Abstract
Free full text

Altered Runx1 Subnuclear Targeting Enhances Myeloid Cell Proliferation and Blocks Differentiation by activating a miR-24/MKP-7/MAP Kinase Network
Abstract
Disruption of Runx1/AML1 subnuclear localization, either by a single amino acid substitution or by a chromosomal translocation (e.g. t(8;21)), is linked to the etiology of acute myeloid leukemia (AML). Here we show that this defect induces a select set of micro-RNAs (miRs) in myeloid progenitor cells and AML patients with t(8;21). Both Runx1 and the t(8;21)-encoded AML1-ETO occupy the miR-24-23-27 locus and reciprocally control miR-24 transcription. miR-24 directly downregulates MAPK Phosphatase-7 and enhances phosphorylation of both JNK and p38 kinases. Expression of miR-24 stimulates myeloid cell growth, renders proliferation independent of IL3 and blocks granulocytic differentiation. Thus, compromised Runx1 function induces a miR-dependent mechanism that, through MAPK signaling, enhances myeloid proliferation but blocks differentiation, key steps that contribute to leukemia.
Introduction
Runx1 (also designated AML1) is a key factor controlling the hematopoietic lineage and is required for definitive hematopoiesis. Cell biological, biochemical and in vivo geneticstudies demonstrate that Runx1 initiates and sustains activation of the hematopoietic transcriptional program(1–4). Runx1 is directed to distinct nuclear microenvironments by a unique and obligatory subnuclear targeting signal, and subnuclear targeting of Runx1 is necessary for biological activity in vivo and ex vivo(5–7). The Runx1 locus is frequently rearranged in myeloid leukemia disrupting transcriptional function and/or intranuclear localization of the protein (2, 8). The AML1-ETO protein, encoded by the 8; 21 translocation, lacks the Runx1 carboxyl terminus and associated functions, including transcriptional activation and subnuclear targeting (2, 9–12). Instead, the runt homology domain of Runx1 is fused in frame with the ETO protein that interacts with co-suppressors and contains intrinsic subnuclear targeting signals (2, 9, 13–17). The chimeric protein occupies Runx1 target gene promoters resulting in suppression(2, 9, 13–17)and is directed to nuclear microenvironments that are distinct from those where Runx1 resides. However, mechanisms that contribute to the leukemic properties of AML1-ETO are not completely understood.
Micro RNAs (miRs) are small RNA molecules that post-transcriptionally regulate gene expression(18–21). miRs have been linked to control of cell proliferation and lineage commitment (22, 23). In hematopoiesis, miRs are implicated in both normal hematopoiesis and leukemogenesis (24–26). For example, miR 27 regulates megakaryocytic differentiation(27). Similarly, translocation of miR 142in B cell leukemia is directly linked to development and progression of disease(28). However, a role for miRs in myeloid differentiation and acute myeloid leukemias has remained elusive.
In this study, we report that Runx1 transcriptionally represses the highly conserved miR-24 and abrogation of Runx1 subnuclear targeting by a point mutation or by chromosomal translocation enhances miR-24 expression. Upregulation of miR-24 inhibitsa MAP kinase phosphatase (MKP-7; also designated dual specificity phosphatase 16), activates downstream signaling and alters myeloid cell proliferation and differentiation. Thus we have identified a novel network that functionally links AML1-ETO with miR-mediated activation of MAP kinase signaling with implications for leukemogenesis.
Materials and Methods
Human patient samples
Total cellular RNA was isolated from bone marrow cells of AML patients. These patients were initially diagnosed as myelodysplastic syndrome (MDS) patients, and later developed the M2-type AML (French-American-British nomenclature system). Presence of the t(8;21) in all patients was confirmed by fluorescence in situ hybridization. Histochemical staining revealed that more than 90% cells were leukemic blasts.
Cell culture, growth curves and differentiation assay
Human erythroleukemia K562 cells were maintained in RPMI (Invitrogen) supplemented with 10% FBS, 2 mmol/L L-glutamine, 100 units/mL penicillin G, and 100 μg/mL streptomycin, while patient-derived Kasumi-1 cells were grown in RPMI (Invitrogen) supplemented with 20% FBS, 2 mmol/L L-glutamine, 100 units/mL penicillin G, and 100 μg/mL streptomycin. Murine 32D cells were maintained in RPMI (Invitrogen) supplemented with 10% FBS, 2 mmol/L L-glutamine, 100 units/mL penicillin G, and 100 μg/mL streptomycin, supplemented with IL-3 (proliferation medium) or G-CSF (differentiation medium). Live cell count to assess growth curves of cells transfected with different miRs was carried out of different samples using trypan blue staining. 32D cells were differentiated into granulocytes by replacing IL-3 with G-CSF. Cells were grown in differentiation medium for 2 days and stained with Giemsa-Wright staining to determine nuclear lobulation.
