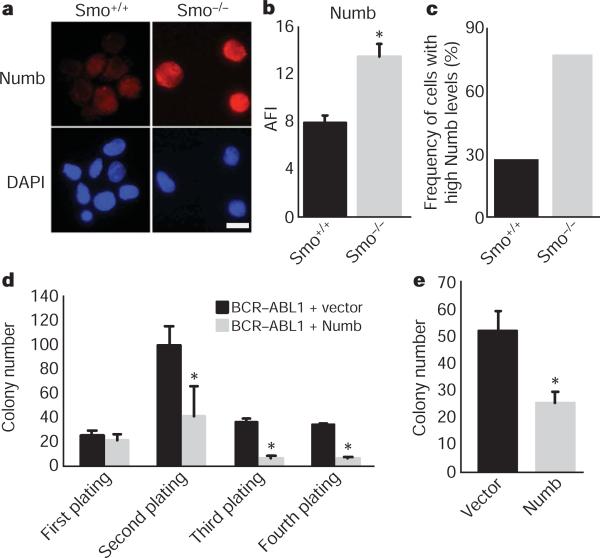
a, CML stem cells from Smo+/+ and Smo−/− leukaemias were stained with Numb (red, top panel) and 4,6-diamidino-2-phenylindole (DAPI; blue, bottom panel). Scale bar, 10 μM. b, The average fluorescence intensity (AFI) was determined by dividing the overall mean fluorescence intensity by the area of the cell (P = 0.002). c, The frequency of cells with high expression levels of Numb was calculated by designating cells above a mean fluorescence intensity value of 1,000 as high expressors (n = 3, using either CML KLS or CML c-Kit+ cells). d, KLSF cells were infected with MSCV-BCR-ABL1-IRES-GFP and either vector MSCV-IRES-YFP or MSCV-Numb-IRES-YFP. GFP and YFP double-positive cells were plated in methylcellulose, colonies were counted and cells were serially replated (n = 2, *P < 0.006). e, CML KLS cells were infected with viruses expressing control vector or Numb IRES-YFP. Infected cells were sorted and plated in methylcellulose (n = 2, P = 0.03). Error bars in b–e are s.e.m.
Our genetic data showing that CML stem cells depend on Hh pathway activity raised the possibility that these cells might be targeted by pharmacological blockade of this pathway. We therefore tested the effect of cyclopamine, which inhibits Hh signalling by stabilizing Smo in its inactive form16. Exposure of CML stem cells to cyclopamine at a dose determined to minimize off-target effects and toxicity (Supplementary Fig. 6) led to a twofold inhibition of colony growth (Fig. 4a). This could be recapitulated using the Hh-blocking antibody 5E1 (Supplementary Fig. 7), suggesting that Hh pathway activation in CML may be ligand dependent17,18. We also delivered cyclopamine to animals transplanted with BCR–ABL1-infected HSCs; whereas all of the control animals succumbed to CML within 4 weeks, 60% of the cyclopamine-treated mice were still alive after 7 weeks (Fig. 4b and Supplementary Fig. 8). Furthermore cyclopamine-treated mice had up to a 14-fold reduction in the CML stem cell population (Fig. 4c, d); consistent with this depletion, leukaemias from cyclopamine-treated mice were unable to propagate disease effectively when transplanted (Supplementary Fig. 9). These data suggest that cyclopamine can target the CML stem cell compartment critical for propagation of CML. Notably cyclopamine was most effective when initiated at early stages after CML establishment (Supplementary Fig. 10).
