Abstract
Free full text

HMP Binding Protein ThiY and HMP-P Synthase THI5 are Structural Homologues†,‡
Abstract
The ATP-binding cassette transporter system ThiXYZ transports N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine (FAMP), a thiamin salvage pathway intermediate, into cells. FAMP is then converted to 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) and recycled into the thiamin biosynthetic pathway. ThiY is the periplasmic substrate binding protein of the ThiXYZ system and delivers the substrate FAMP to the transmembrane domain. We report the crystal structure of Bacillus halodurans ThiY with FAMP bound at 2.4 Å resolution determined by single-wavelength anomalous diffraction phasing. The crystal structure reveals that ThiY belongs to the group II periplasmic binding protein family. The closest structural homologues of ThiY are periplasmic binding proteins involved in alkanesulfonate/nitrate and bicarbonate transport. ThiY is also structurally homologous to thiamin binding protein (TbpA) and to thiaminase-I. THI5 is responsible for the synthesis of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate in the thiamin biosynthetic pathway of eukaryotes and shares approximately 25% sequence identity with ThiY. A homology model of Saccharomyces cerevisiae THI5 was generated based on the structure of ThiY. Many features of the thiamin pyrimidine binding site are shared between ThiY and THI5 suggesting a common ancestor.
Thiamin pyrophosphate (ThDP) is an essential cofactor in all living systems and is involved in branched-chain amino acid synthesis and carbohydrate metabolism. ThDP is implicated in the stabilization of an acyl carbanion intermediate formed in a variety of enzymatic reactions. Bacteria and Archaea, as well as certain lower eukaryotes, synthesize thiamin. Higher eukaryotes cannot produce thiamin and require it as a dietary supplement. Deficiency of thiamin in humans causes beriberi and Wernicke-Korsakoff syndrome (1). In all thiamin biosynthetic pathways, the thiazole and pyrimidine moieties of thiamin are synthesized separately and linked to form thiamin monophosphate (ThMP). The thiamin biosynthetic pathway is well studied in prokaryotes and the details of the eukaryotic pathway are beginning to emerge (2-5). In prokaryotes, the thiazole ring is synthesized from 1-deoxy-D-xylulose 5-phosphate, cysteine, and glycine (or tyrosine in Gram negative bacteria) by five gene products. The pyrimidine ring is synthesized by ThiC, a radical SAM enzyme, from 5-aminoimidazole ribonucleotide (4). In eukaryotes, the thiazole ring is formed from nicotinamide adenine dinucleotide, glycine and cysteine by one enzyme, THI4. The mechanistic details for the formation of the thiazole moiety are beginning to emerge and several steps of the reaction have been biochemically and structurally characterized (5, 6). The pyrimidine moiety is synthesized from histidine and pyridoxal 5′-phosphate by THI5 (7-9). The THI5 family of enzymes has a conserved CCCXC motif; however, no mechanistic or structural details are available for this complex reaction.
In addition to biosynthesis, most organisms have a thiamin uptake system (10-12). In Escherichia coli, thiPBQ encodes an ABC transporter that imports thiamin, thiamin monophosphate (ThMP) and ThDP into the cell (13, 14). The crystal structure of the periplasmic thiamin binding protein (TbpA), with ThMP bound, reveals that the protein belongs to the group II periplasmic binding protein (PBP) family. In addition, the structural similarity of TbpA with thiaminase-I, a thiamin-degrading enzyme, suggests that these proteins evolved from a common ancestor (15).
Binding proteins in Gram-negative bacteria are found in the periplasm; however, in Gram-positive bacteria, which lack an outer membrane, binding proteins are normally tethered to the cell membrane through acylation of a cysteine residue in a lipoprotein signal sequence (16, 17). ThiY contains a cysteine residue at position 19, which is preceded by a hydrophobic N-terminus. Cys19 is conserved in ThiY orthologs from other Bacillus species and is believed to provide the membrane anchor for ThiY. The hydrophobic lipoprotein signal sequence was removed for structural studies.
Thiamin is degraded in basic soil to N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine (FAMP) (18). In Bacillus halodurans, FAMP is transported into the cell by the ABC transporter encoded by thiXYZ where it is deformylated and hydrolyzed to HMP, which is then incorporated into the thiamin biosynthetic pathway (Scheme 1). ThiY is the periplasmic FAMP binding component of the ABC transporter. Here we report the 2.4 Å resolution crystal structure of the B. halodurans ThiY/FAMP complex and compare the structure with those of the thiamin related enzymes TbpA and thiaminase-I. We also report a homology model for THI5, the HMP-phosphate (HMP-P) biosynthetic protein from yeast, based on the structure of ThiY. The modeling studies predict that the pyrimidine binding site is conserved in ThiY and THI5, suggesting that these proteins evolved from a common ancestor. The model also provides some preliminary insights into HMP-P biosynthesis in eukaryotes.