miRNA expression profiling
Microarray and bioinformatic analyses, as well as significance analysis of microarrays, were done as described before (29). Results from three independent experiments are expressed as log2 of fluorescence and as a hierarchical clustering of the average values by dCHIP software.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation assays (ChIPs) were performed by crosslinking asynchronously growing cells with 1% formaldehydein RPMI for 10 minutes at room temperature. Crosslinking was quenched by adding glycine to a final concentration of 250 mM for 10 minutes. Cells were collected and washed twice with PBS. Cell pellets were resuspended in 2.5 ml lysis buffer (150 mMNaCl, 50 mM Tris-HCl pH 8.0, 1% NP-40, 25 μM MG-132, and1x Complete® protease inhibitor cocktail (Roche). After10minutes on ice, cells were sonicated to obtain DNA fragments of 500 bp as determined by agarose gel electrophoresis with ethidium bromide staining. Protein-DNA complexes were isolated by centrifugation at 15,000 rpm for 20 minutes. Supernatants with protein-DNA complexes were incubated for 16 hours with3 μg rabbit polyclonal antibody directed against each protein. The following primary antibodies were used: ETO rabbit polyclonal (Santa Cruz Biotechnology)and AML1 rabbit polyclonal (Active Motif). Antibody-protein-DNA complexes were further incubated with 50–60 μl of 30% protein A/G beads (Santa Cruz Biotechnology) to isolate antibody-bound fractions of chromatin. Immunocomplexes were washed with the following buffers: low salt (20 mM Tris-HCl pH 8.1, 150 mM NaCl, 1% Triton X-100, 2mM EDTA, 1x complete protease inhibitor), high salt (20 mM Tris-HCl, pH 8.1, 500 mM NaCl, 1% Triton X-100, 2 mM EDTA), LiCl (10 mMTris-HCl, pH 8.1, 250 mM LiCl, 1% deoxycholate, 1% NP-40, 1mM EDTA) and twice in TE (10 mM Tris-HCl pH 8.1, 1 mM EDTA). Protein-DNA complexes were eluted in 1% SDS and 100 mM NaHCO3. Crosslinks of pull-down fractions and inputs (2% of total IP fraction) were reversed by incubation overnight in elution buffer and 0.2 M NaCl. DNA then was extracted, purified, precipitated and resuspended in TE for qPCR. Following primers were used to amplify promoter region of the miR 24-23-27 cluster:
Forward primer: 5/GATCACTTGTGCCCAGGAAT 3/
Reverse primer: 5/GCTTACAGGCACCCACCAC 3/
These primers amplify a 126 bp region of the Chromosome 19 in the promoter region of the miR cluster that contains 2 Runx binding sites and is located at the position 13825K on the p arm of the human Chromosome 19.
Northern blot analysis and RT-qPCR
Total cellular RNA was prepared using TRIzol reagent following the manufacturer’s instructions (Invitrogen). Total RNA (15 μg) was separated by electrophoresis in 0.5x Tris-borate EDTA [TBE; 44.5 mmol/L Tris, 44.5 mmol/L sodium borate, and 1 mmol/L EDTA (pH 8.3)] through denaturing 12% urea-polyacrylamide (0.75 mm × 16 cm × 16 cm) gels at 15 to 20 W. RNA was electrotransferred to a Zeta-Probe GT membrane (Bio-Rad) in a semidry apparatus in 0.5x TBE for 1 h at 300 mA. RNA was fixed to the membrane by UV irradiation (optimal cross-linking setting, Spectrolinker XL-1000 UV cross-linker, Spectronics Corporation) and baked at 80°C for 30 minutes under vacuum. Membranes were prehybridized at 42°C for 2 h in buffer (7% SDS/0.2 mol/L sodium phosphate, pH 7.2) and then hybridized with oligonucleotide DNA probes radiolabeled with T4 polynucleotide kinase overnight at 42°C in the same buffer. Membranes were washed for 30 minutes at 42°C twice each time in wash buffer 1 (5% SDS/20 mmol/L sodium phosphate, pH 7.2) and wash buffer 2 (1% SDS/20 mmol/L sodium phosphate, pH 7.2), respectively. One microgram of total cellular RNA was subjected to reverse transcription and quantitative PCR using primers sets specific for tested gene transcripts.
Luciferase constructs and reporter gene assays
Genomic DNA from human WI-38 normal diploid fibroblasts was isolated following a standard protocol. MKP7 3′UTR-Luc plasmid was constructed by cloning segments of the 3′ UTRs spanning the miRNA recognition sites into pMIR-REPORT miRNA Expression Reporter Vector (Ambion)by PCR from WI-38 genomic DNA.
