Abstract
Free full text

U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation
Associated Data
Summary
In higher eukaryotes, U1 snRNP forms spliceosomes in equal stoichiometry with U2, U4, U5 and U6, however its abundance far exceeds that of the other snRNPs. Here, we used antisense morpholino oligonucleotide (AMO) to U1 snRNA for functional U1 snRNP knockdown in HeLa cells and identified accumulated unspliced pre-mRNAs by genomic tiling microarrays. Remarkably, in addition to inhibiting splicing, U1 snRNP knockdown caused premature cleavage and polyadenylation (PCPA) in numerous pre-mRNAs at cryptic polyadenylation signals (PASs), frequently in introns near (< 5 kb) the start of the transcript. This did not occur when splicing was inhibited with U2 snRNA AMO or the U2 snRNP inactivating drug, spliceostatin A, unless U1 AMO was also included. We further show that U1 snRNA-pre-mRNA base pairing was required to suppress PCPA from nearby cryptic PASs located in introns. These findings reveal a critical splicing-independent function for U1 snRNP in protecting the transcriptome, which we propose explains its overabundance.
Messenger RNAs in eukaryotic cells are produced from primary transcripts (pre-mRNAs) by extensive post-transcriptional processing, including 5′ end capping, removal of introns by splicing, and 3′ end cleavage and polyadenylation1-4. Each splicing reaction is carried out by a spliceosome, a large RNA-protein complex comprised predominantly of small nuclear RNPs (snRNPs)5-8. The U1, U2, U4, U6 and U5 snRNPs are components of the major (U2-type) spliceosome, whereas a much less abundant (~1%) minor (U12-type) spliceosome is comprised of U11, U12, U4atac, U6atac and U5 snRNPs5,9-11. The snRNPs, aided by specific RNA-binding proteins, recognize, by snRNA:pre-mRNA base pairing, canonical sequences within pre-mRNAs that define the major- and minor-class introns, including the intron/exon junctions at the 5′- and 3′-splice sites. U1 snRNP plays an essential role in defining the 5′ splice site by RNA:RNA base pairing via the 5′ nine nucleotide sequence of U1 snRNA. To form the catalytic core of the spliceosome, the snRNPs come together in 1:1 stoichiometry as a modular machine5. However, the abundance of the various snRNPs in cells does not reflect their equimolarity in the spliceosomes. This is particularly striking for U1 snRNP which, at an estimated copy number of ~106 molecules per human cell (HeLa), is much more abundant than the other snRNPs in higher eukaryotes12. The potential role of the different amounts of the snRNPs is not known.
Our interest in exploring a potential function for cellular snRNP abundance arose from earlier observations that deficiency in the survival of motor neurons (SMN) protein, a key component in snRNP biogenesis13-17, perturbs the normal abundance of snRNPs in cells (the snRNP repertoire)18,19 and causes widespread splicing abnormalities19. The possible effect of snRNP abundance changes on splicing and the molecular consequences of SMN deficiency in general are of importance because SMN deficiency is the cause of spinal muscular atrophy (SMA), an often fatal motor neuron degenerative disease20-22. However, the snRNP repertoire changes that occur in an SMN-deficient SMA mouse model vary in different tissues and are not uniform for all the snRNPs18,19, including both down- and up-regulation in the levels of several snRNPs simultaneously, making them difficult to recapitulate. To circumvent this, we investigated the effect of functional reduction of individual snRNPs on the transcriptome using antisense morpholino oligonucleotide (AMO). Our experiments revealed an unexpected function for U1 snRNP in protecting pre-mRNAs from premature cleavage and polyadenylation (PCPA), distinct from its role in splicing.
Functional knockdown of U1 snRNP with AMO
To decrease the amount of functional U1 snRNP, we designed an AMO that covers the 5′ end of U1 snRNA (U1 AMO) to block its binding to 5′ splice sites. To confirm the binding of U1 AMO to U1 snRNP and determine the amount required to inhibit it in cells, we performed an RNase H protection assay. Extracts from cells transfected with a scrambled control AMO23,24 or various concentrations of U1 AMO were incubated with RNase H and an antisense DNA oligonucleotide probe also complementary to U1 snRNA’s 5′ end sequence (Figure 1a). A dose-dependent decrease in the amount of cleaved U1 snRNA was observed as the amount of transfected U1 AMO was increased (Figure 1a), indicating that the U1 AMO prevented the antisense DNA oligo probe from binding and eliciting RNase H digestion. Complete or near complete interference with U1 snRNA 5′ base pairing in cells was observed with 7.5 μM of U1 AMO (Figure 1a). In addition, we used in situ hybridization with a LNA probe complementary to U1 snRNA’s 5′ sequence (nt 1-25) to determine if the U1 AMO was bound to the same sequence in cells. The images (Figure 1b) demonstrate that the U1 AMO indeed shields U1 snRNA’s 5′ sequence in a dose dependent manner and that this sequence is completely inaccessible at 7.5 μM U1 AMO, the concentration that was used in all subsequent U1 AMO transfection experiments. To confirm that the U1 AMO inhibited the activity of U1 snRNP directly, we tested its effect on the in vitro splicing. As shown in Figure 1c, U1 AMO, but not control AMO, strongly decreased the amount of spliced product for Ad2ΔIVS. Thus, U1 AMO functionally inactivated U1 snRNP both in vivo and in vitro.
