Abstract
Free full text

Contribution of Lysine 11-linked Ubiquitination to MIR2-mediated Major Histocompatibility Complex Class I Internalization*
Abstract
The polyubiquitin chain is generated by the sequential addition of ubiquitin moieties to target molecules, a reaction between specific lysine residues that is catalyzed by E3 ubiquitin ligase. The Lys48-linked and Lys63-linked polyubiquitin chains are well established inducers of proteasome-dependent degradation and signal transduction, respectively. The concept has recently emerged that polyubiquitin chain-mediated regulation is even more complex because various types of atypical polyubiquitin chains have been discovered in vivo. Here, we demonstrate that a novel complex ubiquitin chain functions as an internalization signal for major histocompatibility complex class I (MHC I) membrane proteins in vivo. Using a tetracycline-inducible expression system and quantitative mass spectrometry, we show that the polyubiquitin chain generated by the viral E3 ubiquitin ligase of Kaposi sarcoma-associated herpesvirus, MIR2, is a Lys11 and Lys63 mixed-linkage chain. This novel ubiquitin chain can function as an internalization signal for MHC I through its association with epsin1, an adaptor molecule containing ubiquitin-interacting motifs.
Introduction
The functions of many membrane proteins, including various signal-transducing receptors, are regulated by internalization, following which the receptors are either recycled or degraded in lysosomes (1). Dysregulation of this process is thought to underlie many disorders, including cancer and autoimmunity (2,–5); consequently, it is important to gain an understanding of the molecular mechanisms of receptor internalization in vivo. In this context, polyubiquitination is an important signal for the internalization of mammalian membrane proteins (6,–10). However, it is still unclear how the presence of a polyubiquitin chain regulates the internalization step because it has been reported that monoubiquitination is sufficient to induce protein internalization in yeast (11, 12).
Monoubiquitination creates an isopeptide bond between a lysine of the target protein and the C-terminal glycine of ubiquitin. Polyubiquitination then results from the sequential linkage of additional ubiquitin moieties via specific lysine residues on ubiquitin, a process catalyzed by E3 ubiquitin ligase (E3) (13). All seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) on ubiquitin can be utilized for the formation of polyubiquitin chains (14). Among them, Lys48 and Lys63 have been classically reported as the acceptor lysine residues for the generation of the polyubiquitin chain. The Lys48-linked and Lys63-linked polyubiquitin chains are well established inducers of proteasome-dependent degradation and signal transduction, respectively (15). Recently, it has been reported that lysine residues other than Lys48 and Lys63 can be utilized for chain formation (14). For example, Lys29-linked polyubiquitination of Deltex by Itch/AIP4 and Lys27-linked polyubiquitination of Jun were reported to be important for their lysosomal degradation (16, 17). In addition, Lys11-linked polyubiquitin chains were reported to be involved in cell cycle control by the anaphase-promoting complex and in endoplasmic reticulum-associated degradation of misfolded proteins (18, 19).
Among the several types of polyubiquitin chains, the Lys63-linked chain has been reported as an inducer of protein internalization (6, 8, 10). Recent structural analysis has demonstrated that the adaptor protein epsin1 recognizes Lys63-linked chains through its ubiquitin-interacting motif (UIM)2 (20). Epsin1 contains several other conserved domains in addition to the UIM: an epsin N-terminal homology domain involved in the interaction with phosphatidylinositol 4,5-bisphosphate (21), DPW (Asp-Pro-Trp) repeat domains that include adaptor protein complex AP-2 and clathrin binding sites, and NPF (Asn-Pro-Phe) repeat domains located in the C-terminal region (22). Because epsin1 contributes to clathrin-mediated internalization, it is thought to be involved in polyubiquitination-induced internalization in vivo (23, 24).
To examine how polyubiquitination regulates protein internalization, we previously generated a tetracycline-inducible (Tet-on) system for a member of the MIR family (25, 26). MIR family members are membrane-bound E3 ubiquitin ligases and have been reported to induce ubiquitination-dependent internalization in vivo (25, 26). Use of this Tet-on system allowed us to visualize the detailed status of polyubiquitin chains that induce internalization in vivo (25). Recently, Gygi and colleagues have also successfully developed a detection system for polyubiquitin chains generated in vivo that uses mass spectrometry (27, 28). In the study reported here, we have combined our Tet-on expression system for MIR2 with quantitative mass spectrometry to address the issue of how the polyubiquitin chain contributes to receptor internalization. We report here that the polyubiquitin chain generated by MIR2 is a novel Lys11 and Lys63 mixed-linkage chain and that this mixed polyubiquitin chain is required for efficient epsin1-dependent internalization of major histocompatibility class I (MHC I) molecules.
EXPERIMENTAL PROCEDURES
Materials
Mutants were constructed by overlapping PCR as described previously (25). Each cDNA was subcloned into the p3×FLAG-CMV (Sigma), pcDNA3 (Invitrogen), or pEGFP-N1 (Clontech) vectors. The hemagglutinin-tagged ubiquitin-expressing plasmid and the enhanced green fluorescent protein (EGFP)-tagged ubiquitin-expressing plasmid were gifts from Dr. N. Tanaka and Dr. K. Umebayashi, respectively (29). We generated T-REx-MIR2 as described previously (25). Briefly, the His-Myc-tagged MIR2-expressing cassette used in our previous study (30) was transferred into the pcDNA5/FRT/TO vector. T-REx-MIR2 cells were generated using the Flp-InTM T-RExTM Core kit (Invitrogen). The T-REx-MIR2 cells were transfected with each CD8-MHC I chimera-expressing plasmid constructed in the p3×FLAG-CMV vector and selected by incubation with 2 mg/ml G418 (Sigma) for 3 weeks. The CD8 chimera-expressing population was sorted with the FACSVantage S.E. Turbo.