Western blot Analysis
Total protein extracts were prepared in direct lysis buffer [50 mmol/L Tris-HCl (pH 6.8), 100 mmol/L DTT, 2% SDS, 10% glycerol, 1x Complete (Roche), and 0.2% bromophenol blue]. Cells were boiled for 5 minutes and centrifuged at 16,100 x g for 5 minutes. Total proteins were loaded onto 10% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride Immobilon-P membrane (Millipore) for 30 minutes at 10 V in a semidry transfer apparatus (model HEP-1, Owl Separation Systems). Immunodetection was done using an appropriate dilution of specific antibodies with the western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences).
Transient transfections
K562 cells (5 × 104 cells per well) were transfected in 12-well plates with Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Cells were transfected with 800 ng of the luciferase constructs MKP-7 3′ UTR-Luc and 5 ng of Renilla luciferase plasmid and cotransfected with human pre-miR-23miRNA precursor or pre-miR-24miRNA precursor (30 nmol/L) obtained from Ambion. After 48 hours, cells were harvested and lysates were measured for luciferase activity and normalized for transfection efficiency to Renilla luciferase (phRL-null) activity (dual-luciferase reporter assay system, Promega). The relative reporter activity was obtained by comparison to a control oligonucleotide arbitrarily taken as 100 (pre-miR miRNA precursor negative control 2, Ambion). 32D cells were transfected with the indicated miRs using Amaxa nucleofection according to the manusfacturer’s instructions. K562 or 32Dcells were transfected with human miR-23, miR-24, or negative control (NS) oligonucleotides (Ambion or Dharmacon) using Oligofectamine following the manufacturer’s instructions. The miRNA oligonucleotide concentrations used ranged from 30 to 150 nmol/L, with or without a 5-fold excess of anti-miRNA inhibitor. Protein extracts were prepared and analyzed by western blot as described above. Cell cycle distribution was monitored by FACS analysis.
Results
Modified Runx1 Subnuclear Targeting by a point mutation or leukemia-related translocation transcriptionally deregulates the miR 24-23-27 cluster
We carried out genome wide micro RNA (miR) profiling to identify mechanisms that contribute to AML1-ETO-mediated leukemogenesis. Samples were analyzed from four AML patients with the 8; 21 translocation as well as from mouse myeloid progenitor cells stably expressing either wild type Runx1 or a single-point mutant that confers altered subnuclear targeting (referred to mSTD – subnuclear targeting defective mutant -Runx1 hereafter)(Figure 1)(30). Of 900 miRs examined (29), expression of 175 miRs was significantly altered in response to Runx proteins in both human patient samples and in mouse 32D cell lines. We sought to initially identify and focus on miRs that were affected by disrupted subnuclear targeting of Runx1 in both the human AML patient samples as well as in murine myeloid progenitor cell lines stably expressing the mSTD mutant of Runx1. Statistical analyses of the results combined with hierarchical clustering of mouse and human miR profiles revealed a group of 10 miRs, including two highly conserved clusters (miR-24-23-27 and miR 181) that was significantly upregulated when intranuclear localization of Runx1 was modified, either by a point mutation as is the case in 32D mSTD cell line, or by chromosomal translocation in human AML patients(Figure 1A). Initially, we assessed the expression of all 10 miRs in the group by quantitative PCR analysis. As shown in Figure 1B, the endogenous expression of all miRs, in 32D cell lines expressing the WT or the mSTD mutant Runx1 or a negative control(EV), was consistent with the data obtained from genome wide miR analysis. All miR transcripts analyzed were upregulated when the subnuclear targeting of Runx1 was compromised. We then focused on the miR cluster 24-23-27, which was consistently one of the most affected miR genes by compromised Runx1 subnuclear targeting. Expression of the miR cluster was further confirmed by Northern blot analysis of total cellular RNA isolated from the 32D stable cell lines (Figure 1C). Our results verified upregulation of these miRs in response to altered Runx1 subnuclear targeting in murine cells. Analysis of the miR-24-23-27 locus revealed that several Runx binding sites are present (Figure 1D, also see Supplementary Figures 1S and 2S). Runx proteins have been shown to directly regulate this locus transcriptionally (27)(Hassan et al, unpublished data; data not shown). To explore the biological relevance of Runx mediated regulation of this cluster, we first carried out chromatin immunoprecipitation analysis to assess whether either wild type Runx1 or AML1-ETO interact with miR clusters in vivo. Our results show that in AML patient-derived Kasumi-1 cells, both the wild type and the chromosomally translocated Runx1proteinsoccupy the miR clusters (Figure 2A). The specificity of these observations is further demonstrated in Jurkat B-cell lymphoma where only Runx1 is expressed endogenously or in cervical carcinoma HeLa cells, where neither Runx1 nor ETO is expressed.