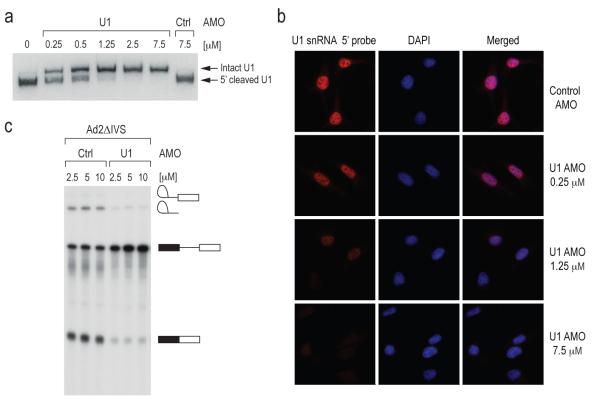
(a) HeLa cells were transfected with the indicated concentrations of control and U1 AMO for 8 hrs. RNase H protection assay was performed using total cell extracts and U1 snRNA was detected by Northern blotting.
(b) In situ hybridization was performed on HeLa cells transfected with varying concentrations of U1 AMO as indicated for 8 hrs using a biotin-labeled LNA probe to the U1 snRNA (left panels) followed by fluorescent Alexa Fluor 594 streptavidin conjugate detection. Nuclei were visualized with DAPI (middle panels) and merged images are shown (right panels).
(c) [α–32P] UTP-labeled Ad2ΔIVS pre-mRNA was spliced in vitro in the presence of control or U1 AMOs at the indicated concentrations. Splicing product identities are depicted to the right of the gel.
Accumulation of unspliced pre-mRNAs after splicing inhibition
To obtain a high resolution global picture of the transcriptome changes that occurred upon U1 snRNP knockdown, including effects on introns and exons, we analyzed total RNA prepared from HeLa cells transfected with either U1 or control AMOs using Affymetrix GeneChip® Human Tiling 2.0R E Arrays. This high-density genomic tiling array includes tiled probes (25-mer oligonucleotides spaced at 10 nt) covering the entire genomic sequence of chromosomes 5, 7 and 16, which are estimated to contain ~3,600 genes. All experiments were carried out as separate biological triplicates and treatments were for 8 hr to allow transfected cells to recover and for sufficient signals to accumulate above background. As a reference, we treated cells in parallel with the potent and general splicing inhibitor, spliceostatin A (SSA), which targets the splicing factor SF3b, a component of U2 snRNP25. As the amount of each pre-mRNA is typically very small and difficult to detect, large increases in intron signals provided the most definitive evidence for accumulation of unspliced pre-mRNAs, ensuring that the corresponding sequences were actively transcribed and their unspliced transcripts are sufficiently stable to be scored. Statistical analysis was performed to identify significant changes that exceeded the following thresholds: fold-change ≥ 2, p-value < 0.01 and length of affected region ≥ 100 nt (corresponding to 3 or more consecutive probes). This identified 319 genes that showed accumulation typically of one or more introns in either U1 AMO (211 genes) or SSA (216 genes). From the outset we expected two patterns due to splicing inhibition. A general reduction in all signals from a transcript could reflect that unspliced pre-mRNA is less stable and rapidly degraded, however, we did not include them in our analysis as they could also potentially result from transcriptional down-regulation. We also expected that U1 AMO and SSA would show similar patterns of intron accumulation throughout the transcript as a result of splicing inhibition in general, but this was observed for only 98 genes (30.7%), as exemplified by ETF1 (Figure 2). A small number of genes (41 genes; 12.9%) showed other profiles but did not exhibit any coherent pattern. Unexpectedly, however, the majority of the genes (180 genes; 56.4%) showed different patterns for U1 AMO and SSA (HSPA9, SRRM2 and CBFB in Figure 2).