Internalization Assay and Antibodies
The internalization assay was performed as reported previously (25). In brief, cells were stained with anti-CD8 Ab for 30 min at 4 °C, cultured for the indicated times at 37 °C, and then stained with phycoerythrin-conjugated goat anti-mouse IgG Ab followed by flow cytometry analysis at each time point. The percentage of the CD8 chimera remaining at the cell surface was determined by dividing the values obtained at each time point by the value obtained at time zero. In the case of the expression by ubiquitin mutants, transfected cells were stained as described above, and the GFP-positive cells were analyzed as the exogenous ubiquitin-expressing population. For immunofluorescence microscopy, FITC-labeled anti-CD8 mAb was added to the medium of T-REx-MIR2 cells, and incubation was continued for 10 min at 37 °C. After washing, the cells were analyzed by immunofluorescence microscopy. The mAbs W6/32 for MHC I, RPA-T8 for CD8, and TU36 for MHC II used for FACScan analysis were obtained from BD Biosciences. M2 anti-FLAG Ab (Sigma), F7 anti-HA Ab (Santa Cruz Biotechnology), FK2 anti-ubiquitin Ab (AFFINITI Research Products), Apu3 anti-Lys63 Ab (Millipore), and anti-V5 Ab (Invitrogen) were used for immunoprecipitation and/or immunoblot analysis.
Detection of Ubiquitinated Surface CD8 Chimeras and Immunoprecipitation
To examine the ubiquitination status of CD8 chimeric molecules that preexist at the plasma membrane before initiation of the ubiquitination process by MIR2, surface molecules of CD8-chimera-expressing T-REx-MIR2 cells were biotinylated followed by incubation with doxycycline (Dox) as described previously (25). The cell pellet was boiled in 1% SDS-containing radioimmune precipitation assay buffer (10 mm Tris (pH 7.5), 1% Nonidet P-40, 0.1% sodium deoxycholate, 0.15 m NaCl, 1 mm EDTA (pH 8.0)) and diluted 10-fold with SDS-free radioimmune precipitation assay buffer. CD8 chimeric molecules were precipitated with M2 anti-FLAG Ab-conjugated Sepharose (Sigma) and then eluted with FLAG peptide (150 μg/ml). The eluted CD8 chimera was further purified with streptavidin-agarose (Pierce). For immunoprecipitation, cells were harvested and lysed with Nonidet P-40 buffer (0.15 m NaCl, 1% Nonidet P-40, and 50 mm HEPES buffer (pH 8.0)) containing protease inhibitors. Immunoprecipitation was performed with the indicated antibody together with 30 μl of protein A/G-agarose beads (Santa Cruz Biotechnology). A GST pulldown assay is described in the supplemental “Experimental Procedures”.
Mass Spectrometric Analysis of MHC I Ubiquitination
T-REx-MIR2 cells (12 × 107) were incubated with or without Dox (1 μg/ml) for 5 h in 10% FBS-containing RPMI 1640 medium (Sigma) to induce ubiquitination of MHC I in the presence of bafilomycin A1 (2 μm). The cells were lysed with Nonidet P-40 lysis buffer containing 200 mm NaCl, protease inhibitors, and 10 mm N-ethylmaleimide. Ubiquitinated CD8-MHC chimeras were purified by a two step method: the first purification was performed with anti-FLAG M2 Ab-conjugated agarose, and the second with FK2-conjugated agarose. The ubiquitinated CD8-MHC I chimeras were eluted with 0.1 m glycine-HCl (pH 2.7) and concentrated by precipitation with methanol/chloroform. To ensure that each sample contained equivalent amounts of chimeric protein, 1% of the sample purified by anti-M2 agarose affinity chromatography was examined by Western blotting with HRP-conjugated M2 antibody. In the case of analysis by SDS-PAGE, specific bands obtained from 4 × 108 T-REx-MIR2 cells were visualized in the gels by staining with Coomassie Blue, and the proteins in the excised gel slices were digested with trypsin. The resulting peptides were labeled with iTRAQ114 of multiplex iTRAQ reagents (iTRAQ reagent kit; Applied Biosystems). In the case of the analysis without SDS-PAGE, purified ubiquitinated samples from Dox(+)- and Dox(−)-treated cells were directly digested with trypsin and labeled with iTRAQ115 and iTRAQ114, respectively. To quantify the amount of each peptide derived from all types of polyubiquitin chains, diglycine signature peptides corresponding to the seven ubiquitin chain linkages were synthesized and labeled with iTRAQ117 for use as internal standards (Peptide Institute, Inc.). Labeled internal standards (1 pmol or 200 fmol) were added to the digested and labeled samples and desalted with EmporeTM Extraction Disk C18 Tips. Desalted samples were separated on a MALDI plate by a direct nano-LC system equipped with a nano-LC/MALDI spotting device (DiNa Map System; KYA Technologies). MALDI plates were analyzed using the ABI 4800 MALDI-TOF/TOF Proteomics Analyzer. To acquire each MS spectrum, signature-ion peaks corresponding to 114.1, 115.1, and 117.1 were obtained by using Protein Pilot software 2.0 (Applied Biosystems). The amount of peptide corresponding to each type of polyubiquitin chain was calculated based on the signals from the internal standards.
