| PMC full text: | Gastroenterology. Author manuscript; available in PMC 2010 Oct 15. Published in final edited form as: Gastroenterology. 2009 May; 136(5): 1701–1710. Published online 2009 Jan 14. doi: 10.1053/j.gastro.2009.01.009 |
Figure 6
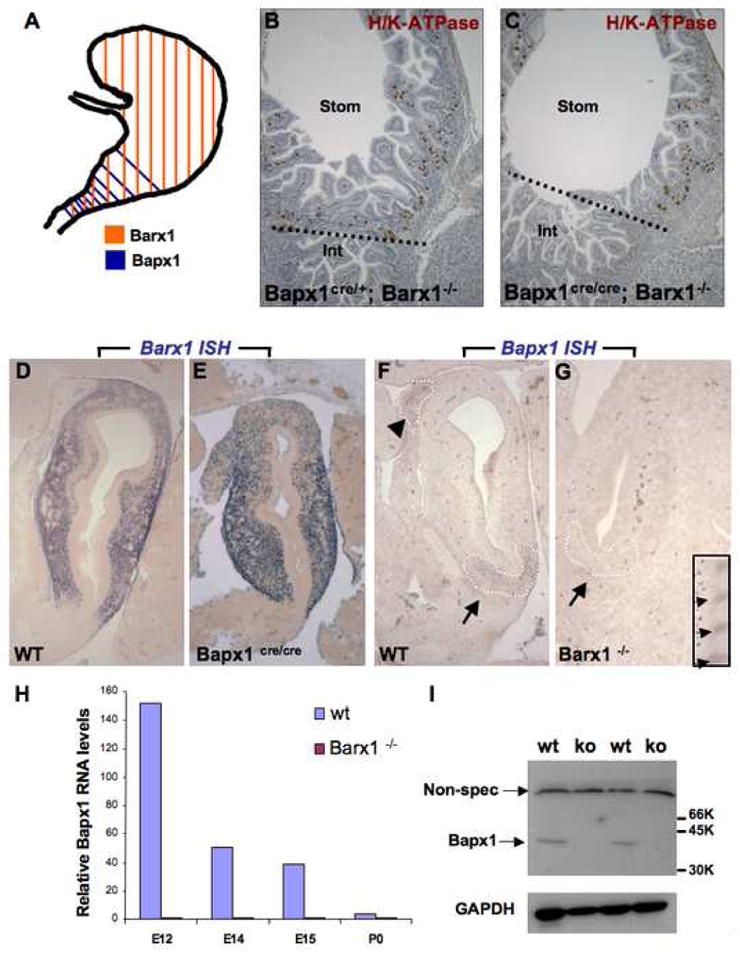
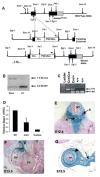
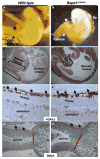
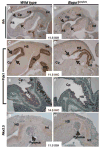
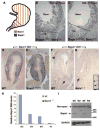
(A) Schematic depiction of Barx1 and Bapx1 expression in mid-gestation mouse stomach mesenchyme. Early Bapx1 expression is restricted to the antrum and pylorus (blue) and overlaps with Barx1 expression, which extends throughout the stomach (orange). (B,C) Further loss of Bapx1 does not worsen the stomach phenotype in Barx1−/− mice. Stomachs from Barx1−/−; Bapx1Cre/+ (B) and Barx1−/−;Bapx1Cre/Cre (C) neonates are shown. H/K-ATPase immunostaining reveals that a defined parietal cell-depleted region representing the antrum is absent with loss of Barx1 regardless of whether Bapx1 is functional or not. Other markers such as Cdx2 and Alcian blue staining gave similar results (data not shown). (D–G) In situ mRNA analysis confirms the overlapping schema depicted in panel A and reveals an expression hierarchy between Barx1 and Bapx1. Barx1 mRNA, which is expressed throughout fetal stomach mesenchyme, is unaltered in level or distribution with loss of Bapx1 (D–E), whereas Bapx1 expression in the embryonic hindstomach is severely compromised with Barx1 loss (F,G; arrows and dashed outlines mark hindstomach mesenchyme). Bapx1 expression was also observed in wild-type spleen (F, arrowhead and dashed outline), but loss of this signal occurred selectively in hindstomach; somites, another prominent site of Bapx1 expression (G inset, small arrows), were unaffected. (H,I) Confirmation of loss of Bapx1 expression by qRT-PCR and immunoblotting. RNA was isolated from stomachs dissected at multiple developmental stages and normalized to Gapdh levels (H) or Bapx1 protein levels were examined in E13.5 stomachs by immunoblot analysis (I). Bapx1 expression was virtually undetectable by either method in Barx1−/− stomachs.