Materials and Methods
Protein Expression and Purification
The construction of the B. halodurans ThiY overexpression strain was described previously (18). In this construct the N-terminal 20 amino acids were deleted to remove a hydrophobic membrane anchor and increase protein solubility. The gene was cloned into the pET16b plasmid and transformed into a methionine auxotrophic strain B834(DE3) of E. coli. A starter culture (10 mL) was grown overnight at 37 °C in LB medium containing 100 mg/mL ampicillin. The starter culture was harvested by centrifugation at 2000 rpm for 15 min, resuspended in 50 mL of minimal medium, and then transferred to 1 L of minimal medium. The minimal medium contained M9 salts supplemented with 40 mg/mL of each amino acid (except for methionine, which was replaced with L-selenomethionine), 100 mg/mL ampicillin, 0.4 % glucose, 2 mM MgSO4, 25 mg/mL FeSO4·7H2O, 0.1 mM CaCl2 and 1 % MEM vitamin solution. The cells were grown to an OD600 of 0.6 at 37 °C. Protein expression was induced by addition of 1 mM isopropyl-1-thio-β-D-galactopyranoside and the temperature was reduced to 15 °C. Cells were harvested after 15 h and stored at -80 °C.
The cell pellet was thawed and resuspended in wash buffer (20 mM tris(hydroxymethyl)aminomethane (Tris), pH 7.5, 150 mM NaCl, 10 mM imidazole and 2 mM β-mercaptoethanol). The cells were lysed by sonication and centrifuged at 24000g for 30 min to separate the lysate from the cell debris. The lysate was passed through a Ni-NTA column pre-equilibrated with wash buffer. The column was washed with 150 mL wash buffer followed by 50 mL of wash buffer containing 35 mM imidazole. The protein was eluted with wash buffer containing 200 mM imidazole and passed through a Sephadex G-75 column pre-equilibrated with 20 mM Tris, pH 7.5, 150 mM NaCl and 3 mM dithiothreitol. Fractions containing protein were identified by UV absorbance at 280 nm and were collected, pooled, concentrated to 10 mg/mL, and stored in aliquots at -80 °C.
Crystallization Conditions
The protein was thawed and incubated with 3-4 molar excess of FAMP for 2 h prior to crystallization. The crystals were grown using the hanging drop vapor diffusion method at 18 °C in 3 M NaCl, 0.1 M sodium acetate, pH 4.5 and 0.1 M lithium sulfate. Clusters of very thin plates with dimensions of 0.15 mm × 0.1 mm × 0.005 mm grew in a week.
Data Collection and Processing
All data were collected at NE-CAT beamline 24-ID-E at the Advanced Photon Source. Crystals were transferred to well solution containing 25% glycerol and flash frozen under liquid nitrogen prior to data collection. The crystals were very thin and were sensitive to radiation damage upon exposure to the X-ray beam. Partial data sets were obtained by exposing each of two crystals multiple times at various spots on the crystal (11 total positions). The partial datasets were merged to obtain a complete, 2.8 Å resolution dataset, which was used for initial structure determination. A second higher resolution (2.4 Å) dataset was obtained later by exposing a larger plate at six different positions. A total of 110° was covered with an exposure time of 1 s per frame and a crystal to detector distance of 300 mm. This dataset was used for final refinement. All data were indexed, integrated and scaled using the HKL2000 program suite (19). Data collection statistics are summarized in Table 1.