Knockdown by RNAi
Inhibition of E2 ubiquitin-conjugating enzymes was performed by retroviral transduction of shRNAs targeting UbcH5b/c as described previously (25). For knockdown of epsin1 and clathrin heavy chain, HeLa, HeLa-MIR2, and T-REx-MIR2 cells were transfected three times at 24-h intervals with 10 pmol of siRNA/well with HiPerFect Transfection Reagent (Qiagen) and analyzed 48 h after the third transfection. Epsin1 (5′-GAACTGGCGTCACGTTTAC-3′) and clathrin heavy chain (5′-GCAATGAGCTGTTTGAAGA-3′) mRNA target sequences were purchased from Qiagen. AllStars Negative Control siRNA was purchased as a control siRNA from Qiagen.
RESULTS
Effective Internalization of MHC Class I by Polyubiquitination in T-REx-MIR2 Cells
To examine how the polyubiquitin chain contributes to protein internalization, we used the MIR2 protein of Kaposi sarcoma-associated herpesvirus because MIR2 is known to reduce the surface expression of MHC class I (MHC I) proteins by inducing their ubiquitination-mediated internalization (31, 32). The Tet-on system for MIR2 expression, designated T-REx-MIR2, was generated as reported previously (25). To facilitate a detailed analysis of the ubiquitination and internalization of MHC I, we expressed a FLAG-tagged CD8 chimera containing the transmembrane and cytoplasmic regions of the MHC I α chain, designated CD8-MHC I, in T-REx-MIR2 cells as described previously (30). We selected these particular regions of the class I α chain for inclusion in the chimeric protein because it has been reported that CD8 chimeras containing both the transmembrane and cytoplasmic regions of MIR E3 ubiquitin ligase substrates are internalized and ubiquitinated just like the authentic endogenous substrate molecules. This seems to be the case because the transmembrane region of the substrate is thought to interact with MIRs, whereas the cytoplasmic region of the substrate contains the MIR ubiquitination site (25, 31, 32). The Dox-induced expression of MIR2 suppressed the surface expression of MHC I proteins but not of MHC class II proteins (Fig. 1A). As previously shown by the analysis of endogenous MHC I with MIR2 (33), the MIR2-mediated down-regulation of CD8-MHC I was mediated through the ubiquitination of cytoplasmic lysine residues; a mutant chimeric protein in which all class I cytoplasmic lysine residues were mutated to arginine was not ubiquitinated or down-regulated by MIR2 (Fig. 1, B and C). These results demonstrate that the CD8-MHC I chimera is a valid surrogate for native MHC I.

Ubiquitination is required for MHC I down-regulation in T-REx-MIR2 cells. A, after being incubated with or without Dox for 24 h, the expression level of MHC I and MHC II on T-REx-MIR2 cells was examined by flow cytometry. Data from the cells incubated with (open histograms) and without (shaded histograms) Dox are shown. B, the expression level of the indicated CD8 chimeras on T-REx-MIR2 cells was examined as in A. C, whole cell lysates extracted from the indicated CD8 chimera-expressing T-REx-MIR2 cells that had been cultivated with (+) or without (−) Dox for 6 h were incubated with an anti-FLAG Ab. Precipitated (IP) samples were probed (IB) with anti-FLAG M2 Ab (FLAG) or anti-ubiquitin FK2 Ab (Ubi). D, surface CD8-MHC I was labeled with anti-CD8 Ab after the cells were incubated with (+) or without (−) Dox for 6 h. After cultivation at 37 °C for the indicated times, the expression of the remaining surface CD8-MHC I was examined by staining with phycoerythrin-conjugated goat anti-mouse IgG. At each time point, the percentage of remaining CD8-MHC I was calculated relative to the value of labeled CD8-MHC I at 0 min (upper panel). In the lower panel, T-REx-MIR2 cells were incubated with Dox for 6 h, and FITC-conjugated anti-CD8 Ab was added for the last 10 min of culture. Internalized CD8-MHC I was observed by fluorescence microscopy. E, surface molecules on T-REx-MIR2 cells were biotinylated, and the cells were incubated with (+) or without (−) Dox for 6 h in the presence of 2 μm of bafilomycin A1. The biotinylated CD8-MHC I was sequentially purified with anti-FLAG Ab and streptavidin-agarose. Each sample was probed with FK2 Ab or M2 Ab.
Next, we examined whether surface-expressed MHC I is ubiquitinated and down-regulated by MIR2. The fate of surface-expressed CD8-MHC I was examined by surface biotinylation and staining with antibody as performed in our previous studies (25). Compared with the slow turnover of surface MHC I molecules in noninduced cells, Dox-induced MIR2-expressing cells showed a much more rapid reduction (Fig. 1D). Concomitantly, the Dox-induced cells showed a marked increase in the ubiquitination of surface-expressed CD8-MHC I (Fig. 1E). To examine the relationship between ubiquitination and down-regulation of surface-expressed CD8-MHC I further, we used small interfering RNA to inhibit expression of UbcH5b/c, reported to be an E2 enzyme of the MIR family of E3 ligases (25, 31). Although ubiquitination could not be completely blocked, down-regulation of surface-expressed CD8-MHC I was significantly inhibited (supplemental Fig. S1). At present, we cannot explain why incompletely blocked ubiquitination could still significantly inhibit the down-regulation of CD8-MHC I. UbcH5b/c may support the ubiquitination of other molecules that contribute to the down-regulation of CD8-MHC I. Indeed, several adaptor molecules involved in ubiquitination-mediated internalization, e.g. β-arrestin, have been reported to be monoubiquitinated (34). Thus, these results strongly suggest that surface-expressed CD8-MHC I is down-regulated by MIR2-mediated ubiquitination in T-REx-MIR2 cells.