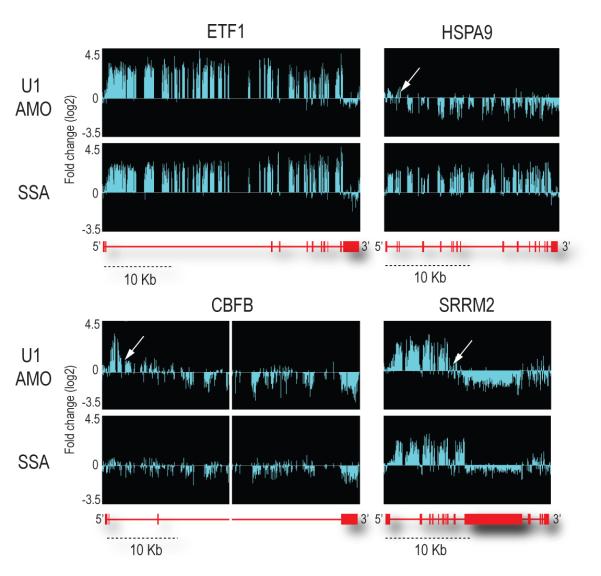
RNA samples prepared from control or U1 AMOs-transfected cells (7.5 μM, 8 hrs) or SSA (100 ng/ml, 8 hrs)-treated cells were analyzed using genomic tiling array. Fold-changes (log2) of signal intensities of U1 AMO-transfected and SSA-treated cells compared to control cells are shown above the corresponding structure of each gene. With a scale shown below, gene structures are depicted in red, with horizontal lines indicating introns and boxes indicating exons. The middle part of CBFB gene (~35 kb) was removed. White arrows indicate points showing abrupt drop of the signal (inflection points).
U1 snRNP knockdown causes PCPA
Most remarkably, the majority of the affected genes in the U1 AMO-transfected cells showed a similar pattern consisting of strong intron signals that terminated prematurely relative to SSA, for example in HSPA9, SRRM2 and CBFB as indicated by arrows in Figure 2. This usually occurred in introns in the first quarter of the gene with a strong 5′ bias, as shown for several functionally diverse genes in Figure 3. Notably, in U1 AMO-transfected cells there was an abrupt decrease in unspliced intron signals frequently within 3-5 kb from the start of the transcript. From the point at which a sharp drop in the signals in U1 AMO-treated cells was observed, all downstream signals, including exons, were consistently lower than those of control and the SSA-treated samples. To define the termination point, we characterized cDNA fragments from this region (indicated by arrows in Figure 3), including 3′ RACE products for transcripts of several genes that showed this pattern from U1 AMO-transfected cells, of which NR3C1 and STK17A were sequenced (Figure 4a). Surprisingly, this revealed that the transcripts had a poly(A) sequence at the 3′ end that is not found in the genomic sequence and which was added post-transcriptionally ~20 nucleotides downstream of a potential polyadenylation signal (PAS), typically AAUAAA (Figure 4a). These findings suggest that pre-mRNAs are prematurely cleaved and polyadenylated within an intron when the levels of functional U1 snRNP are reduced.

Representative examples of genes differentially affected by U1 AMO and SSA. The sudden drop in signals in U1 AMO-transfected cells is indicated by green arrow heads. With a scale shown below, gene structures are depicted in red with horizontal lines indicating introns and boxes indicating exons.
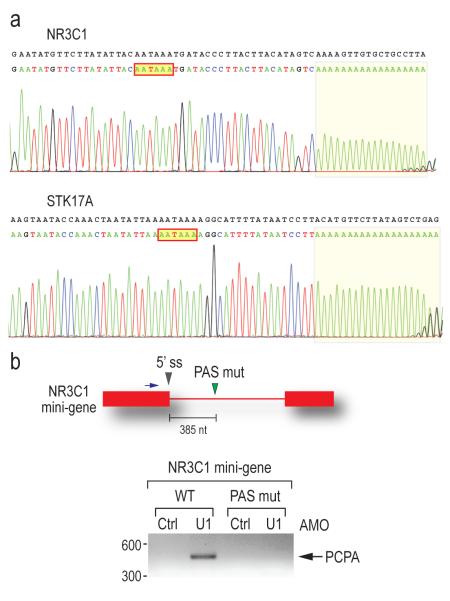
(a) 3′ RACE using nested PCR was performed to detect polyadenylated mRNAs using total RNA from U1 AMO (7.5 μM, 8 hrs)-transfected cells. Sequencing results of the 3′ RACE product for the NR3C1 and STK17A genes are shown with the corresponding genomic sequence in black. The poly(A) tails are shaded and the putative PASs are indicated in red line boxes.