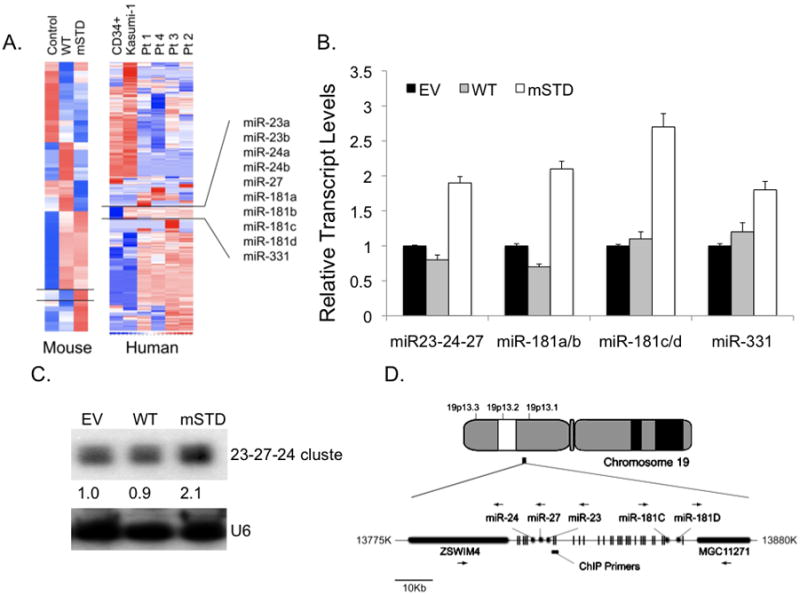
Total cellular RNA from murine myeloid progenitor 32D cells, stably expressing normal Runx1 (WT) or a subnuclear targeting defective mutant (mSTD), or from human AML patients (Pt1-4) and AML patient derived Kasumi-1 cells were subjected to genome wide micro RNA analysis. (A) The data were obtained as log2 of the control (Control lane for 32D cells, and CD34+ normal myeloid cells for human samples) and represented as hierarchical clusters using web-based dChIP analysis. A group of 10 miRs that were upregulated upon defective subnuclear targeting of Runx1 by a single amino acid substitution (mSTD in 32D cell samples) or by chromosomal translocation (Kasumi-1 and Pt1-4 samples). (B) Total cellular RNA from the indicated 32D stable cell lines was subjected to either quantitative RT-PCR for all the 10 miRs in the selected group (B)or to Northern blot analysis (C)using probes for either miR-24-23-27 cluster or for U6 RNA (as a loading control). The intensities of the bands were semi-quantified by using Image J software (http://rsbweb.nih.gov/ij/). (D) Genomic location and sequence analysis of the miR 24-23-27 cluster. The cluster is present on Chromosome 19 and have numerous Runx binding sites (vertical lines). The horizontal bars represent the region amplified in ChIP assay, while the black arrows indicate the direction of gene transcription.
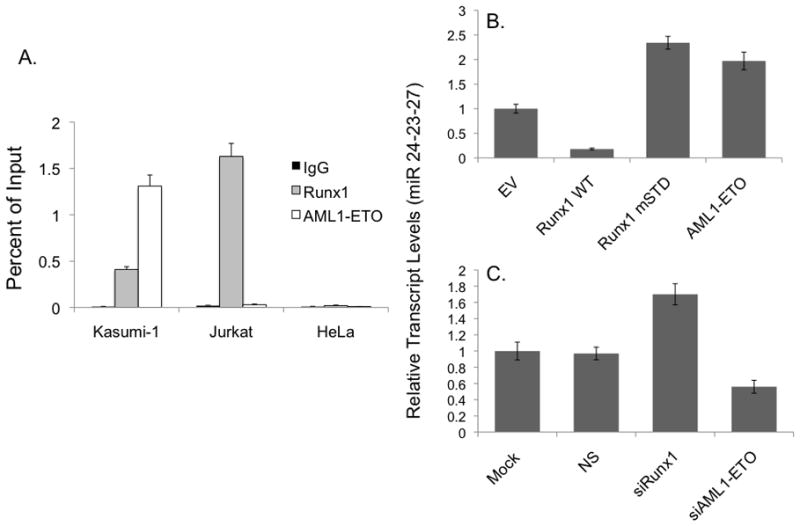
(A) AML patient derived Kasumi-1 cells that express both Runx1 and AML1-ETO, B-cell lymphoma Jurkat cells that express only Runx1, or cervical carcinoma HeLa cells that do not express either protein were subjected to ChIP assay using antibodies specific for Runx1 (light gray bar) or AML1-ETO (white bar). Immuno-precipitated DNA was subjected to qPCR using locus specific primers and the results were represented as bar graphs of % input. IgG was used as a negative control for the immunoprecipitation. (B) K562 cells were infected with retroviruses carrying either Runx1WT, Runx1 mSTD or AML1-ETO cDNAs. Total cellular RNA was isolated from cells 48 hours after the infection and was subjected to qPCR using primers specific for miR 24-23-27 transcript. Bar graphs represent transcript levels in various experimental groups relative to empty vector (EV). (C) Kasumi-1 cells were transfected with siRNA against Runx1 or AML1-ETO. Cells were subjected to total cellular RNA isolation and qPCR 48 hours post-transfection. Data were represented as bar graphs of transcript levels relative to mock transfected cells.