We next examined the sites on MHC I that are responsible for the MIR2-mediated down-regulation. MHC I has three lysine residues (Lys335, Lys340, and Lys364) in its cytoplasmic tail, therefore we mutated these candidate ubiquitination sites to arginine as shown in Fig. 2A. Each CD8-MHC I mutant was stably expressed in T-REx-MIR2 cells, and the ubiquitination status and down-regulation of the surface-expressed chimeric proteins were analyzed. Although there was some background ubiquitination of Lys340 and Lys364 in the absence of Dox, presumably due to Dox-independent MIR2 expression, Dox-induced MIR2 ubiquitinated Lys335 and Lys340 but only minimally ubiquitinated Lys364 (Fig. 2B). MHC I was only down-regulated when Lys335 was ubiquitinated (Fig. 2C). To confirm the contribution of Lys335 to the ubiquitination-mediated down-regulation of MHC I, we made another mutant in which Lys335 was mutated to arginine but the other lysines were left intact (K335R). As shown in supplemental Fig. S2, this mutant was not significantly down-regulated or ubiquitinated by MIR2. These results demonstrate that ubiquitination at Lys335 is important for the down-regulation of MHC I by MIR2.

Ubiquitination at Lys335 is a critical event for MHC I down-regulation by MIR2. A, protein sequences of the cytoplasmic region of each CD8 mutant chimera are aligned. B, the indicated CD8-mutant chimera-expressing T-REx-MIR2 cells were incubated with Dox for 6 h in the presence of 2 μm bafilomycin A1, and the ubiquitination status of the chimeras was examined as in Fig. 1E. C, after being incubated with or without Dox for 9 h, the expression level of the indicated CD8 chimeras on T-REx-MIR2 cells was examined by flow cytometry. Data from the cells incubated with (open histograms) and without (shaded histograms) Dox are shown (upper left). In the upper right panel, each surface-expressed CD8 chimera was labeled with anti-CD8 Ab after the cells were incubated with (+) or without (−) Dox for 6 h. After cultivation at 37 °C for the indicated times, the expression level of the CD8 chimeric proteins remaining on the cell surface was examined as in Fig. 1D. At each time point, the percentage of remaining CD8 chimera was calculated relative to its level at 0 min. In the lower panel, the indicated CD8 mutant chimera-expressing T-REx-MIR2 cells were incubated with Dox for 6 h, and FITC-conjugated anti-CD8 Ab was added for the last 10 min of culture. The internalized CD8 chimera was observed by fluorescence microscopy.
These results strongly suggest that surface-expressed CD8-MHC I is internalized as a result of ubiquitination. To confirm this, we repeated the FACS analysis as performed in Figs. 1D and and22C, but within a shorter time frame, 10 min, which is before the endocytic events take place. Also, we examined whether polyubiquitination or monoubiquitination could induce internalization of CD8-MHC I more efficiently. To make a monoubiquitinated molecule, we constructed a ubiquitin-fused CD8 chimera (termed K-less-UbiK0), in which a lysine-less ubiquitin (UbiK0) was fused to the C-terminal portion of CD8-MHC I-K-less (Fig. 3A). As shown in Fig. 3B, surface-expressed Lys335 was significantly down-regulated within the first 5 min, but only when MIR2 was expressed. By contrast, surface-expressed CD8-MHC I-K-less was not down-regulated at all within the first 5 or 10 min even in the presence of MIR2. The monoubiquitinated CD8-MHC I (K-less-UbiK0) was only slightly down-regulated by 10 min, and moreover this down-regulation occurred to an equivalent extent in the presence or absence of Dox (Fig. 3B). These data indicate that polyubiquitination serves as a better internalization signal than monoubiquitination in the context of MIR2. Therefore, T-REx-MIR2 cells and the CD8-MHC I chimera together provide a suitable experimental system to study how MHC I internalization is regulated by its polyubiquitination.

Polyubiquitination is necessary for efficient CD8-MHC I internalization. A, schematic representation of the CD8-chimera fusion proteins used in this study is shown. The extracellular region of CD8 (CD8-Ex) and the transmembrane (TM) and cytoplasmic regions of MHC I (MHC I-Cyto) are indicated. B, surface CD8 chimeras were labeled with anti-CD8 Ab after the cells were incubated with (+) or without (−) Dox for 24 h. After cultivation at 37 °C for the indicated times, the expression of the remaining surface CD8 chimera was examined as in Fig. 1D. At each time point, the percentage of remaining CD8 chimera was calculated relative to the value at 0 min (lower panel). The values are mean ± S.E. (error bars) (n = 3); *, p < 0.05 compared with 335K-Dox(−), K-less-Dox(− or +) and K-less-UbiK0-Dox(− or +). **, p < 0.05 K-less-UbiK0-Dox(+) was compared with K-less-Dox(+).