Table 1
Data collection statistics for ThiY
| Dataset 1 | Dataset 2 | |
|---|---|---|
| Wavelength (Å) | 0.9792 | 0.9792 |
| No. of crystals | 2 | 1 |
| Space Group | P212121 | P212121 |
| a (Å) | 60.0 | 59.9 |
| b (Å) | 97.8 | 97.0 |
| c (Å) | 105.1 | 104.9 |
| Resolution (Å) | 2.80 | 2.40 |
| Total/Unique reflections | 135105/29248 | 94804/23837 |
| Redundancy | 4.6(4.0) | 4.0(2.9) |
| Completion (%) | 99.8(98.6) | 96.5(87.6) |
| I/σ | 10.3(3.6) | 7.7(2.2) |
| aRsym(%) | 16.8(39.8) | 19.2(42.2) |
| Matthews no. (Å3/Da) | 2.24 | 2.21 |
| Solvent content (%) | 43.1 | 43.4 |
Values in parenthesis are for the highest resolution shell
Structure Determination and Refinement
Analysis of the unit cell contents suggested two monomers of ThiY in the asymmetric unit, with six methionine residues per monomer excluding the N-terminal methionine residues. Ten initial Se sites were obtained using the program SHELXC (20, 21). The sites were input to the AUTO_SOL program in PHENIX (22); however, the resulting map was of poor quality because of the very thin crystals and because of pseudotranslational symmetry resulting from the twofold noncrystallographic symmetry. Therefore, the Fold and Function Assignment system server (23) was used to identify SsuA, an alkanesulfonate/nitrate binding protein, from Xanthomonas axonopodis (PDB ID 3E4R) as the closest structural homologue of ThiY, and a truncated model of SsuA was used for molecular replacement. The SAD phases and molecular replacement phases were combined using the program PHASER_EP (24) and the program RESOLVE (25) was used for density modification. These phases were used to locate the two remaining Se sites. The 12 Se site SAD phases and molecular replacement phases were combined, improved by density modification and non-crystallographic symmetry averaging, and used to calculate an interpretable electron density map. Model building was performed using the program COOT (26) and the model was refined using the CNS program suite (27). The model obtained from 2.8 Å SAD phasing was refined against the 2.4 Å dataset, which became available after the initial model building. The refinement process included successive rounds of simulated annealing, minimization, B-factor refinement, calculation of composite omit maps, difference Fourier maps and model building. A Fo- Fc difference Fourier map was used to identify FAMP bound to the enzyme. The ligand was added to the model followed by another round of refinement and identification of water molecules. The parameter and the topology files for FAMP were generated using the HIC-UP server (28). The final refinement statistics are given in Table 2.
Table 2
Refinement statistics for ThiY
| Resolution (Å) | 2.4 |
| aR | 20.3 |
| bRfree | 26.4 |
| No of non-H atoms | |
| Protein | 4648 |
| Ligand | 24 |
| Water | 259 |
| B-factors | |
| Protein (Å2) | 22.9 |
| Ligand (Å2) | 13.0/15.2 |
| rms deviations | |
 bonds (Å) bonds (Å) | 0.007 |
 angles angles | 1.3 |
 dihedrals dihedrals | 22.5 |
| Ramachandran plot | |
 Most favored region (%) Most favored region (%) | 91.9 |
 Additional favored region (%) Additional favored region (%) | 8.1 |
 Generously allowed region (%) Generously allowed region (%) | 0 |
 Disallowed region (%) Disallowed region (%) | 0 |
Homology Modeling of THI5
The sequence alignment of Saccharomyces cerevisiae THI5 and B. halodurans ThiY was obtained using the program ClustalW (29). The sequence alignment showed three main areas of gaps/insertions: (1) THI5 contains an N-terminal deletion of about two dozen residues corresponding to the lipoprotein signal sequence of ThiY, (2) THI5 contains an insertion of nine residues near the conserved CCCXC motif, which is absent in ThiY, and (3) ThiY contains a deletion of about two dozen residues near the C-terminus; however, this deletion is difficult to position because of low sequence identity. The program MODELLER 9v3 was used to generate five homology models followed by energy minimization of each model based on spatial restraints (30). The model with the lowest energy was considered for further analysis. In this model, the conserved CCCXC motif was located in a cavity near the predicted THI5 active site, suggesting a [4Fe-4S] cluster or some other metal center. A [4Fe-4S] cluster was manually placed in the cavity and residues Cys195, Cys197, and Cys199 were reoriented to make covalent bonds between the sulfur and Fe atoms. The resulting structure was energy minimized to a gradient of 0.01 kJ/mol in vacuo using the AMBER* force field in the program Macromodel (31-33). The minimization used a TNCG minimization technique and a distance dependant 4r dielectric treatment (34). All the residues located within 5 Å of the [4Fe-4S] cluster were allowed to move during minimization and the rest of the protein remained frozen.
Results
Overall Structure
ThiY crystallized in the space group P212121 with unit cell dimensions a = 59.9 Å, b = 96.5 Å, c = 104.9 Å and one dimer per asymmetric unit. The ThiY protein used for crystallization had an N-terminal 20 amino acid deletion. In addition, residues 21-29 were disordered in the crystal structure. Electron density was present for all other amino acid residues, which were included in the final model. The crystal structure reveals that ThiY belongs to the group II PBP family. The monomer is comprised of two domains. Domain 1 is a three layer αβα sandwich comprised of a mixed β-sheet of five β-strands with topology ↑β4↓β10↑β3↑β1↑β2. The β-sheet is flanked by ten α-helices and one 310-helix. Domain 2 is a three layer αβα sandwich comprised of a mixed β-sheet containing five β-strands with topology ↑β7↓β6↑β8↑β5↑β9. This β sheet is flanked by three α-helices and four 310-helices. The two domains are connected by crossover loops between strands β4 and β5 and between helix η5 and strand β10. A ribbon drawing of the ThiY monomer and the topology diagram showing connections between the secondary structural elements are shown in Figure 1.