To address whether Runx1 and AML1-ETO transcriptionally regulate miR clusters23-24-27, we exogenously expressed each protein in human K562 erythro-leukemia cells. Cells infected with Runx1 showed a downregulation of miR 24-23-27transcripts (Figure 2B). In contrast, AML1-ETO as well as mSTD Runx1 upregulates these transcripts, consistent with our genome wide miR profiling of human patient samples(see Figure 1A). We then transfected Kasumi-1 cells with small interfering RNA molecules (siRNA) targeting either wild type Runx1 or the leukemic AML1-ETOprotein. ThemiR-24-23-27transcript was upregulated when Runx1 was depleted, while AML1-ETO depletion down-regulated these miRs(Figure 2C, also see Supplementary Figure S4). Together, these findings demonstrate that both the normal and leukemic Runx1 proteins directly interact with regulatory elements in the miR-23-24-27 and miR-181 clusters but have opposite effects on the expression of these miRs.
miR-24 downregulates MAP kinase phosphatase 7 and activates MAP kinase signaling
One of the two clusters that we have identified includes onco-miRs 181 C and D that have been previously linked to myeloid leukemia (31), but the biological roles of the miRs encoded by the 24-23-27 cluster have been under-explored. We focused on miR-24, as it is responsive to both the wild type and the leukemic Runx1 factors. One of the predicted targets of miR-24 identified by the web-based TargetScan and miRGen algorithms is the mitogen activated protein kinase phosphatase 7(MKP-7; also designated dual specificity phosphatase 16) (Figure 3, also see Supplementary Figure 3S). MKP-7 attenuates myeloid cell proliferation by negatively regulating the mitogen activated p38 and JNK kinases (32, 33). To determine whethermiR-24directly regulates MKP-7, we cloned the 3′ untranslated region (UTR)of the MKP-7 mRNA, which containsamiR-24 target sequence, downstream of a luciferase reporter gene(Figure 3A, please also see Supplementary Figure 3 for additional details of the sequence and other miRs with the potential to target MKP-7 3′-UTR). Reporter activity was significantly reduced by miR-24, but not by a non-specific miR or miR-23, which is expressed from the same cluster but does not interact withMKP-7(Figure 3B). This effect was reversed by anti-miR-24. These results establish that the MAPK attenuator MKP-7is a direct and specific downstream target of miR-24in hematopoietic cells.
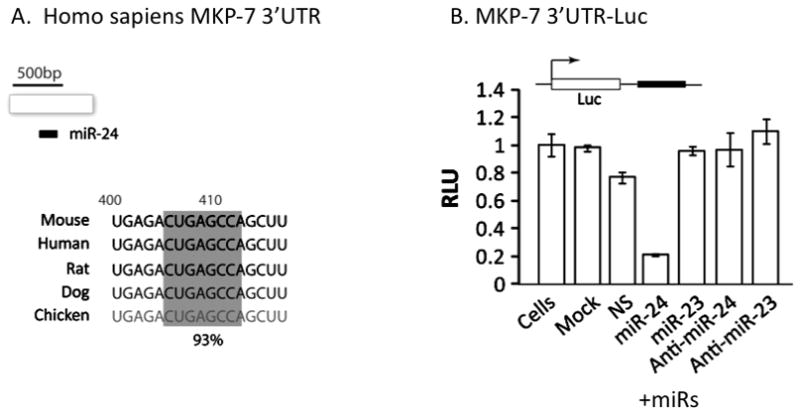
(A) A sequence analysis of the 3′ UTR of MKP-7of several organisms revealed the presence of seed sequence for miR-24 (gray box). (B) The 3′ UTR of MKP-7was cloned downstream of a luciferase gene and used in reporter promoter assays. K562 cells were transfected with the reporter plasmid and indicated miRs or anti-miRs. Luciferase activity in the cell lysates expressing indicated combinations of plasmid and miRs or anti-miRs was assessed using a luminometer.