Epsin1 Is an Adaptor Molecule for MHC I Internalization by MIR2
We next examined how MIR2 induces the internalization of MHC I. Recently, several adaptor molecules (epsin1, EPS15, and EPS15R) have been shown to mediate the interaction between ubiquitinated cargo molecules and clathrin, which is an inducer of internalization (7, 9). We first examined the contribution of clathrin to MIR2-mediated internalization. As shown in supplemental Fig. S3, siRNA-mediated clathrin depletion blocked the down-regulation of CD8-MHC I even though CD8-MHC I was still ubiquitinated. Next, the binding of epsin1, EPS15, and EPS15R to ubiquitinated MHC I was examined. As shown in Fig. 4A and supplemental Fig. S3F, the CD8-MHC I molecules internalized as a result of MIR2-mediated ubiquitination associated efficiently with epsin1, but not with EPS15 or EPS15R. To confirm the association between the UIM domains of epsin1 and the ubiquitinated MHC I, several epsin1 mutants were coexpressed in HeLa cells with MIR2 and CD8-MHC I containing only the Lys335 ubiquitination site in its cytoplasmic tail (CD8-MHC I 335K). All UIMs-containing epsin1 molecules interacted with polyubiquitinated CD8-MHC I (Fig. 4C), and similar results were obtained with endogenous MHC I (supplemental Fig. S4). Because K-less-UbiK0 showed some slight internalization as seen in Fig. 3B, we also examined whether epsin1 could associate with this monoubiquitinated CD8-MHC I and found that it did not (supplemental Fig. S5). These results suggest that epsin1 interacts with polyubiquitinated MHC I more strongly than with monoubiquitinated MHC I and functions as an adaptor molecule for MIR2-mediated internalization.
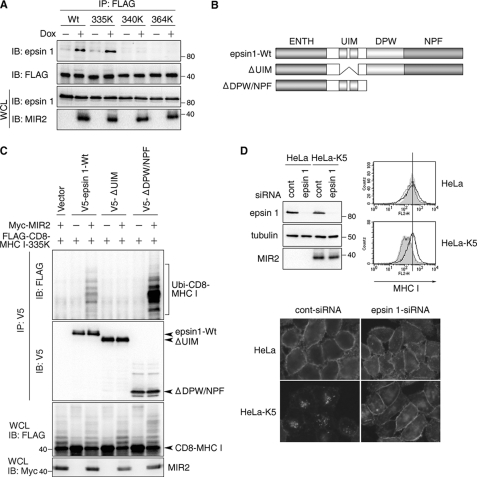
Epsin1 is an adaptor molecule for MIR2-mediated MHC I internalization. A, after incubation with or without Dox for 6 h, protein complexes that included the indicated CD8-MHC I chimeras were isolated from the indicated CD8-MHC I-expressing T-REx-MIR2 cells. The purified protein complexes were examined with anti-epsin1 Ab or M2 Ab. B, schematic representation of the epsin1 mutants used in this study is shown. The UIM, DPW domain, NPF domain, and epsin N-terminal homology domain (ENTH) are indicated. C, each indicated V5-tagged epsin1 was coexpressed with CD8-MHC I 335K and MIR2 in HeLa cells. Protein complexes that included each exogenous epsin1 were purified with anti-V5 Ab and analyzed by Western blotting with M2 and V5 Ab. The expression of CD8-MHC I 335K and MIR2 was examined in whole cell lysates with M2 Ab and anti-Myc Ab, respectively. D, control siRNA (scrambled) or siRNA for epsin1 was transfected into HeLa or MIR2-expressing HeLa cells. Whole cell lysates from the indicated cells were analyzed with anti-epsin1 and anti-tubulin Abs. The expression of MHC I in the indicated HeLa cells was examined by flow cytometry. The gray histogram indicates MHC I expression in nontransfected HeLa cells. The dotted line indicates the MHC I expression in control siRNA-transfected HeLa cells. The solid line indicates the MHC I expression in epsin1 siRNA-transfected HeLa cells. In the lower panel, the indicated transfected cells were cultivated with anti-MHC I Ab (W6/32) for 10 min. Internalized MHC I was observed by fluorescence microscopy as in Fig. 1D.
To test this hypothesis further, we inhibited the expression of epsin1 by siRNA in MIR2-expressing HeLa cells (HeLa-MIR2; Fig. 4D), which have low level expression of MHC I because of its enhanced internalization. The siRNA-mediated inhibition of epsin1 rescued the surface expression of MHC I by blocking its internalization (Fig. 4D), and the same result was obtained with CD8-MHC I 335K (supplemental Fig. S6). To confirm that the association between epsin1 and ubiquitinated MHC I is necessary for MHC I internalization, we attempted to disrupt this association by using epsin1 dominant negative mutants. We reasoned that if the association requires the epsin1 UIM, then expression of a UIM-deletion mutant of epsin1 (ΔUIM) might disrupt it. As expected, the association between ubiquitinated CD8-MHC I 335K and endogenous epsin1 was inhibited, and the down-regulation of CD8-MHC I was prevented by ΔUIM (supplemental Fig. S7). Taken together, these results strongly suggest that recognition of ubiquitin chains by the epsin1 UIMs is important for polyubiquitination-enhanced MHC I internalization.