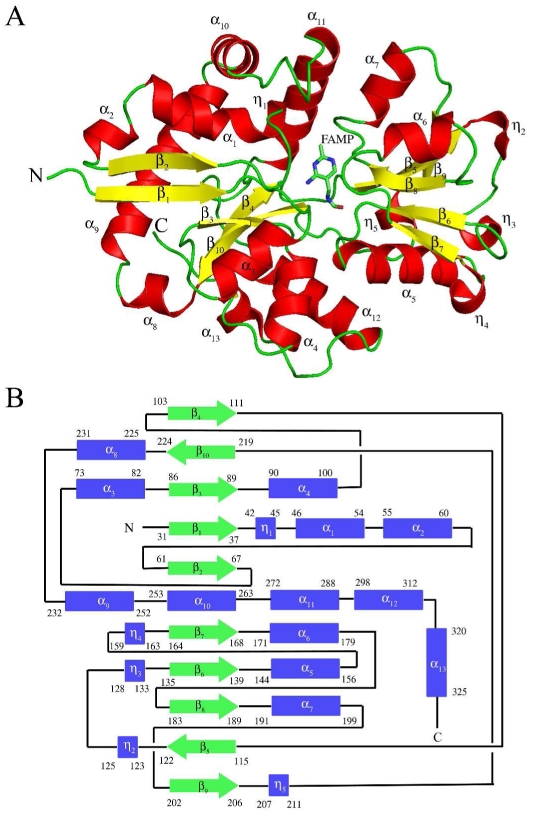
Monomeric structure of ThiY. (A) The cartoon diagram of ThiY with the secondary structure elements labeled. The 310 helices are labeled as η. FAMP is shown as sticks. Carbon atoms in FAMP are colored green, nitrogen atoms are colored blue and oxygen atoms are colored red. (B) The topology diagram of ThiY showing the domain organization.
Dimeric Structure
The enzyme crystallizes as a tightly packed dimer with monomers related by noncrystallographic twofold symmetry (Figure 2). The dimer interface is primarily hydrophilic and stabilized by extensive hydrogen bonding between residues Thr70-Asp167′, Asn71-Tyr139′, Thr136-Asn268′, Asp167-Phe170′, Arg180-Asn268′, Arg180-Phe170′ (where ′ denotes a residue from the other monomer). The dimer buries 1810 Å2 of surface area and the formation of the dimer seals off the entry/exit path for the ligand.
Ligand Binding Site
The ligand binding site of ThiY is located in a cleft between domain 1 and domain 2. FAMP was added during crystallization and was bound in the crystal structure. A stereoview of FAMP in the binding site is shown in Figure 3A and a schematic of interactions of FAMP in the binding site is shown in Figure 3B. The pyrimidine ring of FAMP is sandwiched between the side chains of Trp39 and Tyr188. Residues Phe170 and Phe269 interact edge-to-face with Trp39. Residue Tyr40 interacts edge-to-face with Phe170. The N2 atom of FAMP is hydrogen bonded to Glu192 and the N1 atom is hydrogen bonded to a water molecule. The water molecule is hydrogen bonded to the side chains of Asp38 and Asn42. The side chain of Glu192 is stabilized by hydrogen bonds to the amide group of Tyr188 and a water molecule. The amino group (N3) of FAMP hydrogen bonds to side chain atoms of Asp38 and Tyr90. The carbonyl terminus of FAMP hydrogen bonds to the amide group of Asn145. The hydroxyl group of Tyr188 hydrogen bonds to the N4 atom of FAMP and to the carboxylate group of Glu218. The hydroxyl group of Tyr188 is also at a distance of 2.85 Å from the CE1 atom of Tyr90 in chain B. Residues Trp39 and Phe170 appear to act as a gate covering the entry/exit site for FAMP. Residues Trp39, Phe170, Tyr40 and Phe269 form a hydrophobic pocket shielding the binding site from external solvent.