(b) HeLa cells were transfected with the wild-type and PAS mutant (mutated from AAUAAA to GAAUUC) NR3C1 mini-gene construct followed by either control or U1 AMOs. 3′ RACE was performed as described in (a). The mini-gene structure is depicted to the above of the gel, with a blue arrow indicating the forward primer for 3′ RACE. Gray arrowhead: PAS mutation. PCPA: premature cleavage and polyadenylation. Band sizes (bp) are indicated to the left of the gel.
To determine if these putative PASs are functionally relevant, we constructed a NR3C1 mini-gene consisting of exons 2 and 3 which flank a trancated intron 2 where the cleavage and polyadenylation occurred. An identical plasmid was also constructed with a mutation in the putative PAS in intron 2 inferred from the tiling array results (Figure 4b). Cells expressing each of these plasmids were then transfected with either control or U1 AMOs. Wild type plasmid expressing cells showed PCPA at the cryptic PAS only after U1 AMO treatment (Figure 4b), consistent with the result of the endogenous NR3C1 gene. No cleavage and polyadenylation occurred in the transcript containing a mutant PAS in either control or U1 AMO-transfected cells (Figure 4b), demonstrating that this cryptic PAS is functional and that this process is similar to that which occurs normally at the 3′ ends of mRNAs.
Suppression of PCPA is U1 snRNP-specific and splicing independent
Unlike U1 AMO, tiling arrays with SSA or U2 AMO (which generally showed a tiling array pattern of splicing inhibition similar to that of SSA; Supplementary Figure 1) did not show PCPA. To test this directly, we used an oligo(dT) reverse primer to amplify NR3C1 and STK17A transcripts in control, U1 or U2 AMO-treated cells and SSA-treated cells (Figure 5a). As expected, PCPA was observed for U1 AMO, however none or very little was seen in U2 AMO and SSA-treated cells, which could be due to destabilization of U1 snRNP binding upon U2 snRNP inhibition. Interestingly, PCPA still occurred in cells treated simultaneously with U1 AMO and SSA, indicating that the effect of U1 inhibition is dominant over splicing inhibition (Figure 5b). We conclude that U1 suppresses cleavage and polyadenylation and that this is not a consequence of the splicing inhibition that it causes, as neither U2 AMO nor SSA showed this effect.
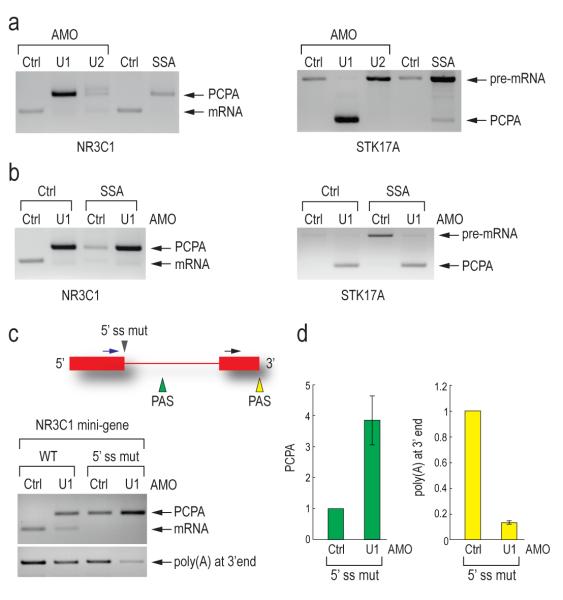
(a) 3′ RACE was carried out as described in Figure 4a on the endogenous NR3C1 and STK17A genes using RNA samples from HeLa cells transfected with control, U1 (7.5 μM, 8 hrs) or U2 AMO (25 μM, 8 hrs), or treated with SSA (100 ng/ml, 8hrs).
(b) 3′ RACE was carried out as described in Figure 4a using RNA samples from HeLa cells transfected with control or U1 AMOs (7.5 μM) with or without SSA (100 ng/ml) for 8 hrs.
(c) HeLa cells were transfected with wild-type and 5′ splice site mutant NR3C1 (mutated from AAGGTAAGA to GTCCATTCA) mini-gene. 3′ RACE was performed as described in Figure 4a. The mini-gene structure is depicted. Blue arrow: forward primer to detect PCPA; Black arrow: forward primer to detect polyadenylation at 3′ end; Gray arrowhead: 5′ splice site mutation; Green and yellow arrowheads: polyadenylation signals in intron and at 3′ end, respectively. An unspliced and normally cleaved product was too large to detect.
(d) Quantitation of PCPA and normal polyadenylation at 3′ end in control and U1 AMO treated cells was performed using real-time PCR. Error bars indicate s.d. (n=3). PCPA in panel a, b, c and d: premature cleavage and polyadenylation.
