Abstract
Rationale
Vascular remodeling in pulmonary arterial hypertension (PAH) involves proliferation and migration of endothelial and smooth muscle cells, leading to obliterative vascular lesions. Previous studies have indicated that the endothelial cell proliferation is quasineoplastic, with evidence of monoclonality and instability of short DNA microsatellite sequences.Objectives
To assess whether there is larger-scale genomic instability.Methods
We performed genome-wide microarray copy number analysis on pulmonary artery endothelial cells and smooth muscle cells isolated from the lungs of patients with PAH.Measurements and main results
Mosaic chromosomal abnormalities were detected in PAEC cultures from five of nine PAH lungs but not in normal (n = 8) or disease control subjects (n = 5). Fluorescent in situ hybridization analysis confirmed the presence of these abnormalities in vivo in two of three cases. One patient harbored a germline mutation of BMPR2, the primary genetic cause of PAH, and somatic loss of chromosome-13, which constitutes a second hit in the same pathway by deleting Smad-8. In two female subjects with mosaic loss of the X chromosome, methylation analysis showed that the active X was deleted. One subject also showed completely skewed X-inactivation in the nondeleted cells, suggesting the pulmonary artery endothelial cell population was clonal before the acquisition of the chromosome abnormality.Conclusions
Our data indicate a high frequency of genetically abnormal subclones within PAH lung vessels and provide the first definitive evidence of a second genetic hit in a patient with a germline BMPR2 mutation. We propose that these chromosome abnormalities may confer a growth advantage and thus contribute to the progression of PAH.Free full text

Somatic Chromosome Abnormalities in the Lungs of Patients with Pulmonary Arterial Hypertension
Abstract
Rationale: Vascular remodeling in pulmonary arterial hypertension (PAH) involves proliferation and migration of endothelial and smooth muscle cells, leading to obliterative vascular lesions. Previous studies have indicated that the endothelial cell proliferation is quasineoplastic, with evidence of monoclonality and instability of short DNA microsatellite sequences.
Objectives: To assess whether there is larger-scale genomic instability.
Methods: We performed genome-wide microarray copy number analysis on pulmonary artery endothelial cells and smooth muscle cells isolated from the lungs of patients with PAH.
Measurements and Main Results: Mosaic chromosomal abnormalities were detected in PAEC cultures from five of nine PAH lungs but not in normal (n = 8) or disease control subjects (n = 5). Fluorescent in situ hybridization analysis confirmed the presence of these abnormalities in vivo in two of three cases. One patient harbored a germline mutation of BMPR2, the primary genetic cause of PAH, and somatic loss of chromosome-13, which constitutes a second hit in the same pathway by deleting Smad-8. In two female subjects with mosaic loss of the X chromosome, methylation analysis showed that the active X was deleted. One subject also showed completely skewed X-inactivation in the nondeleted cells, suggesting the pulmonary artery endothelial cell population was clonal before the acquisition of the chromosome abnormality.
Conclusions: Our data indicate a high frequency of genetically abnormal subclones within PAH lung vessels and provide the first definitive evidence of a second genetic hit in a patient with a germline BMPR2 mutation. We propose that these chromosome abnormalities may confer a growth advantage and thus contribute to the progression of PAH.
Pulmonary arterial hypertension (PAH) is characterized by vascular remodeling of the pulmonary arterioles, including formation of plexiform and concentric lesions comprised of proliferative endothelial cells and myofibroblasts (1–3). Together with vasoconstriction, this remodeling leads to a sustained elevation in pulmonary artery pressure. The primary triggers are largely unknown. Treatments such as nitric oxide, prostanoids, and endothelin receptor antagonists (4–6) target pathophysiological changes and have improved survival, but PAH remains a potentially life-threatening disease that may require lung transplantation. Heterozygous germline mutations of the bone morphogenetic protein receptor type-II (BMPR2) gene are identifiable in approximately 75% of familial PAH cases and in approximately 20% of idiopathic cases (7–9). However, a BMPR2 mutation alone is not sufficient to cause PAH; the penetrance averages only 20%, suggesting that other genetic or environmental factors play a role.