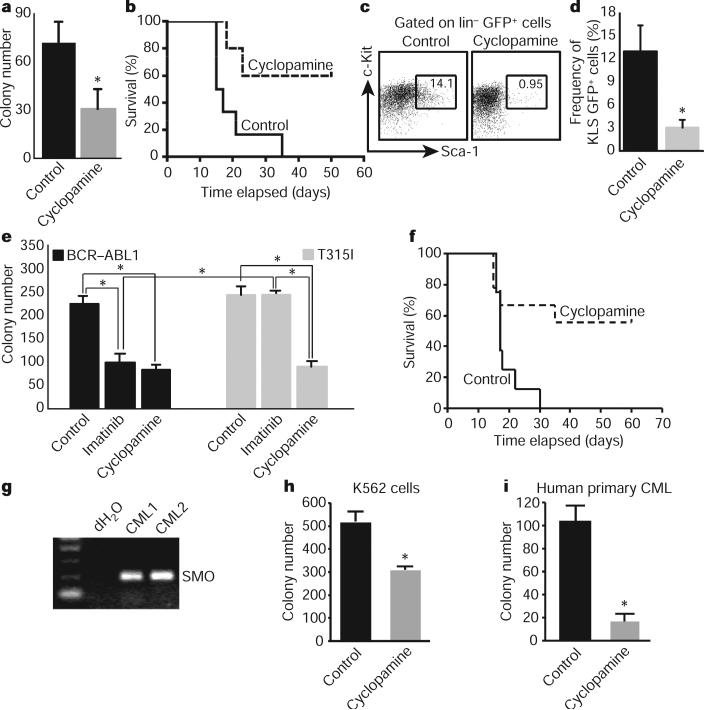
a, CML stem cells were plated in methylcellulose in the presence of 1 μM tomatidine (control) or cyclopamine (n = 2, *P = 0.005). b, Survival curve of mice transplanted with BCR–ABL1-infected KLS cells in the presence of vehicle or cyclopamine (n = 3 with 32 mice, P = 0.02). c, Representative example of CML stem-cell frequency in vehicle- or cyclopamine-treated CML. d, Average CML stem cell frequency in vehicle-or cyclopamine-treated mice (n = 4, *P = 0.03). e, KLSF cells were infected with wild-type BCR–ABL1 or the imatinib-resistant T315I mutant BCR–ABL1, and plated in methylcellulose with imatinib (2 μM) or cyclopamine (1 μM) (n = 2, *P < 0.004). f, Survival curve of mice receiving T3151 BCR–ABL1-infected KLSF cells and either vehicle or cyclopamine, 7 days after transplantation (n = 2 with 16 mice, P = 0.021). g, SMO PCR on primary uncultured human CML samples. h, Imatinib-resistant K562 blast crisis CML cells were plated in methycellulose in the presence of 1 μM tomatidine or cyclopamine (n = 2, *P < 0.01). i, CD34+ cells were sorted from primary uncultured human blast crisis CML samples and plated in methylcellulose in the presence of 1 μM tomatidine or cyclopamine (n = 3, *P < 0.05). Error bars in all bar graphs are s.e.m.
One rationale for identifying new pathways involved in CML is that targeting these pathways may be used to prevent or overcome imatinib resistance19. To test whether cyclopamine could impair the growth of imatinib-resistant CML, we infected cells with virus encoding either wild-type BCR–ABL1 or the T315I mutant4,6,20 that is resistant to imatinib and other tyrosine kinase inhibitors used at present. Cells infected with mutant BCR–ABL1 were unresponsive to imatinib (Fig. 4e), but their growth was reduced 2.5-fold by cyclopamine (Fig. 4e). Progression of drug-resistant CML in vivo was also significantly impaired in the presence of cyclopamine (Fig. 4f) as well as in the genetic absence of Smo (Supplementary Fig. 11). These data indicate that Hh pathway activity is required for maintenance of CML stem cells, and raise the possibility that small molecule inhibitors of the pathway could be useful for targeting both normal and imatinib-resistant CML.
Finally we tested whether targeting this pathway could inhibit the growth of human CML. Freshly isolated primary human blast crisis CML samples expressed SMO (Fig. 4g). To determine whether SMO may functionally contribute to human CML growth, we tested the effect of cyclopamine on both a human CML cell line and patient samples in a methylcellulose assay. Cyclopamine decreased colony formation in an imatinib-resistant human CML cell line (Fig. 4h) and in three independent primary patient samples (Fig. 4i and Supplementary Fig. 12), but not in untransformed cord blood cells (Supplementary Fig. 13). Consistent with this, inhibition of SMO by the delivery of a new SMO antagonist also impaired the propagation of imatinib-resistant human CML cells in xenografts (A.A., M.M., T.V.A. and C.H.J., manuscript in preparation). Cumulatively, these data suggest that Hh signalling is important for human CML growth and that it may continue to have a role even as the disease progresses into blast crisis phase.