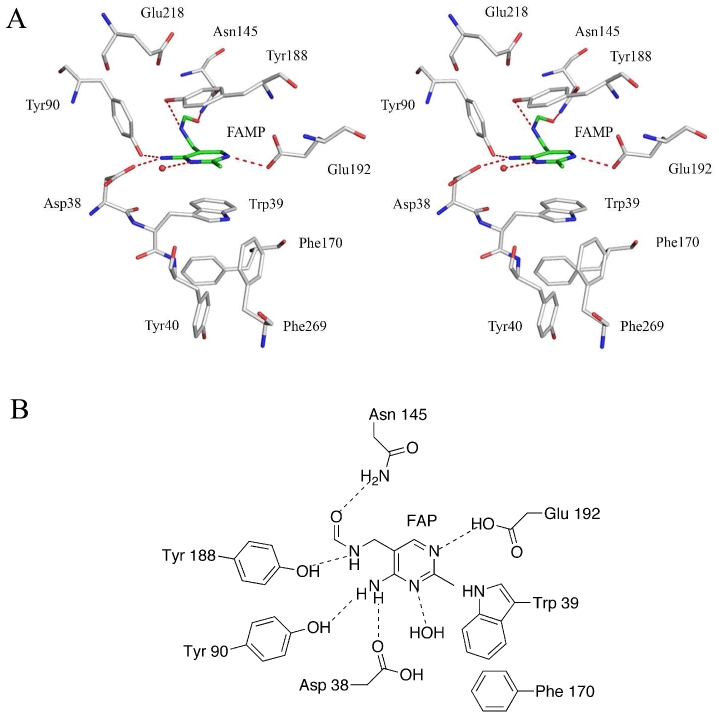
FAMP binding site in ThiY. (A) Stereoview of the FAMP binding site. FAMP has carbon atoms colored green. Water molecules are shown as red spheres and hydrogen bonds are shown as dashed lines. (B) Schematic representation of the binding site showing the key hydrogen bonding and stacking interactions.
Homology Modeling of THI5
A homology model of yeast THI5 was generated based on its sequence similarity to ThiY. The sequences are 25% identical and 47% similar. The sequence alignment of ThiY and THI5 shows that the residues involved in binding the pyrimidine ring of FAMP in ThiY are partially conserved in THI5. A superposition of THI5 and ThiY/FAMP suggests that the HMP-P binding residues in THI5 are Trp12, Asn11, Gln165, His18, and Thr15. Residues Asn11 and Trp12 in THI5 align with Asp38 and Trp39 of ThiY, respectively. Residue Gln165 in THI5 aligns with Glu192 of ThiY, which is involved in hydrogen bonding to the N1 atom of FAMP. Residue His45 is hydrogen bonded to Asp38 in ThiY and is also conserved in THI5. These residues are critical to binding FAMP in ThiY and are predicted to correspond to the HMP-P binding site in THI5.
The homology model of THI5 is similar to the group II PBPs with two distinct domains connected by two crossover loops. The conserved CCCXC motif is located in one of the crossover loops between domain 1 and domain 2 and is located 15 -20 Å away from Trp12. A [4Fe-4S] cluster was built in the cavity between the domains facing the active site. The cavity between Trp12 and the [4Fe-4S] cluster is lined with residues Asn11, Gln164, Gln165, Phe117, His66, Glu161, and Gln121.
Discussion
PBPs and ThiY
ATP-binding cassette (ABC) transporters are responsible for the transportation of a variety of ligands across the cell membrane. The importers and the exporters are believed to have diverged very early from a common ancestor. The importers are generally found in prokaryotes and are comprised of a periplasmic substrate binding domain and two cytosolic ATP binding domains. The exporters consist of a transmembrane domain and an ATP binding domain and recruit their ligands directly from the cytoplasm (35). The role of PBPs is to deliver small molecules and ions to the membrane-bound transporter for entry into the cell.
The crystal structures of many of the PBPs have been determined and strong structural homology suggests a common ancestor for this family of proteins. The PBPs are comprised of two domains connected by variable linker regions. This two domain architecture, which is believed to arise from gene duplication, is present in all PBPs. The PBPs have evolved into three different classes characterized by the number of crossovers between the two domains and the topological arrangement of the central β-sheet in the domains. Group I PBPs have two α/β type domains with the β sheet topology of β2β1β3β4β5 and three crossovers between the domains. Group II PBPs have two domains with the β sheet topology of β2β1β3βnβ4 with two crossovers between the domains. The strand βn represents a strand immediately after the crossover from the other domain. Group III PBPs have one crossover between the domains and little is known about the topological arrangement of the β-sheets in the domains. The substrate binding site in all PBPs is located between the domains and involves residues from both domains. These domains undergo a hinge-like bending motion (“Venus flytrap mechanism”) during ligand binding (36, 37).