The functional PAS from which cleavage and polyadenylation occurred in the NR3C1 mini-gene upon U1 snRNP depletion is less than 400 nt downstream of the 5′ splice site and it thus seemed likely that U1 snRNP base paired to this 5′ splice site suppresses utilization of this cryptic PAS. To test this, we mutated the 5′ splice site, which inactivated splicing as evidenced by the lack of mRNA (Figure 5c). While the wild type mini-gene showed PCPA only upon U1 AMO treatment, in 5′ splice site mutant transfected cells, PCPA was observed even with control morpholino treatment. This suggests that U1 snRNP base paired to the 5′ splice site is able to suppress the cryptic PAS (Figure 5c). However, treatment of the 5′ splice site mutant with U1 AMO resulted in ~4-fold increase in PCPA from the same cryptic PAS and concomitantly ~8-fold decrease in polyadenylation from the normal PAS at the transcript’s 3′ end (Figure 5c, d). Therefore, U1 snRNP base paired to sequences other than the 5′ splice site also suppresses PCPA. The large number of potential U1 snRNP binding sites in introns precluded identification of other sites from which U1 snRNP might suppress premature utilization of this PAS (Supplementary Figure 2). These findings suggest that the PCPA results from loss of U1 snRNA’s 5′ end base pairing with the pre-mRNA, indicating a U1 snRNP function other than and independent of its known function in splicing.
Discussion
Our experiments here revealed an unexpected function for U1 snRNP in protecting transcripts from PCPA in addition to and independent of its role in splicing. As a reference for U1 AMO, the general splicing inhibitor, SSA25, which inactivates the U2 snRNP component SF3b5,26, allowed identification of introns that were stable enough and accumulate to significantly detectable levels when their splicing was inhibited. As expected, the patterns observed for U1 AMO and SSA (which is similar to that of U2 AMO) showed that both efficiently inhibited splicing. However, U1 snRNP functional reduction had an additional and striking effect, resulting in the failure to produce full-length pre-mRNA from the majority of genes in our dataset. We showed that this was due to premature cleavage and polyadenylation from a cryptic PAS, typically in an intron and frequently within the first few kilobases (< 5 kb) from the start of RNA polymerase II transcripts. A non-splicing role for a snRNP has been previously shown for U2 snRNP in the 3′ end formation of histone mRNAs27,28.
The mechanism by which U1 snRNP suppresses PCPA is not presently known. However, as it occurs from canonical PASs, it is reminiscent of previous observations on the capacity of tethered U1 snRNP to regulate normal 3′ end cleavage and polyadenylation from the natural PASs in the last exon29, and may have features in common with it. For example, the U1 snRNP protein U1-70K can interact directly with the poly(A) polymerase (PAP)30,31 and inhibit polyadenylation. Targeting 5′-mutated U1 snRNAs with complementarity to sequences in the vicinity (within < 500 nt) of the natural PAS at 3′-terminal exon results in degradation of the transcript because cleavage occurs without addition of a poly(A) tail, leaving the transcript vulnerable to 3′ exonucleases32. A considerable number of genes we surveyed, but were not included in our analysis, showed a decrease in exon signals or in both introns and exons throughout the transcript in U1 AMO-treated cells. It is possible that in these cases cleavage occurred without subsequent polyadenylation, and the transcript was therefore rapidly degraded. Alternatively, cleavage and polyadenylation may have occurred very close to the transcription start site, making these transcripts difficult to detect. These scenarios are nevertheless consistent with a role for U1 snRNP in suppressing cleavage and polyadenylation throughout the entire pre-mRNA by a similar machinery that until now was thought to only process the 3′ end of mRNA, in an even larger number of genes than our dataset presents.
Stochastically, canonical PASs (most frequently AAUAAA or AUUAAA) occur every 2,000 nucleotides, though in several of the genes we studied, including NR3C1, STK17A and BASP1, cryptic PASs are found every 500-800 nt. The strong 5′ bias with which PCPA occurred in these genes upon U1 snRNP functional knockdown suggests that one of the first few cryptic PASs is utilized. Up to the point at which PCPA occurred, these transcripts also contained many cryptic 5′ splice sites (Supplementary Figure 2). We propose the following model to explain our observations. Pre-mRNA processing factors, including splicing factors, hnRNP proteins, snRNPs and 3′ end cleavage and polyadenylation factors co-transcriptionally associate with nascent transcripts33-37. Direct association of cleavage/polyadenylation factors with the CTD of RNA pol II in the transcription elongation complex has been demonstrated36. U1 snRNP associates with nascent transcripts, by base pairing with cognate sequences on the nascent pre-mRNA, including 5′ splice sites and cryptic 5′ splice sites and inhibits the cleavage/polyadenylation machinery from attacking the pre-mRNA at cryptic PASs. We envision that when U1 snRNP’s base pairing is prevented, as is the case in U1 AMO-transfected cells, cleavage and polyadenylation occurs co-transcriptionally at the first actionable PAS that the transcription elongation complex encounters. By actionable PAS, we mean one that has the necessary hexanucleotide consensus and is in an RNP context that makes it accessible and susceptible to attack by the cleavage/polyadenylation machinery unless U1 snRNP base paired in the vicinity is able to protect it. We suggest that under normal circumstances, this encounter happens after the last strong U1 binding site (5′ splice site or a cryptic 5′ splice site) in the 3′ UTR of the terminal exon because a sufficient density of U1 snRNP base paired throughout protects the transcript up to that point. The likelihood of normal or premature termination may be enhanced by the presence of pausing sites38.