The studies discussed earlier demonstrate a dependence of normal and neoplastic stem cells on Hh signalling. Previously, mammalian Hh proteins have been reported to preserve and increase the short-term repopulating capacity of human HSCs21. Our genetic loss-of-function studies indicate that intact Hh signalling is needed for HSC renewal. This requirement may be shared with the nervous system, in which progenitor development is dependent on Hh pathway activation22,23. Just as Hh signalling seems to be required for stem cells, its aberrant activation has been associated with the development of several solid cancers17,18,24,25. In the haematopoietic system, Hh signalling has been implicated in the growth of B cell lymphoma26 and multiple myeloma27. Our observations provide critical genetic evidence for a role of Hh signalling in myeloid leukaemia, and together with previous work, suggest that this pathway may have a broader role in haematological malignancies. Our data are also unique in demonstrating that loss of endogenous cancer stem cells impairs in vivo cancer progression, supporting the notion that the leukaemia stem cells identified by transplantation are the relevant drivers of endogenous cancer growth28–30. The real impetus for identification of new pathways that drive cancer is the opportunity to inhibit these pathways instead of, or in addition to, conventional therapies. This is especially true for CML, in which imatinib is incapable of eradicating the disease, not only necessitating its continual use but also increasing the likelihood of resistance and relapse. Our findings that the pharmacological inhibition of Hh signalling can impair growth of CML driven by wild-type and imatinib-resistant BCR–ABL1, as well as human CML, raise the possibility that Hh antagonists may be useful as a therapy for normal and drug resistant CML.
METHODS SUMMARY
For CML assays, HSCs were infected with MSCV-BCR-ABL1-IRES-GFP or MSCV-IRES-GFP (control) and transplanted into recipient mice. CML was assessed by flow cytometry and histopathology. Pharmacological inhibition of Hh was initiated 6–8 days after transplantation by delivering 25 mg kg−1 of cyclopamine twice a day by oral gavage. Treatment was continued for five consecutive days, stopped for two days and then continued once a day until the mice were analysed after signs of leukaemia. For human CML studies, peripheral blood CD34+ cells were sorted and plated in methylcellulose in tomatidine or cyclopamine.
Acknowledgements
We wish to thank R. Wechsler-Reya for critical advice, D. Kioussis for Vav-Cre transgenic mice, A. M. Pendergast, A. Means and B. Hogan for review of the manuscript, S. Li for advice on mouse CML experiments, G. Daley, M. Azam and S. Li for the T315I BCR–ABL1 construct, B. Harvat for cell sorting, and J. Harris, B. Zimdahl, N. D'Amato and S. Honeycutt for experimental help. T.R. is a recipient of a CRI Investigator Award, an EMF New Scholar award and a Leukemia and Lymphoma Society Scholar Award, M.F. is supported by the Duke Molecular Cancer Biology Training grant and C.H.J. is funded by the California Institute of Regenerative Medicine and a sponsored research agreement with Pfizer. This work was also supported by National Institutes of Health grants DK63031, DK072234, AI067798 and the Lisa Stafford Memorial Prize to T.R.
METHODS
Mice
The Smo−/− and Smo+/+ mice used were in a mixed 129X1/SvJ and C57BL/6J background. Transplant recipients (C57BL/6 CD45.1) were 8–10 weeks of age. All mice were bred and maintained on acidified, antibiotic water in the animal care facility at Duke University Medical Center. All animal experiments were performed according to protocols approved by the Duke University Institutional Animal Care and Use Committee.