ThiY has two crossover connections between domain 1 and domain 2 revealing that it belongs to the group II PBP family. The topology of the central β sheet in both the domains is in accordance with the group II PBP family. The ThiY construct lacks the first 20 residues (hydrophobic membrane anchor) and an additional nine N-terminal residues were not visible in the electron density. The enzyme binds FAMP in the binding site located between domain 1 and domain 2. FAMP makes extensive stacking and hydrogen bonding interactions with the protein and is shielded from the environment by residues Trp39, Phe170, Tyr40, and Phe269. ThiY crystallizes as a dimer in the asymmetric unit and this dimerization closes the entry/exit path for the ligand. A similar dimeric arrangement was observed in the thiamin binding protein TbpA. In this case the dimerization also occurs in solution and is dependent on the concentration of the ligand (15). The physiological relevance of the TbpA dimerization is unclear. The dimerization of ThiY may be related to the mechanism of delivery of substrate to the ABC transporter or may be an artifact of crystallization.
Substrate Specificity of ThiY
The ThiXYZ transporter is found in a gene cluster involved in the salvage of FAMP, which is hydrolyzed to HMP for thiamin biosynthesis. The role of ThiY is delivery of FAMP to the ABC transporter and the equilibrium binding constant for FAMP is 200 nM (18). Our structure provides the atomic details of this binding. Tyr188 and Asn145 are involved in binding the formylamino group of FAMP through hydrogen bonding. Tyr188 is also involved in a stacking interaction with the pyrimidine ring. A sequence alignment of diverse ThiY homologues reveals that Tyr188 and Asn145 are not strictly conserved. The tyrosine residue is sometimes replaced by a phenylalanine residue, and the asparagine residue is replaced with a variety of residues such as serine, aspartate and glutamate. Residues Glu192, Trp39, Asp38 and Tyr90 that interact with the pyrimidine ring of FAMP are strictly conserved. The sequence variability in the ligand binding site suggests that the ThiXYZ system may have evolved to transport a variety of different thiamin degradation products with intact pyrimidines into the cell and therefore suggests the existence of other degradation and salvage pathways for thiamin. In addition to ThiXYZ, other transporters such as YkoEDC and CytX are also likely to be involved in the uptake of HMP or HMP precursors (10).
Comparison of ThiY to Other PBPs
A DALI (38) search identified several structural homologues of ThiY. The closest homologues are SsuAs from X. axonopodis (XaSsuA; PDB ID 3E4R) and E. coli (EcSsuA; PDB ID 2X26) (39), which are alkanesulfonate/nitrate binding proteins, with a Z scores of 25.1 and 24.2, respectively. SsuA belongs to the type II PBP superfamily and XaSsuA has a molecule of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) bound in the active site. A superposition of ThiY and XaSsuA is shown in Figure 4A. Despite low sequence idenity, XaSsuA was successfully used as a molecular replacement model in combination with SAD during initial phasing. The Cα trace of XaSsuA also served as helpful reference during ThiY model building. Other structural homologues of ThiY are hypothetical phosphate binding protein (PDB ID 1ZBM; Z = 21.4), nitrate binding protein (PDB ID 2G29; Z = 22.6) (40), and bicarbonate binding protein (PDB ID 2I4B; Z = 22.2) (41). Most of the top ThiY structural homologues are group II PBPs involved in transport of small ligands; however, MqnD, which is an enzyme involved in menaquinone biosynthesis, shows up in the top ten (PDB ID 3A3U; Z = 22.7) (42).

Structural homologues for ThiY. Comparisons of ThiY with (A) SsuA, (B) thiaminase-I and (C) TbpA. ThiY carbon atoms are colored green and carbon atoms of the compared structure are colored cyan. The ligands are shown in ball and stick. (D) Superposition of the binding sites of ThiY and TbpA. Hydrophobic residues involved in stacking of TMP in TbpA are shown.
Comparison of ThiY with TbpA
Both ThiY and TbpA are transport proteins involved in thiamin salvage. FAMP is the preferred ligand for ThiY, while the preferred ligands for TbpA are thiamin, ThMP and ThDP. A DALI search showed that ThiY and TbpA are structural homologues (PDB ID 2QRY, Z = 10.7) (15). A superposition of ThiY and TbpA (Figure 4C) shows that the ligand binding sites of both proteins are in a similar position. The structure of TbpA was determined with bound ThMP (15) and the binding site residues interacting with ThMP in TbpA are shown in Figure 4D. The thiazole ring stacks between residues Tyr27 and Tyr215 and is positioned edge-to-face from Trp197. The pyrimidine ring is buried in a pocket formed by Trp197, Trp280, Tyr221 and Tyr201 and the phosphate group is hydrogen bonded to Ser161, Trp197, Gly60 and Asp59. A comparison between thiamin and FAMP binding shows that the pyrimidine ring of FAMP superposes on the thiazole moiety of thiamin bound to TbpA and π-stacking is a common feature in the binding of both ligands. The formyl terminus of FAMP is in a similar position to the phosphate of thiamin.