U1 snRNP bound to 5′ splice sites may thus serve a dual purpose – in splicing and suppression of PCPA. The perimeter of U1 snRNP’s protective zone is not known, but its binding to 5′ splice site alone is unlikely to be able to protect the majority of introns, which in humans average ~ 3.4 kb in length39. Furthermore, if suppression of actionable PASs was provided only from U1 snRNP bound to 5′ splice sites, 5′ splice site mutations would be expected to cause premature termination, as opposed, for example, to exon skipping, which would be extremely deleterious and, to our knowledge, has not been observed. Additional U1 snRNP binding sites, including cryptic 5′ splice sites, may function as tethering sites for its activity in suppression of cleavage and polyadenylation in introns. Viewed from this perspective, sequences referred to as cryptic 5′ splice sites may serve a non-splicing purpose to recruit U1 snRNP to protect introns. It is also reasonable to consider that modulating U1 snRNP levels or its binding at sites that protect actionable PASs could be a mechanism for regulating gene expression, including down regulation of the mRNA or switching expression to a different mRNA produced from a prematurely terminated pre-mRNA. We suggest that the vulnerability to PCPA would be expected to increase with increasing intron size if U1 snRNP and cognate base-pairing sites are not available to protect it. We propose that the large excess of U1 snRNP over what is required for splicing in human cells serves an additional critical biological function, to suppress PCPA in introns and protect the integrity of the transcriptome.
Methods Summary
Antisense moroholino oligonucleotide (AMO) transfection was performed by electroporation. The sequences of the U1 and control AMOs (Gene Tools) are 5′-GGTATCTCCCCTGCCAGGTAAGTAT-3′ and 5′-CCTCTTACCTCAGTTACAATTTATA-3′, respectively23,24. RNase H protection assay was carried out using AMO-transfected cell extracts and antisense DNA oligo for U1 snRNA (5′-CAGGTAAGTAT-3′). After RNase H treatment, RNA samples were purified and analyzed by Northern blotting with an U1 snRNA probe (5′-CAAATTATGCAGTCGAGTTTCCCACATTTG-3′). In situ hybridization of U1 snRNA was performed with a biotin-labeled LNA probe (5′-GGTATCTCCCCTGCCAGGTAAGTAT-3′). Nuclei were stained by DAPI. For in vitro splicing, [α–32P] UTP labeled Ad2ΔIVS pre-mRNA was prepared as previously described40. In vitro splicing reactions were carried out in 293T whole cell extracts prepared as previously described41. Splicing products were resolved on denaturing PAGE, and gels were autoradiographed. For tiling array, labeled cDNA targets were prepared and applied to Affymetrix® GeneChip® Human tiling 2.0R E arrays. Arrays were scanned to produce .CEL files. The .CEL files were analyzed using the Affymetrix® Tiling Analysis Software (TAS) to produce .BED files of signal intensity and p-value. Overlapping regions of two datasets were chosen using Galaxy (http://galaxy.psu.edu/)42. We produced .BAR files from the .CEL files using TAS to visualize on the Integrated Genome Browser (Affymetrix). For 3′ RACE, cDNA was synthesized from total RNA using an oligo dT18-XbaKpnBam primer. The first and second (nested) PCR reactions were performed using gene specific forward primers and the XbaKpnBam reverse primer. For 3′ RACE of NR3C1 mini-gene, pcDNA3.1-5′ primer was used as the first primer to distinguish mini-gene RNA from endogenous NR3C1 RNA. To construct the NR3C1 mini-gene, DNA fragments of NR3C1 intron 1-exon 2-intron 2 and NR3C1 intron 2-exon 3 were amplified and subcloned into pcDNA3.1 vector. The poly(A) site and 5′ splice site were mutated in this construct where indicated. Sequences of all primers are listed in Supplementary Table 1.