HSC isolation and analysis
Isolation of HSCs from bone marrow and their transplantation for in vivo analysis of function were performed as described12. For analysis of lineage markers, bone marrow cells from control or Smo−/− mice were incubated with antibodies to murine Ter119 (also known as Ly76), Mac-1 (Itgam), B220 (Ptprc) and CD3 (Cd3e) (eBiosciences), and analysed by FACS. For the reconstitution assays, bone marrow cells from control, Smo−/− or SmoM2 mice were stained and analysed for the KLSF cells12, and 500 control or Smo−/− KLSF cells were injected along with 200,000 competing bone marrow cells into lethally irradiated CD45.1 recipients. Multilineage repopulation was assessed at 20 weeks. Secondary repopulation assays were carried out by isolating whole bone marrow from primary recipients originally transplanted with control or Smo−/− KLSF cells. Recipient mice were killed and analysed for donor chimaerism at 24 weeks.
Generation and analysis of diseased mice
HSCs from Smo−/−, SmoM2 or control mice were isolated and cultured overnight in X-vivo with 10% fetal bovine serum (FBS), 100 ng ml−1 stem cell factor (SCF, also known as KITLG) and 20 ng ml−1 thrombopoietin (THPO) in a 96-well U-bottom plate (50,000 per well). Subsequently, cells were infected with MSCV-BCR-ABL1-IRES-GFP or MSCV-IRES-GFP as a control. After 48 h, different cell doses ranging from 15,000–20,000 Smo−/− or 5,000–8,000 limiting SmoM2 cells were transplanted retro-orbitally along with 200,000 whole bone marrow cells into lethally irradiated allelically mismatched recipients. After transplantation, recipient mice were evaluated daily for signs of morbidity, weight loss, failure to groom and splenomegaly. Premorbid animals were killed and relevant tissues were collected and analysed by flow cytometry and histopathology. For flow cytometric analysis of CML stem cells, leukaemic cells were stained for KLS and analysed on FACS-Vantage (BD Biosciences) and Flowjo software (Tree Star, Inc.).
Cyclopamine treatment of diseased mice
Mice were given 25 mg kg−1 of cyclopamine twice a day by oral gavage beginning 6–8 days after transplantation. Treatment was continued for five consecutive days, stopped for two days, and then continued once a day until the end of the monitoring period. Mice were visually assessed daily for signs of distress. In mice that showed signs of distress treatment was stopped and continued when they showed recovery.
Methylcellulose colony formation assays
For the colony formation assays CML KLS GFP+ cells, or KLSF cells infected with wild-type BCR–ABL1 or T315I BCR–ABL1 were sorted into a 24-well plate with complete methylcellulose medium (Stem Cell Technologies) containing cyclopamine (Toronto Research Chemicals) and/or imatinib (Seqouia Research Chemicals). Colonies were counted 8–10 days after plating. For the serial replating assay, cells were collected and counted, and 10,000 cells were replated into 12-well plates.
Immunofluorescence staining
Primary CML cells collected from bone marrow and spleen were sorted for KLS and GFP+ cells by FACS. Cells were cytospun, airdried and fixed in 4% paraformaldehyde. The primary antibody used was goat anti-Numb (Novus Biologicals), and the secondary antibody used was donkey anti-goat-Alexa Fluor 594 (Molecular Probes). DAPI (Molecular Probes) was added as a nuclear counterstain. Slides were viewed on the Axio Imager (Zeiss) at ×630 magnification. Fluorescence intensity analysis was quantified using Metamorph software (Molecular Devices).
Real-time PCR analysis
RNA was isolated using RNAqueous-Micro (Ambion) and converted to cDNA using Superscript II (Invitrogen). cDNA concentrations were measured with a fluorometer (Turner Designs) using RiboGreen reagent (Molecular Probes). Quantitative rtPCR was performed using an iCycler (BioRad) by mixing equal amounts of cDNAs, iQ SYBR Green Supermix (BioRad) and gene specific primers. Primer sequences are available on request. All real-time data was normalized to actin.