Generation of a Lys11 and Lys63 Mixed-linkage Ubiquitin Chain at Lys335
Several atypical ubiquitin chains have been reported to be novel signal mediators (14, 15, 19); therefore, we examined the composition of the ubiquitin chain(s) that induce the internalization of MHC I. Because we found that epsin1 is a functional adaptor for the polyubiquitin chain generated on MHC I, we purified ubiquitin chains associated with epsin1 from denatured CD8-MHC I 335K and analyzed them by mass spectrometry. As shown in Fig. 5A, among the seven possible linkages, the Lys63 linkage was most strongly detected, but interestingly, the Lys11 linkage was also readily detectable. Similar results were obtained with endogenous MHC I polyubiquitinated by MIR2 (supplemental Fig. S8). Based on these results, two possible scenarios can be envisioned: either each MHC I molecule receives a pure Lys11-linked or a pure Lys63-linked polyubiquitin chain at Lys335, or each MHC I molecule receives a single polyubiquitin chain consisting of both Lys11 and Lys63 linkages.

Generation of the Lys11 and Lys63 mixed-linkage ubiquitin chain at Lys335. A, CD8-MHC I 335K and V5-tagged epsin1 were coexpressed in HeLa cells with (MIR2(+)) or without MIR2 (MIR2(−)). First, the protein complex containing ubiquitinated CD8-MHC I 335K and epsin1 was purified with an anti-V5 Ab and denatured by boiling in 2% SDS buffer. The denatured, CD8-MHC I 335K was then purified with an anti-FLAG M2 Ab. After confirmation by Western blot analysis (left panel), each sample was analyzed by mass spectrometry (right panel). B, each indicated EGFP-tagged ubiquitin was coexpressed with CD8-MHC I 335K and MIR2 in HeLa cells. The ubiquitinated form of CD8-MHC I 335K was purified with the FK2 Ab and examined with the M2 Ab. Equal amounts of CD8-MHC I 335K were analyzed in all samples. C, the ubiquitinated forms of CD8-MHC I were sequentially purified from T-REx-MIR2 cells with M2 Ab and FK2 Ab and separated by SDS-PAGE. After confirmation by Western blot analysis with the indicated Abs, the peptides obtained from the bands indicated by the rectangle were analyzed by LC-MS/MS. D, each indicated EGFP-tagged ubiquitin was coexpressed with CD8-MHC I 335K and MIR2 in HeLa cells. The ubiquitinated form of CD8-MHC I 335K was purified with the M2 Ab and examined with the Lys63 linkage-specific Ab, Apu3. Equal amounts of CD8-MHC I 335K were analyzed in all samples.
To discriminate between these two possibilities, we examined how mutant ubiquitin modifies the formation of the polyubiquitin chains generated at Lys335. The indicated mutant ubiquitin was coexpressed with CD8-MHC I 335K and MIR2 in HeLa cells (Fig. 5B). To monitor the expression of the mutant ubiquitin, the wild-type or mutant protein was fused to the N terminus of GFP. Each GFP-tagged ubiquitin was cleaved at its junction with GFP, and the resultant free GFP was used as an indicator of ubiquitin expression. Ubiquitin in which Lys11 and/or Lys63 was mutated to arginine inhibited the generation of polyubiquitin chains (Fig. 5B). By contrast, ubiquitin in which Lys48 or Lys29 was mutated to arginine had no effect on this process. Ubi-K11R significantly inhibited the generation of polyubiquitinated bands with molecular weights greater than that of Ubi4-CD8-MHC I, and the inhibitory effect of this mutant depended on its expression level (supplemental Fig. S9). However, Ubi-K63R significantly inhibited the generation of polyubiquitinated bands with molecular weights greater than that of Ubi2-CD8-MHC I (Fig. 5B). The significance of these data was confirmed statistically (supplemental Fig. S10). These results suggest that a single Lys11 and Lys63 mixed-linkage chain is generated on MHC I and that in this mixed chain, Lys63 linkages and Lys11 linkages are present in the proximal and distal regions, respectively.
To test this hypothesis further, we examined two ubiquitinated CD8-MHC I 335K bands of different lengths: Ubi2-CD8-MHC I and Ubi4-CD8-MHC I (Fig. 5C). As expected, Ubi2-CD8-MHC I mainly consisted of Lys63 linkages, whereas Ubi4-CD8-MHC I mainly consisted of Lys11 and Lys63 linkages. A monoclonal antibody is now available that specifically detects Lys63 linkages (35). Therefore, we used Western blot analysis to confirm that both Ubi-K11R and Ubi-K63R inhibit the formation of Lys63 linkages. Ubi-K11R inhibited the generation of Lys63-linkage-containing polyubiquitinated bands with molecular weights greater than that of Ubi4-CD8-MHC I, and Ubi-K63R inhibited the generation of Lys63-linkage-containing polyubiquitinated bands with molecular weights greater than that of Ubi2-CD8-MHC I (Fig. 5D). Taken together, these data provide strong evidence that a Lys11 and Lys63 mixed-linkage ubiquitin chain is generated at Lys335 of MHC I and that this unusual chain contributes to the internalization of MHC I by MIR2.