Because the predominant mechanism of miR action in metazoans is translational inhibition, we investigated the effect ofmiR-24, a non-specific miR (NS) andmiR-23on endogenous MKP-7protein. We find that the MKP-7protein is selectively downregulated in the presence of miR-24 (Figure 4A). MKP-7negatively regulates MAP kinase signaling by specifically removing phosphate groups from MAP kinases(32, 33). Because miR-24 caused significant down-regulation of MKP-7transcript and protein, we directly addressed its role in modulating the MAP kinase signaling pathway. We assessed the phosphorylation of JNK and p38 kinases, the two most prominent MAP kinase pathways that play a major role in hematopoietic cell biology (34, 35). There was significant increase in the phosphorylated form of p38 kinase in the presence of miR-24, while a slight increase in JNK phosphorylation was also observed(Figure 4B). In contrast, phosphorylation of AKT, an un-related kinase that is not targeted by MKP-7, remained unaltered(Figure 4C). Together, our findings establish that miR-24 modulates MAP kinase signaling in hematopoietic cells by directly targeting MAP kinase phosphatase 7.
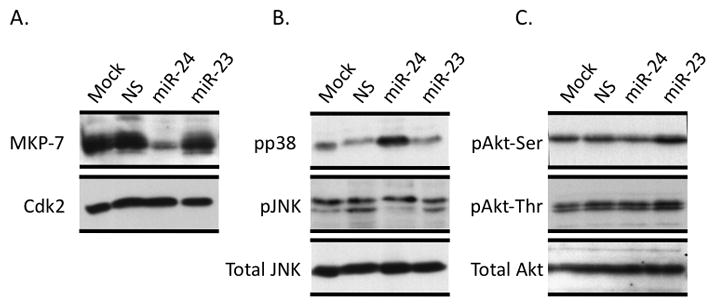
Human erythroleukemia K562 cells were transfected with the indicated miRs for 48 hours and total cellular proteins were subjected to western blot analysis. K562 cells transfected with the indicated miRs were subjected to western blot analysis. Specific antibodies were used to detect MKP-7. Cdk2 levels were assessed for protein loading control (A). Phospho-specific antibodies were used to detect activation of JNK, p38 (B) and AKT kinases (C). Antibodies against total JNK and AKT kinases were used for protein loading control
miR-24 induced growth factor independence of myeloid progenitor cells accelerates proliferation and blocks granulocytic differentiation
Sustained proliferation is a hallmark of blast cells in leukemia(36). Our results show that miR-24 expression is increased in AML patients and is transcriptionally upregulated by the AML1-ETO fusion protein (see Figures 1 and and2).2). We experimentally addressed whether the accelerated proliferation of leukemia cells expressing AML1-ETO (37)is mediated by miR-24. Increased cell growth was observed when miR-24 was expressed (Figure 5A). It is well established that leukemic blast cells are also growth factor independent(38). We directly addressed this possibility in 32D myeloid progenitor cells that require the cytokine interleukin-3 (IL-3)for normal growth. In the presence of miR-24,32D cells continued proliferating following IL-3withdrawal while this effect was not observed in cells treated with control miRs (Figure 5B). These results indicate that, like AML1-ETO, miR-24 renders cells growth-factor independent.
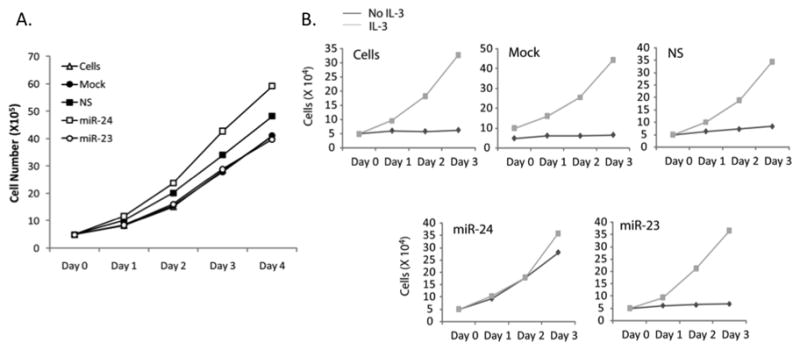
(A) Live cell count of K562 cells transfected with indicated miRs was carried out using Tryphan blue staining and is represented here as number of cells in each group for 4 days post-transfection. (B) Myeloid progenitor 32D cells were transfected with the indicated miRs in the presence (gray line) or absence (black line) of IL3. Live cells were counted every day for 3 days post transfection. A representative from 2 independent experiments have been shown.