Similarity Between ThiY and THI5
In eukaryotes, HMP-P is synthesized from PLP and histidine in a reaction catalyzed by THI5, the yeast pyrimidine synthase (Scheme 2). The activity of THI5 has not been reconstituted in solution and its mechanism has not been established. Iterated BLAST searches (43) starting with ThiY identified various and proteins annotated as possible periplasmic binding proteins, as well as a few proteins identified as HMP-P synthases. However, all of the proteins identified by this search as HMP-P synthases lacked the conserved CCCXC motif of THI5 and are likely misannotated. Iterated BLAST searches starting with S. cerevisiae THI5 identifed orthologs with the conserved CCCXC motif but also periplasmic binding proteins. The S. cerevisiae THI5 shows 25% sequence identity and 47% sequence similarity to the B. halodurans ThiY, suggesting that ThiY and THI5 are structural homologues. In addition, ThiY binds FAMP, which is structurally similar to HMP-P, the product of THI5. Therefore, we undertook homology modeling in hopes that the THI5 model might provide useful clues for the successful reconstitution of the THI5-catalyzed reaction.
The sequence alignment of ThiY and THI5 shows that the residues involved in binding the pyrimidine ring in ThiY (Asp38, Trp39, Glu192, and His45) are conserved in THI5. In addition, Ser79, Ser82, Tyr113, Asp155, Asn271, Trp304 and Phe209, conserved in diverse sequences of THI5, are also conserved in ThiY. These residues are outside the active site of ThiY and may be involved in maintaining the structural integrity of the protein.
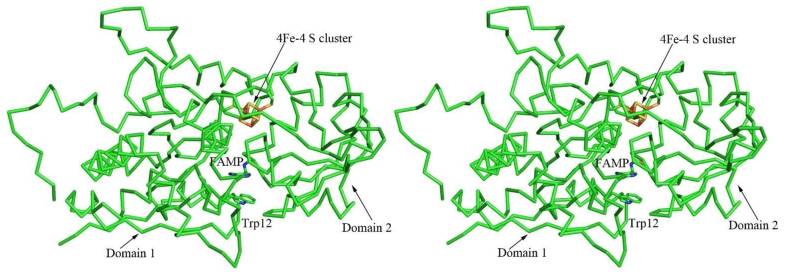
THI5 has a conserved CCCXC motif. Related cysteine rich sequences (e.g., CX3CX2C) bind [4Fe-4S] clusters involved in electron transfer or radical reactions (44) and are also known to serve as Lewis acids (45). The CCCXC motif was included in the homology model to evaluate if THI5 might contain a [4Fe-4S] cluster (or other metal center). The location of the HMP binding site was also predicted to identify residues potentially important for binding or catalysis. The homology model shows that the CCCXC motif is located in a deeply buried cleft between domain 1 and domain 2 (Figure 5). In this model, Cys195, Cys197 and Cys199 serve as ligands for the [4Fe-4S] cluster. The active site (identified from the location of FAMP in the superimposed ThiY) is present ~15 Å away from the cleft. The space between the putative [4Fe-4S] and the HMP-P binding site could be occupied by the histidine and PLP substrates. The [4Fe-4S] cluster is ~13 Å away from Trp12, which is proposed to stack against the pyrimidine ring of the product. While the [4Fe-4S] cluster has reasonable geometry, its occurrence has not been experimentally verified. It is also possible that the cysteine motif could be involved in binding smaller iron sulfur clusters. The residues lining the proposed active site cavity (Asn11, Gln164, Phe117, His66, Glu161, and Gln121) are strictly conserved in THI5 and may play a role in the reaction mechanism. These predictions will be used in developing a strategy to reconstitute the THI5 catalyzed reaction in vitro.
Comparison of ThiY and Thiaminase-I
DALI searches show that ThiY is structurally homologous to thiaminase-I, an enzyme involved in thiamin degradation (PDB ID 4THI) (46), but with a much lower Z score of 5.6 for the full length protein. The homology is comparatively better when only domain I is used with a Z score of 7.1. The superposition of ThiY and thiaminase-I is shown in Figure 4B. The crystal structure of thiaminase-I determined with the mechanism based inhibitor 2,5-dimethylpyrimidin-4-amine shows the pyrimidine moiety covalently attached to Cys113 in the active site (46). The comparison of the structures shows that the binding site for the pyrimidine is in a similar location for both ThiY and thiaminase-I. A closer look at the binding conformations of the substrates reveals that the pyrimidine ring of FAMP is orthogonal to the pyrimidine ring of the inhibitor in thiaminase-I; however, this may result from the covalent attachment to Cys113. The centers of the pyrimidine rings are separated by ~4 Å in the superposition. TbpA and thiaminase-I are structural homologues and believed to have a common ancestor (15). The structure of ThiY described here demonstrates that the thiamin metabolic proteins ThiY, TbpA, thiaminase-I and THI5 are (1) all members of the group II PBP family, and (2) all have binding sites for a pyrimidine moiety derived from thiamin. These similarities suggest that all four proteins may have evolved from a common ancestor.