Methods
Cell culture and antisense morpholino oligonucleotide transfection
HeLa PV cells were cultured as previoiusly described. HeLa cells (~107 cells per transfection) were trypsinized, washed twice with PBS and resuspended in 400 μl of DMEM without serum. After mixing cells with morpholino oligo, they were transferred to a 0.4 cm gap cuvette (Bio-rad). Electroporation was performed using a Bio-Rad Gene pulser at 960 μF and 280 V. After electroporation, cells were cultured for 8 hours in 6 well plates with 2 ml DMEM. The sequence of the 25-mer U1 AMO is 5′-GGTATCTCCCCTGCCAGGTAAGTAT-3′, which is complementary to nucleotides 1-25 in human U1 snRNA. The sequence of the 25-mer U2 AMO is 5′-TGATAAGAACAGATACTACACTTGA-3′, which is complementary to nucleotides 27-51 in human U2 snRNA. The 25-mer scrambled sequence control AMO is 5′-CCTCTTACCTCAGTTACAATTTATA-3′, as previously described. AMOs were obtained from Gene Tools, LLC.
RNA preparation and 3′ RACE
Total RNA was extracted from cultured cells using TRIzol (Invitrogen). cDNA was synthesized using SuperScript® III reverse transcriptase (Invitrogen) using oligo dT18-XbaKpnBam primer for 3′ RACE according to the manufacturer’s directions. 3′ RACE was carried out using the first and second (nested) forward primers and XbaKpnBam reverse primer.
For 3′ RACE of NR3C1 mini-gene, pcDNA3.1-5′ primer was used as a forward primer to distinguish mini-gene RNA from endogenous NR3C1 RNA. PCR products were cut with HindIII to distinguish PCR products of prematurely polyadenylated RNA from mRNA spliced and polyadenylated at the canonical PAS at the 3′ end. Quantitation of PCPA and polyadenylation at the canonical PAS at the 3′ end in control and U1 AMO treated cells (Figure 5d) was performed using real-time PCR. Primer sequences are listed in Supplementary Table 1.
Tiling array target preparation, hybridization and data analysis
Total RNAs were prepared from HeLa cells transfected with control or U1 AMOs (7.5 μM), or SSA (100 ng/ml) or methanol for 8 hrs. Labeled cDNA targets were prepared using the GeneChip® WT amplified Double-Stranded cDNA Synthesis Kit and the GeneChip® WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix) according to the manufacturer’s directions. The end-labeled cDNA targets were applied to GeneChip® Human tiling 2.0R E arrays (Affymetrix). Hybridization was performed using F450-001 fluidics wash and stain script on the Affymetrix GeneChip Fluidics Station 450. Arrays were scanned using the Affymetrix GCS 3000 7G GeneChip Operating Software (GCOS) to produce .CEL files.
For tiling array analysis, we used .CEL files and the Affymetrix® Tiling Analysis Software (TAS) Version 1.1 to produce .BED files of the following signal intensity and p-value (BW = 50, Min. Run = 100, Max. Gap = 100, fold change ≥ 2-fold, P-value < 0.01). Overlapping regions of two files of signal intensity and p-value were chosen using Galaxy (http://galaxy.psu.edu/). We produced .BAR files from the .CEL files using TAS to visualize on the Integrated Genome Browser (IGB) (Affymetrix).
In vitro splicing
[α–32P] UTP labeled Ad2ΔIVS pre-mRNA was prepared as previously described44. In vitro splicing reactions were carried out in 293T whole cell splicing extracts prepared as previously described. Increasing amounts of control and U1 AMOs were added to reactions that were incubated for 90 min at 30°C. Splicing products were purified with TRIzol, resolved on denaturing PAGE, and gels were autoradiographed.
RNase H protection assay and Northern blotting
HeLa cells were transfected with AMOs and incubated for 8 hrs. Cells were harvested and total cell extract was prepared using RSB-100 buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2). The cell extract was incubated with 1.5 U of RNase H (Promega) and 5 μM antisense DNA oligo in a 20 μl reaction for 25 min at 30°C. Antisense DNA oligo for U1 snRNA is 5′-CAGGTAAGTAT-3′. After RNase H treatment, RNA samples were purified and analyzed by Northern blotting with [γ–32P] ATP labeled U1 snRNA probe, the sequence of which is 5′-CAAATTATGCAGTCGAGTTTCCCACATTTG-3′.