In addition to enhanced proliferation, a block in differentiation is characteristic of acute myeloid leukemia(36). We experimentally addressed the hypothesis that miR-24 prevents myeloid differentiation (Figure 6). Myeloid progenitor 32D cells can be differentiated into the granulocytic lineage by replacing IL-3 with G-CSF. We transfected 32D cells with miR-24 or control miRs and then induced differentiation with G-CSF. Cells were harvested and histochemically stained to identify nuclear lobulation, a prominent morphological feature of granulocytic differentiation(Figure 6A). Differentiation was quantified by assessing segmentation of stained nuclei. In contrast with control miRs (NS and miR-23), miR-24 significantly reduced differentiation of myeloid cells into granulocytes; approximately 80%cells expressing miR-24 remained undifferentiated when compared to mock or non-specific siRNA transfected cells where majority of cells were differentiated i.e., 90% (Figure 6B). Consistent with these results, miR-24 resulted in decreased expression of granulocytic maturation markers (i.e., Granzyme B, CD11b and myeloperoxidase (MPO), although only Granzyme B and CD11b exhibited a significant decrease (p=0.05, Figure 6C). Biological relevance of these observations was further demonstrated by the reciprocal expression of Runx1 and the miR-23-24-27 cluster during granulocytic differentiation of 32D myeloid progenitor cells; Runx1 was decreased upon granulocytic differentiation, while miR-23-24-27 transcript levels were increased. Together, these findings establish that miR-24 enhances growth-factor independent proliferation and blocks granulocytic differentiation of myeloid cells.
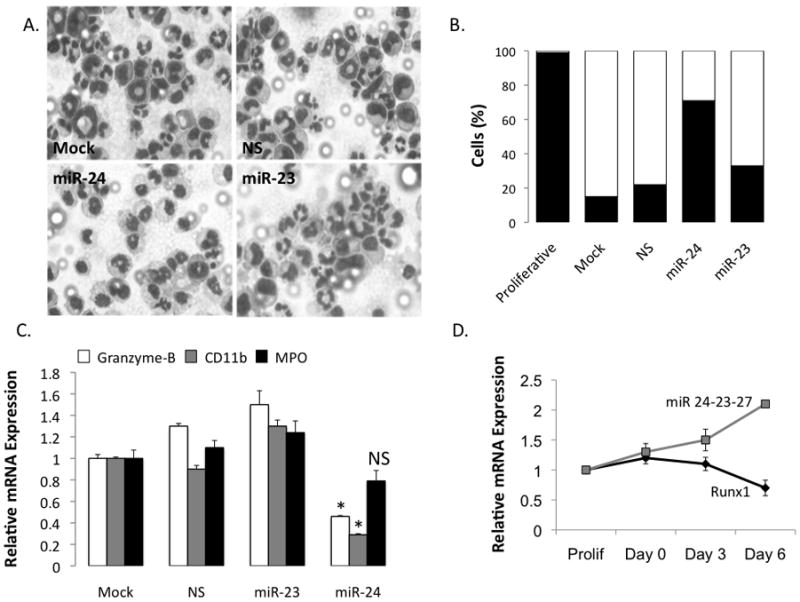
32D cells were transfected with the indicated miRs and subjected to granulocytic differentiation by replacing IL-3 with G-CSF in the growth medium 24 hours post-transfection. Cells were allowed to differentiate for an additional 48 hours and harvested for Giemsa-Wright staining (A). Nuclear lobulation, a hallmark of granulocytic differentiation, was scored in all the experimental groups and represented as % cells with segmented nuclei (light gray bars) or with blast nuclei (dark bars) (B). (C) Total cellular RNA was subjected to RT-qPCR to assess the expression of granulocyte differentiation markers. Primers specific for mRNAs of early (Granzyme-B) as well as late (CD11b and MPO) granulocyte markers were used. Bar graphs represent mRNA expression of each of the markers in each experimental set relative to that in mock-transfected cells. Expression of GAPDH in each sample was used as internal control. Only Granzyme B and CD11b showed statistically significant down-regulation (p=0.05, indicated by an asterisk), while MPO was not changed significantly (marked as non-significant; NS) (D) Murine myeloid 32D cells were subjected to granulocytic differentiation by replacing IL-3 with G-CSF and cells were harvested for RNA isolation at Days 0, 3, and 6 of differentiation. Actively proliferating cells were included as a control. Total cellular RNA was isolated and subjected to qRT-PCR using primers specific for Runx1 or miR 23-24-27 transcripts. The expression of the transcripts was normalized with GAPDH internal control.
Discussion
Initiation, progression and maintenance of the myeloid leukemic phenotype is a multistep process that includes chromosomal translocation, generation of the chimeric AML1-ETO protein and disruption of the Runx1-mediated hematopoietic transcriptional program, as well as re-localization of the Runx1 DNA binding domain to different nuclear microenvironments(1–3, 5–8, 10–14, 14–17, 39, 40). However, both mouse genetic models and human AML patients with the 8;21 translocation indicate that AML1-ETO is not sufficient to initiate the disease (40, 41). Additional mechanisms are required.