MHC I Internalization by Lys11 and Lys63 Mixed-linkage Ubiquitin Chains
To examine whether a Lys11 and Lys63 mixed-linkage chain functions as internalization signal for MHC I, we expressed GFP-tagged ubiquitin mutants in MIR2-expressing BJAB cells. MIR2-expressing cells express lower levels of cell surface MHC I compared with the parental BJAB cells (Fig. 6A). As we expected, ubiquitins in which Lys11 and/or Lys63 were mutated to arginine inhibited the down-regulation of MHC I in cells in which transfected ubiquitin was highly expressed, as assessed by EGFP expression (shown by the rectangles in the upper right quadrants of Fig. 6A), whereas the wild type and the Lys48 ubiquitin mutants did not (Fig. 6A). To evaluate this phenomenon more quantitatively, we measured the surface expression levels of MHC I in low and high GFP-expressing cell populations and calculated the ratios of these two values (Fig. 6B, upper panel). The significance of the inhibition of the down-regulation of MHC I surface expression by each ubiquitin mutant was confirmed statistically (Fig. 6B, lower panel). As expected, neither the Lys29 nor the Lys33 ubiquitin mutants rescued MIR2-mediated MHC I down-regulation (Fig. 6A). The same results were obtained with CD8-MHC I 335K (supplemental Fig. S11). Next, we examined whether blocking Lys11- or Lys63-linked ubiquitination suppressed the internalization of MHC I in MIR2-expressing BJAB cells. First, we analyzed the down-regulation of surface-expressed MHC I in cells that highly expressed the indicated ubiquitin molecules at the indicated times and found that both Ubi-K11R and Ubi-K63R inhibited the down-regulation of surface-expressed MHC I (Fig. 6C). To examine whether this down-regulation was due to inhibition of internalization, we analyzed the down-regulation of surface-expressed MHC I within the first 10 min as performed in Fig. 3B. As shown in Fig. 6D, both Ubi-K11R and Ubi-K63R inhibited the down-regulation of surface-expressed MHC I within this time frame, indicating that a Lys11 and Lys63 mixed-linkage polyubiquitin chain is required for MHC I internalization.

Contribution of the Lys11 and Lys63 mixed-linkage ubiquitin chain to protein internalization. A, each indicated EGFP-tagged ubiquitin was expressed in constitutively MIR2-expressing BJAB cells. Two days after the introduction of each exogenous ubiquitin by electroporation, the expression level of MHC I was examined by flow cytometry. B, the surface expression levels of MHC I in low GFP-expressing and high GFP-expressing cell populations were measured as indicated in the upper panel, and the ratios of these two values are shown in the lower panel. The values are mean ± S.E. (error bars) (n = 3); *, p < 0.01 compared with wild-type ubiquitin (Wt). C, after each EGFP-tagged ubiquitin was expressed as in A, the down-regulation of surface MHC I at the indicated time points was examined in the cell population in which EGFP was highly expressed, as in Fig. 1D. The values are mean ± S.E. (n = 3); *, p < 0.01 compared with wild-type ubiquitin. D, the same experiment as in C was performed at a different time point (10 min). The values are mean ± S.E. (n = 3); *, p < 0.01 compared with wild-type ubiquitin.
The next important question was how do these Lys11 and Lys63 linkages contribute to the interaction between the mixed polyubiquitin chain and epsin1. After confirmation of the association between a Lys11 and Lys63 mixed-linkage chain and epsin1 by inhibiting chain formation with Ubi-K63R (supplemental Fig. S12), we examined the interaction between pure Lys11- or pure Lys63-linked polyubiquitin chains and a glutathione S-transferase (GST) fusion protein containing the UIM domains of epsin1 (GST-UIM). The pure Lys63-linked polyubiquitin chain, but not the pure Lys11-linked polyubiquitin chain, interacted with GST-UIM (supplemental Fig. S13). This result suggests that the Lys11 linkages do not directly contribute to the binding to epsin1.
DISCUSSION
It has been reported that protein ubiquitination is an important signal for receptor internalization from the cell surface (6,–10). However, it is still unclear how a polyubiquitin chain regulates the internalization step because it has been shown that monoubiquitination is sufficient to induce receptor internalization in yeast (11, 12). By use of the Tet-on expression system for MIR2 and quantitative mass spectrometry, we demonstrated that the polyubiquitin chain generated by MIR2 is a Lys11 and Lys63 mixed-linkage chain that is required for efficient epsin1-dependent internalization of MHC I. Given that it has recently been reported that the Lys11-linked polyubiquitin chain is a signal for proteasomal degradation involved in anaphase-promoting complex-mediated cell-cycle control and in endoplasmic reticulum-associated degradation (18, 19), our findings provide further evidence that atypical polyubiquitin chains have specific functions in vivo.
Previous studies of how polyubiquitination contributes to internalization were performed with polyubiquitin chains generated at multiple ubiquitin acceptor sites in the target membrane proteins, making it very difficult to analyze the function of a single polyubiquitin chain. We were able to define a single polyubiquitin chain consisting of mixed Lys11 and Lys63 linkages by using a MHC I mutant with only one lysine, Lys335, as the ubiquitin acceptor site. In addition, we found that both lysine residues (Lys335 and Lys340) in the cytosolic tail in MHC I are ubiquitinated, but only the ubiquitination at Lys335 contributes to efficient internalization. Thus, the experimental system reported in this study may be useful for further investigation of how the quality of ubiquitination (e.g. ubiquitin acceptor site of the substrate, ubiquitin linkage) contributes to internalization in vivo.