Plasmids construction
To construct NR3C1 mini-gene, DNA fragments of NR3C1 intron 1-exon 2-intron 2 and NR3C1 intron 2-exon 3 were amplified from genomic DNA from HEK293T cells using NR3C1 int1 for and NR3C1 int2 rev XhoI, and NR3C1 int2 for XhoI and NR3C1 Ex3 rev XhoI. The NR3C1 intron 1-exon 2-intron 2 fragment was digested with XhoI and EcoRI, and the NR3C1 intron 2-exon 3 fragment was digested with XhoI. These fragments were subcloned into pcDNA3.1 vector (Invitrogen). The intron 2 of NR3C1 was truncated from 80 kb to 2 kb to facilitate cloning, mutagenesis and transfection. To introduce mutations, QuikChange® II Site-Directed Mutagenesis Kits (Stratagene) was used with NR3C1 polyA site mutation for and NR3C1 polyA site mutation rev, and NR3C1 5′ SS mutation for and NR3C1 5′ SS mutation rev primers. Sequences of all primers are listed in Supplementary Table 1.
In situ hybridization
In situ hybridization of U1 snRNA in HeLa cells transfected with control or U1 AMO for 8 hrs was performed with a biotin-labeled LNA probe (5′-GGTATCTCCCCTGCCAGGTAAGTAT-3′) obtained from Exiqon. The protocol was essentially as described by the manufacturer (Exiqon). Hybridization was performed in 50% formamide, 2X SSC, 50 mM sodium phosphate (pH 7), and 10% dextran sulfate, containing 10 nM LNA probe, at ~ 20° C below the melting temperature (Tm) for 1 hr (50° C) in a humidified chamber. Following hybridization, cells were washed in 2X SSC + 0.1 % Triton X-100, followed by detection with a fluorescent Alexa Fluor 594 streptavidin conjugate. Cells were washed 3 × 5 mins at 37° C in 4X SSC + 0.1 % Triton X-100, followed by washes in 2X and 1X SSC, with a final wash in PBS at RT. Nuclei were stained by DAPI.
Supplementary Material
1
2
3
Acknowledgements
We are grateful to the members of our laboratory, especially Drs. Jeongsik Yong and Jennifer Bachorik, for helpful discussions and comments on this manuscript. We thank Dr. Minoru Yoshida for providing spliceostatin A. We also thank Dr. Don A. Baldwin and Hetty Rodriguez at the Microarray Core Facility at the University of Pennsylvania School of Medicine for help with the tiling array. This work was supported by the Association Française Contre les Myopathies (AFM). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Full methods are available in the online version of the paper at www.nature.com/nature
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature09479
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2996489
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature09479
Article citations
Physiological and pathological roles of the transcriptional kinases CDK12 and CDK13 in the central nervous system.
Cell Death Differ, 12 Nov 2024
Cited by: 0 articles | PMID: 39533070
Review
Steering research on mRNA splicing in cancer towards clinical translation.
Nat Rev Cancer, 09 Oct 2024
Cited by: 0 articles | PMID: 39384951
Review
The Arabidopsis U1 snRNP regulates mRNA 3'-end processing.
Nat Plants, 10(10):1514-1531, 23 Sep 2024
Cited by: 1 article | PMID: 39313562 | PMCID: PMC11489095
The MTR4/hnRNPK complex surveils aberrant polyadenylated RNAs with multiple exons.
Nat Commun, 15(1):8684, 17 Oct 2024
Cited by: 0 articles | PMID: 39419981 | PMCID: PMC11487169
PAIP1 binds to pre-mRNA and regulates alternative splicing of cancer pathway genes including VEGFA.
BMC Genomics, 25(1):926, 03 Oct 2024
Cited by: 0 articles | PMID: 39363305 | PMCID: PMC11451205
Go to all (417) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A U1/U4/U5 snRNP complex induced by a 2'-O-methyl-oligoribonucleotide complementary to U5 snRNA.
Science, 272(5263):881-884, 01 May 1996
Cited by: 14 articles | PMID: 8629024
U4 snRNP inhibits premature cleavage and polyadenylation of pre-mRNAs.
Proc Natl Acad Sci U S A, 121(27):e2406710121, 25 Jun 2024
Cited by: 1 article | PMID: 38917004 | PMCID: PMC11228486
The U1 antisense morpholino oligonucleotide (AMO) disrupts U1 snRNP structure to promote intronic PCPA modification of pre-mRNAs.
J Biol Chem, 299(7):104854, 22 May 2023
Cited by: 4 articles | PMID: 37224962 | PMCID: PMC10404622
Principles and correction of 5'-splice site selection.
RNA Biol, 19(1):943-960, 01 Jan 2022
Cited by: 7 articles | PMID: 35866748 | PMCID: PMC9311317
Review Free full text in Europe PMC
Funding
Funders who supported this work.