Acknowledgments
We thank Leslie Kinsland for assistance in the preparation of this manuscript. We thank the NE-CAT staff of the Advanced Photon Source for assistance in data collection and providing beam time. We thank Dr. Jeremiah Hanes for the synthesis of FAMP. We thank Dr. Cynthia Kinsland of the Cornell Protein Production Facility for the clones for ThiY.
Abbreviations
- FAMP
- N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine
- HMP
- 4-amino-5-hydroxymethyl-2-methylpyrimidine
- ABC
- ATP-binding cassette
- PBP
- periplasmic binding protein
- TbpA
- thiamin-binding protein
- SAD
- single-wavelength anomalous diffraction
- NiBP
- nitrate binding protein
- ThMP
- thiamin monophosphate
- ThDP
- thiamin pyrophosphate
- Tris
- tris(hydroxymethyl)aminomethane
- HMP-P
- HMP-phosphate
- PLP
- pyridoxal 5′-phosphate
Footnotes
†This work was supported by NIH grants DK 44083 (T. P. B.) and DK 67081 (S. E. E.). This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. S.E.E. is indebted to the W. M. Keck Foundation and the Lucille P. Markey Charitable Trust.
‡The Protein Data Bank code has been deposited under the accession number 3IX1.
References
Full text links
Read article at publisher's site: https://doi.org/10.1021/bi101209t
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2957805?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1021/bi101209t
Article citations
HbpA from Glaesserella parasuis induces an inflammatory response in 3D4/21 cells by activating the MAPK and NF-κB signalling pathways and protects mice against G. parasuis when used as an immunogen.
Vet Res, 55(1):93, 29 Jul 2024
Cited by: 0 articles | PMID: 39075605 | PMCID: PMC11285476
New chemical and microbial perspectives on vitamin B1 and vitamer dynamics of a coastal system.
ISME Commun, 4(1):ycad016, 10 Jan 2024
Cited by: 2 articles | PMID: 38390520 | PMCID: PMC10881298
Rhizobium determinants of rhizosphere persistence and root colonization.
ISME J, 18(1):wrae072, 01 Jan 2024
Cited by: 1 article | PMID: 38690786 | PMCID: PMC11103875
Thiamine metabolism genes in diatoms are not regulated by thiamine despite the presence of predicted riboswitches.
New Phytol, 235(5):1853-1867, 01 Jul 2022
Cited by: 7 articles | PMID: 35653609 | PMCID: PMC9544697
A bacterial riboswitch class senses xanthine and uric acid to regulate genes associated with purine oxidation.
RNA, 26(8):960-968, 28 Apr 2020
Cited by: 12 articles | PMID: 32345632 | PMCID: PMC7373994
Go to all (13) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (Showing 7 of 7)
-
(1 citation)
PDBe - 2QRYView structure
-
(1 citation)
PDBe - 2X26View structure
-
(1 citation)
PDBe - 2I4BView structure
-
(1 citation)
PDBe - 3IX1View structure
-
(1 citation)
PDBe - 3A3UView structure
-
(1 citation)
PDBe - 4THIView structure
-
(1 citation)
PDBe - 2G29View structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structural similarities between thiamin-binding protein and thiaminase-I suggest a common ancestor.
Biochemistry, 47(5):1346-1357, 05 Jan 2008
Cited by: 26 articles | PMID: 18177053 | PMCID: PMC6040661
Functional characterization of the HMP-P synthase of Legionella pneumophila (Lpg1565).
Mol Microbiol, 115(4):539-553, 03 Nov 2020
Cited by: 1 article | PMID: 33034117 | PMCID: PMC8026769
A new thiamin salvage pathway.
Nat Chem Biol, 3(8):492-497, 08 Jul 2007
Cited by: 56 articles | PMID: 17618314
The genes and enzymes involved in the biosynthesis of thiamin and thiamin diphosphate in yeasts.
Cell Mol Biol Lett, 13(2):271-282, 10 Apr 2008
Cited by: 21 articles | PMID: 18161008 | PMCID: PMC6275658
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCRR NIH HHS (2)
Grant ID: RR-15301
Grant ID: P41 RR015301
NIDDK NIH HHS (6)
Grant ID: R01 DK044083
Grant ID: R37 DK044083
Grant ID: DK 44083
Grant ID: DK 67081
Grant ID: R01 DK067081
Grant ID: R56 DK044083
NIGMS NIH HHS (1)
Grant ID: P41 GM103403