Abstract
Free full text

Transcriptional regulation is only half the story
The central dogma of molecular biology, articulated by Francis Crick, posits a flow of information from DNA to RNA to protein. Although the Human Genome Project has helped elucidate the first step in this cascade, the relationship between mRNA abundance and protein abundance has resisted systematic quantification, especially in higher eukaryotes. In their recent publication in Molecular Systems Biology, Vogel et al (2010) use a combination of microarrays and shotgun proteomics to quantify absolute mRNA and protein levels for over 1000 genes in a human cell line. Their analysis identifies sequence features related to translation and protein degradation that are as important as transcription in determining steady-state protein levels. This work provides an unprecedented, system-wide accounting of how information stored in our DNA determines the eventual state of our cells.
Molecular biologists have traditionally focused on transcriptional regulation as the main determinant of protein levels and, thus, cellular function. This focus is due, in part, to the historical sequence of discoveries following the work of Jacques Monod, and, more recently, to the development of microarray- (Schena et al, 1995) and sequencing-based technologies for large-scale mRNA quantification. The detailed view of transcription that has emerged from high-throughput mRNA measurements, including the inference of global regulatory networks (e.g. Lee et al, 2002), has shaped our understanding of how a healthy cell works, and also how pathologies arise and might be remedied. In fact, our knowledge of transcriptional regulation has improved so rapidly over the past decade that it is easy to forget the diverse array of post-transcriptional processes—including mRNA processing and modification, miRNA modulation, translation initiation, elongation, termination, and protein degradation—that also influence steady-state protein levels.
Not so fast, say Vogel et al (2010). Systematic studies of post-transcriptional processes have come of age, owing again to the development of technologies for high-throughput measurements. In this study, the authors used microarrays to quantify mRNA levels, together with a sophisticated mass-spectrometry-based proteomics method called APEX (Lu et al, 2007) to quantify soluble protein levels in a tumor cell line. The APEX method, originally developed in yeast and Escherichia coli, is the work-horse behind the present study. Under this protocol, proteins are digested into peptides, which are separated by liquid chromatography, and then ionized and sequenced with tandem mass spectrometry. In principle, protein amounts are then quantified simply by counting the numbers of corresponding peptides observed in repeat runs. In practice, the APEX method critically corrects for factors, such as efficiency of ionization, that influence the a priori probability of peptide detection. As a result, APEX provides reliable quantification of protein levels over five orders of magnitude.
Vogel et al. analyzed about 200 sequence features as potential determinants of the steady-state protein levels they measured. The correlates considered include features such as coding-sequence length, amino-acid composition, predicted mRNA structure, putative miRNA target sites, and the presence of upstream start codons. The authors observed a lognormal distribution of protein-per-mRNA ratios—suggesting that many impendent factors together contrive to determine translational efficiency and protein degradation rates. Some of the strongest individual correlates of protein abundance identified in the study are unsurprising: longer coding sequences typically produced less protein, controlling for mRNA levels, consistent with the idea that long transcripts are translated inefficiently and are prone to protein misfolding. Similarly, amino-acid content is also correlated with protein abundance, controlling for mRNA levels, consistent with variable costs associated with the depletion of different amino acids and different propensities for protein misfolding as a function of amino-acid composition. Furthermore, strong 5′ mRNA secondary structure or the presence of upstream start codons both reduced protein levels, again controlling for mRNA. However, several features had a surprisingly small role: codon adaption and miRNA target sites did not significantly influence protein abundance. The most important take-home message, furnished by a non-linear multiple regression, is that features related to post-transcriptional processes, especially those found in the coding sequence, together explained as much variation in protein levels as mRNA levels themselves did (Figure 1). Thus, transcriptional regulation is only half the story.
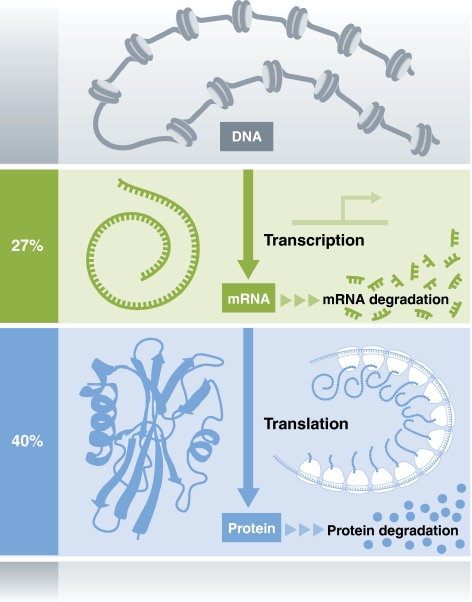
The relative influence of transcriptional and translational regulation on protein abundance. Processes related to translation and protein degradation can explain about 40% of the variation in human protein abundance, similar to the approximately 27% explained by mRNA abundance. Together these factors can explain two-thirds of the variation in human protein abundance.