Our study, for the first time, links the modified Runx1 transcriptional program and subnuclear targeting in AML with specific miRs and perturbed MAP kinase signaling. Like AML1-ETO or a mutant form of Runx1 that mimics the altered subnuclear targeting of AML1-ETO(30), expression of miR-24 renders myeloid progenitor cells hyper-proliferative and growth factor independent and leads to a block in granulocytic differentiation.
Our data further show that miR 24 function is in part mediated by the down-regulation of MKP-7, a negative regulator of MAP kinase signaling(32, 33). MKP-7 is a putative tumor suppressor that attenuates BCR-ABL induced hematopoietic cell transformation (42). These findings suggest that MKP-7 may play a role in myeloid lineage malignancies. Furthermore, selective deletion of individual components of MAP kinase signaling pathways enhances cell proliferation and inhibits differentiation(34, 35). Thus, our results provide the first experimental evidence for a regulatory network in which Runx responsive miRs controlmyeloid cell proliferation and differentiation by targeting a negative regulator of MAP kinase signaling. We propose that down-regulating miR 24 in leukemic blast cells will reverse all or some aspects of the disease phenotype linked toAML1-ETO. Additional studies are required to confirm the link between up-regulation of miR-24 and onset and progression of leukemia in an in vivo setting. However, our results suggest miR-24 is a novel and selective therapeutic target for the treatment of AML.
Supplementary Material
1
2
3
4
Acknowledgments
We thank Pat Jamieson for editorial assistance and members of the participating laboratories for valuable suggestions throughout the study. Studies reported were in part supported by National Institutes of Health grants P01CA082834and DK32520. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-09-1567
Read article for free, from open access legal sources, via Unpaywall:
http://cancerres.aacrjournals.org/content/canres/69/21/8249.full.pdf
Free after 12 months at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/content/full/69/21/8249
Free after 12 months at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/reprint/69/21/8249.pdf
Free to read at cancerres.aacrjournals.org
http://cancerres.aacrjournals.org/cgi/content/abstract/69/21/8249
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1158/0008-5472.can-09-1567
Article citations
MicroRNA target homeobox messenger RNA in HIV induced hematopoietic inhibition.
Front Cell Dev Biol, 12:1382789, 24 Apr 2024
Cited by: 0 articles | PMID: 38721526 | PMCID: PMC11076784
Expression of miRNA1, miRNA133, miRNA191, and miRNA24, as Good Biomarkers, in Non-Small Cell Lung Cancer Using Real-Time PCR Method.
Asian Pac J Cancer Prev, 23(5):1565-1570, 01 May 2022
Cited by: 2 articles | PMID: 35633539 | PMCID: PMC9587880
Versatile role of miR-24/24-1*/24-2* expression in cancer and other human diseases.
Am J Transl Res, 14(1):20-54, 15 Jan 2022
Cited by: 15 articles | PMID: 35173828 | PMCID: PMC8829624
Review Free full text in Europe PMC
RUNX1: an emerging therapeutic target for cardiovascular disease.
Cardiovasc Res, 116(8):1410-1423, 01 Jul 2020
Cited by: 34 articles | PMID: 32154891 | PMCID: PMC7314639
Review Free full text in Europe PMC
Inhibition of the RUNX1-CBFβ transcription factor complex compromises mammary epithelial cell identity: a phenotype potentially stabilized by mitotic gene bookmarking.
Oncotarget, 11(26):2512-2530, 30 Jun 2020
Cited by: 10 articles | PMID: 32655837 | PMCID: PMC7335667
Go to all (73) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
AML1/ETO proteins control POU4F1/BRN3A expression and function in t(8;21) acute myeloid leukemia.
Cancer Res, 70(10):3985-3995, 11 May 2010
Cited by: 13 articles | PMID: 20460523 | PMCID: PMC2883733
An AML1-ETO/miR-29b-1 regulatory circuit modulates phenotypic properties of acute myeloid leukemia cells.
Oncotarget, 8(25):39994-40005, 01 Jun 2017
Cited by: 10 articles | PMID: 28611288 | PMCID: PMC5522207
Thrombopoietin/MPL participates in initiating and maintaining RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling.
Blood, 120(4):868-879, 21 May 2012
Cited by: 37 articles | PMID: 22613795 | PMCID: PMC3412349
Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts.
Blood, 110(3):799-805, 05 Apr 2007
Cited by: 68 articles | PMID: 17412887 | PMCID: PMC1924771
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (3)
Grant ID: P01CA082834
Grant ID: P01 CA082834
Grant ID: P01 CA082834-09
NIAMS NIH HHS (2)
Grant ID: R01 AR039588
Grant ID: R01 AR049069
NIDDK NIH HHS (3)
Grant ID: P30 DK032520
Grant ID: P30 DK032520-25
Grant ID: DK32520