Human CML samples
All experiments on human CML samples were carried out with approval from the Duke University Institutional Review Board. Patient samples were either obtained from the Duke Adult Bone Marrow Transplant Clinic, or purchased from Open Biosystems. Mononuclear cells were isolated from peripheral blood samples using density-gradient centrifugation. RNA was isolated using RNAqueous-Micro (Ambion) and was converted to cDNA using Superscript II (Invitrogen). PCR with reverse transcription (RT–PCR) was performed (DNA Engine DYAD) with primer sequences specific to human SMO. Primer sequences are available on request. For all colony formation assays, samples were sorted for hCD34+ cells and plated into a 12-well plate (100,000 cells per well) with complete methylcellulose medium (Stem Cell Technologies) in the presence of tomatidine or cyclopamine (1 μM, Toronto Research Chemicals). Colony numbers were counted 14–17 days after plating.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature07737
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2946231?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
SKF-96365 Expels Tyrosine Kinase Inhibitor-Treated CML Stem and Progenitor Cells from the HS27A Stromal Cell Niche in a RhoA-Dependent Mechanism.
Cancers (Basel), 16(16):2791, 08 Aug 2024
Cited by: 0 articles | PMID: 39199564 | PMCID: PMC11352811
Exploiting Matrix Stiffness to Overcome Drug Resistance.
ACS Biomater Sci Eng, 10(8):4682-4700, 05 Jul 2024
Cited by: 0 articles | PMID: 38967485 | PMCID: PMC11322920
Review Free full text in Europe PMC
Unravelling the Mysteries of the Sonic Hedgehog Pathway in Cancer Stem Cells: Activity, Crosstalk and Regulation.
Curr Issues Mol Biol, 46(6):5397-5419, 29 May 2024
Cited by: 0 articles | PMID: 38920995 | PMCID: PMC11202538
Review Free full text in Europe PMC
GLI1 polymorphisms influence remission rate and prognosis of young de novo acute myeloid leukemia patients treated with cytarabine-based chemotherapy.
Ann Hematol, 103(6):1967-1977, 27 Apr 2024
Cited by: 0 articles | PMID: 38676765
Non-kinase off-target inhibitory activities of clinically-relevant kinase inhibitors.
Eur J Med Chem, 275:116540, 31 May 2024
Cited by: 0 articles | PMID: 38852338
Review
Go to all (542) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation.
Cancer Cell, 14(3):238-249, 01 Sep 2008
Cited by: 325 articles | PMID: 18772113
Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34+ CML stem/progenitor cells.
Int J Cancer, 133(3):579-589, 13 Mar 2013
Cited by: 58 articles | PMID: 23341351
Tracking molecular relapse of chronic myeloid leukemia by measuring Hedgehog signaling status.
Leuk Lymphoma, 54(2):342-352, 05 Sep 2012
Cited by: 7 articles | PMID: 22762548
The controversial role of the Hedgehog pathway in normal and malignant hematopoiesis.
Leukemia, 25(11):1665-1673, 10 Jun 2011
Cited by: 37 articles | PMID: 21660044 | PMCID: PMC4310480
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (7)
Grant ID: U19 AI067798-050006
Grant ID: U19 AI067798
Grant ID: U19 AI067798-030006
Grant ID: AI067798
Grant ID: U19 AI067798-040006
Grant ID: U19 AI067798-010006
Grant ID: U19 AI067798-020006
NIDDK NIH HHS (18)
Grant ID: DK63031
Grant ID: R01 DK063031-08
Grant ID: R01 DK063031-03
Grant ID: R01 DK063031-07
Grant ID: R01 DK072234-03
Grant ID: R01 DK072234
Grant ID: R01 DK072234-02
Grant ID: R01 DK063031
Grant ID: R01 DK063031-06
Grant ID: DK072234
Grant ID: R01 DK063031-01
Grant ID: R01 DK063031-04
Grant ID: R01 DK063031-05
Grant ID: R01 DK063031-07S1
Grant ID: R01 DK072234-01A1
Grant ID: R01 DK072234-04
Grant ID: R01 DK063031-01S1
Grant ID: R01 DK063031-02