In this report, we demonstrated that a Lys11 and Lys63 mixed-linkage chain generated by MIR2 is required for internalization of MHC I. These results are consistent with the findings recently reported by Boname et al. (36). Furthermore, we found that this novel polyubiquitin chain can function as an internalization signal for MHC I through its association with epsin1, an adaptor molecule containing a UIM domain. Therefore, an important next question is how the Lys11/Lys63 mixed-linkage chain is recognized by the UIM domains. Recently, it has been reported that the UIM domains of epsin1 bind preferentially to Lys63-linked rather than Lys48-linked ubiquitin chains (20) because the inter-UIM region forms a 12-Å-long α-helix. This structure ensures that the UIMs are spatially arranged to allow specific binding to Lys63-linked diubiquitin. Moreover, using molecular modeling of ubiquitin, Lys11-linked chains are predicted to form a closed conformation similar to that in Lys48-linked chains (37), whereas Lys63-linked chains adopt a highly open conformation (38). Consistent with these findings, we observed that the UIM domains of epsin1 associate with Lys63-linked chains, but not with Lys11-linked chains, suggesting that the Lys11 linkages generated by MIR2 may not directly contribute to interaction with epsin1. The most plausible model at this point is that epsin1 binds to a Lys63-linked diubiquitin and longer chains and that the presence of Lys11 (either as a mixed chain, as a cap, or as a side branch) facilitates some events related to Ub-dependent endocytosis. The Lys11 linkages might support the extension of the polyubiquitin chain required for efficient internalization, or they might generate a suitable chain topology to promote the association between Lys63 linkages and epsin1 in vivo. Also, Lys11 linkages may interact with a second ubiquitin-interacting protein that is needed for endocytosis and/or subsequent sorting. Further intensive experiments are needed to test these hypotheses.
Previous studies have focused exclusively on the contribution of the Lys63 linkage to internalization and lysosomal degradation (8, 39, 40), a reasonable approach because no significant amounts of Lys11-linked chains, compared with Lys48- or Lys63-linked chains, had been observed (40, 41). Therefore, we examined whether Lys11 and Lys63 linkages are present in polyubiquitin chains generated on substrates (e.g. B7-2) ubiquitinated by other MIR family members. Quantitative mass spectrometric analysis indicates that indeed there are both Lys11 linkages and Lys63 linkages in the polyubiquitin chains generated on these substrates.3 Although further analysis is required, it now seems plausible that Lys11 and Lys63 mixed-polyubiquitin chains are a common internalization signal utilized by MIR family members.
Acknowledgments
We thank H. Ploegh for HC10 antibody, K. Umebayashi for the EGFP-Ubi-expressing plasmid, P. Burrows for critical reading of the manuscript, and S. Dohi for manuscript preparation.
*This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Japan Society for the Promotion of Science.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. S1–S13.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. S1–S13.
3E. Goto, Y. Yamanaka, A. Ishikawa, M. Aoki-Kawasumi, M. Mito-Yoshida, M. Ohmura-Hoshino, Y. Matsuki, M. Kajikawa, H. Hirano, and S. Ishido, unpublished data.
2The abbreviations used are:
- UIM
- ubiquitin-interacting motif
- Dox
- doxycycline
- EGFP
- enhanced green fluorescent protein
- MIR
- modulator of immune recognition
- Tet-on
- tetracycline-inducible.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m110.112763
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/article/S0021925820468711/pdf
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/content/full/285/46/35311
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/285/46/35311.pdf
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/285/46/35311
Citations & impact
Impact metrics
Citations of article over time
Article citations
Getting to the Root of Branched Ubiquitin Chains: A Review of Current Methods and Functions.
Methods Mol Biol, 2602:19-38, 01 Jan 2023
Cited by: 1 article | PMID: 36446964
Review
ANTH domains within CALM, HIP1R, and Sla2 recognize ubiquitin internalization signals.
Elife, 10:e72583, 25 Nov 2021
Cited by: 4 articles | PMID: 34821552 | PMCID: PMC8648300
E3 ubiquitin ligases: styles, structures and functions.
Mol Biomed, 2(1):23, 30 Jul 2021
Cited by: 95 articles | PMID: 35006464 | PMCID: PMC8607428
Review Free full text in Europe PMC
Ubiquitomics: An Overview and Future.
Biomolecules, 10(10):E1453, 17 Oct 2020
Cited by: 16 articles | PMID: 33080838 | PMCID: PMC7603029
Review Free full text in Europe PMC
Emerging functions of branched ubiquitin chains.
Cell Discov, 7(1):6, 26 Jan 2021
Cited by: 65 articles | PMID: 33495455 | PMCID: PMC7835216
Review Free full text in Europe PMC
Go to all (44) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains.
Traffic, 11(2):210-220, 17 Nov 2009
Cited by: 92 articles | PMID: 19948006 | PMCID: PMC3551259
Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules.
EMBO J, 25(8):1635-1645, 06 Apr 2006
Cited by: 189 articles | PMID: 16601694 | PMCID: PMC1440841
Stabilization of an E3 ligase-E2-ubiquitin complex increases cell surface MHC class I expression.
J Immunol, 184(12):6978-6985, 12 May 2010
Cited by: 13 articles | PMID: 20483773 | PMCID: PMC3551299
Emerging roles for Lys11-linked polyubiquitin in cellular regulation.
Trends Biochem Sci, 36(7):355-363, 07 Jun 2011
Cited by: 55 articles | PMID: 21641804
Review