Aside from generating the largest dataset to date of protein and mRNA concentrations in human cells, this study systematically quantifies the importance of translation and protein degradation regulatory processes, both individually and in aggregate. This work extends similar analyses performed in bacteria (Nie et al, 2006) and yeast (Brockmann et al, 2007; Wu et al, 2008), and it is preferable to analyses that are based on mRNA and protein measurements obtained from separate experiments. Nonetheless, this study is still limited to about 1000 soluble proteins, measured in an asynchronous, log-phase population of a tumor cell line, which contains chromosomal and methylation irregularities. Moreover, the strict separation of sequence features into those that determine steady-state mRNA levels and those that act post-transcriptionally is problematic: some nominally post-transcriptional features, such as those that influence ribosomal initiation, may feed back to influence steady-state mRNA levels as well (Iost and Dreyfus, 1995). Nonetheless, future studies in multiple cell lines, ideally including membrane proteins and synchronized populations, should elucidate how protein levels differ between and, indeed, define alternative cellular states. Such studies will be especially powerful when combined with high-throughput techniques for measuring ribosomal occupancy (Ingolia et al, 2009), allowing us to compare protein levels with direct estimates of translational efficiency, and to quantify protein stabilities as well.
The quantification and analysis of protein levels for 1000 human genes is a remarkable technical feat and is emblematic of the system-wide approach to studying basic questions in molecular biology. Without doubt, the growing literature based on high-throughput mass spectroscopy will continue to inform our understanding of post-transcriptional regulation, much as microarrays revolutionized our understanding of transcriptional regulation. Such measurements performed in relatively natural cellular conditions on endogenous genes will nicely complement manipulative experiments that interrogate protein production using synthetic, heterologous gene constructs (e.g. Voges et al, 2004). Together, these systematic approaches promise to elucidate the operational details of Crick's central dogma.
References
- Brockmann R, Beyer A, Heinisch JJ, Wilhelm T (2007) Post-transcriptional expression regulation: what determines translation rates? PLoS Comput Biol 3: e57. [Abstract] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [Europe PMC free article] [Abstract] [Google Scholar]
- Iost I, Dreyfus M (1995) The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J 14: 3252–3261 [Europe PMC free article] [Abstract] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [Abstract] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25: 117–124 [Abstract] [Google Scholar]
- Nie L, Wu G, Zhang W (2006) Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics 174: 2229–2243 [Europe PMC free article] [Abstract] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [Abstract] [Google Scholar]
- Vogel C, de Sousa RA, Ko D, Le S-H, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [Europe PMC free article] [Abstract] [Google Scholar]
- Voges D, Watzele M, Nemetz C, Wizemann S, Buchberger B (2004) Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem Biophys Res Commun 318: 601–614 [Abstract] [Google Scholar]
- Wu G, Nie L, Zhang W (2008) Integrative analyses of post-transcriptional regulation in the yeast Saccharomyces cerevisiae using transcriptomic and proteomic data. Curr Microbiol 57: 18–22 [Abstract] [Google Scholar]
Articles from Molecular Systems Biology are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/msb.2010.63
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1038/msb.2010.63
Citations & impact
Impact metrics
Article citations
Molecular Signatures of Neurodegenerative Diseases Identified by Proteomic and Phosphoproteomic Analyses in Aging Mouse Brain.
Mol Cell Proteomics, 23(9):100819, 26 Jul 2024
Cited by: 0 articles | PMID: 39069073 | PMCID: PMC11381985
Translational landscape of direct cardiac reprogramming reveals a role of Ybx1 in repressing cardiac fate acquisition.
Nat Cardiovasc Res, 2(11):1060-1077, 16 Oct 2023
Cited by: 0 articles | PMID: 38524149 | PMCID: PMC10959502
Integrative Analysis of Transcriptome, Proteome, and Phosphoproteome Reveals Potential Roles of Photosynthesis Antenna Proteins in Response to Brassinosteroids Signaling in Maize.
Plants (Basel), 12(6):1290, 13 Mar 2023
Cited by: 2 articles | PMID: 36986978 | PMCID: PMC10058427
Effect of Ethanol Consumption on the Placenta and Liver of Partially IGF-1-Deficient Mice: The Role of Metabolism via CYP2E1 and the Antioxidant Enzyme System.
Biology (Basel), 11(9):1264, 25 Aug 2022
Cited by: 1 article | PMID: 36138743 | PMCID: PMC9495332
Small-scale sequencing enables quality assessment of Ribo-Seq data: an example from Arabidopsis cell culture.
Plant Methods, 17(1):92, 24 Aug 2021
Cited by: 2 articles | PMID: 34429136 | PMCID: PMC8386038
Go to all (29) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Report on the tenth international workshop on the identification of transcribed sequences 2000. Heidelberg, Germany, October 28-31, 2000.
Cytogenet Cell Genet, 92(1-2):49-58, 01 Jan 2001
Cited by: 0 articles | PMID: 11306796
Translational control in endothelial cells.
J Vasc Surg, 45 Suppl A:A8-14, 01 Jun 2007
Cited by: 10 articles | PMID: 17544019 | PMCID: PMC1939822
Review Free full text in Europe PMC
Chipping away at the transcriptome.
Nat Genet, 27(3):232-234, 01 Mar 2001
Cited by: 10 articles | PMID: 11242095
The EZH2 polycomb transcriptional repressor--a marker or mover of metastatic prostate cancer?
Cancer Cell, 2(5):349-350, 01 Nov 2002
Cited by: 63 articles | PMID: 12450788
Review