Endothelial cells within the plexiform lesions of idiopathic and appetite suppressant–associated PAH cases were previously demonstrated to be monoclonal (10, 11). Some lesions also showed microsatellite instability, a hallmark of hereditary nonpolyposis colon cancer, and mutations of the apoptosis regulator BAX (12). In culture, cells from PAH pulmonary arteries are hyperproliferative, apoptosis-resistant and predominantly glycolytic, with decreased numbers of mitochondria (13–15). Together, these findings have led to the hypothesis that the pathogenesis of complex vascular lesions in PAH is akin to neoplasia, with the acquisition of multiple genetic mutations (10, 16, 17). BMPRII immunostaining in PAH lung tissue is markedly reduced below the 50% level expected in patients with heterozygous BMPR2 mutations (18), suggesting that BMPR2 may act as a classical tumor suppressor gene, with somatic inactivation of the wild-type allele in affected lungs. However, no evidence of loss of heterozygosity was found at the BMPR2 locus (19), suggesting that any second hits lie elsewhere in the genome. To test this hypothesis, we conducted genome-wide copy number analysis using single-nucleotide polymorphism (SNP) arrays on pulmonary artery endothelial cells (PAEC) and pulmonary artery smooth muscle cells (PASMC) isolated from PAH lungs. We present evidence of somatic chromosomal abnormalities in more than 50% of PAH lungs examined, including one patient with a germline BMPR2 mutation, supporting the hypothesis that multiple genetic hits may underlie the vascular proliferation in this disease. The results of this study have been previously reported in the form of an abstract at national meetings of the American Thoracic Society and American Society of Human Genetics (20).
METHODS
Isolation and Culture of PAEC and PASMC
This study was approved by all relevant institutional review boards, and all patients gave written informed consent. PAEC were isolated as previously described (14, 15) from the lungs of nine PAH patients, eight healthy control subjects, and five patients with cystic fibrosis or chronic obstructive pulmonary disease. Patients with PAH were classified according to the updated World Health Organization guidelines (21). Control cells were isolated from donor lungs unsuitable for transplant (n = 4) or purchased commercially (n = 4) (Lonza, Allendale, NJ). To assess endothelial cell purity by flow cytometry, confluent cultures were trypsinized and stained with anti-human CD31-FITC (Becton Dickinson, San Jose, CA). Isotype-matched irrelevant antibody was used as control. At least 10,000 events were acquired on a FACScan flow cytometer (Becton Dickinson). Data were analyzed using Cell-Quest 3.3 software (Becton Dickinson). Background staining of less than 5% was considered acceptable. PASMC, bronchial smooth muscle cells, and airway epithelial cells were isolated as previously described (22, 23) and analyzed as available (Table 1).
TABLE 1.
SUBJECTS AND CELL TYPES EXAMINED FOR CHROMOSOME ABNORMALITIES
| Subject Code | WHO Class | Age | Sex | Smoking (pack-yr) | BMPR2 Mutation | % PAEC* with Cytogenetic Abnormally | Other Cell Types Tested (normal except where indicated otherwise) |
|---|---|---|---|---|---|---|---|
| PAH-1 | 1.2 | 41 | M | 0 | del exon 1-8 | 20% monosomy 13 | PASMC, BrSMC, AEC, PBMC |
| PAH-2 | 1.4 | 58 | F | 6 | No | 24% monosomy X | PBMC |
| PAH-3 | 1.4 | 45 | F | NK | No | 46% monosomy X | PASMC (del X)†, BrSMC (normal) |
| PAH-4 | 1.1 | 49 | F | NK | No | 41% monosomy X | PASMC, BrSMC |
| PAH-5 | 1.1 | 64 | M | 0 | No | 12% monosomy 8p23.1-p12‡ | PASMC, PBMC |
| PAH-6 | 1.2 | 50 | F | 0 | del exon 4-5 | None | PBMC |
| PAH-7 | 1.1§ | 60 | F | 10 | No | None | PASMC, PBMC |
| PAH-8 | 1.1 | 30 | F | NK | No | None | |
| PAH-9 | 1.1 | 34 | F | NK | No | None | |
| CF-1 | 43 | M | 21 | None | |||
| CF-2 | 7 | F | 0 | None | |||
| COPD-1 | 57 | F | 60 | None | PASMC | ||
| COPD-2 | 34 | F | 0 | None | PASMC | ||
| COPD-3 | 61 | M | 20 | None | |||
| Controls (×8) | NK | Both | NK | None |
Definition of abbreviations: AEC = airway epithelial cells; BrSMC = bronchial smooth muscle cells; CF = cystic fibrosis; COPD = chronic obstructive pulmonary disease; NK = not known; PAEC = pulmonary artery endothelial cells; PAH = pulmonary arterial hypertension; PASMC = pulmonary artery smooth muscle cells; PBMC = peripheral blood mononuclear cells.
DNA Analysis
Cells were harvested by manual scraping, and DNA was extracted using the Qiagen DNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer's recommended protocol. BMPR2 mutation analysis was performed by direct sequencing and multiplex ligation-dependent probe amplification as previously described (8, 24). SMAD9 was analyzed by direct sequencing (conditions available on request). To detect genome-wide copy number changes, DNA was hybridized to Illumina (San Diego, CA) Human CNV370 and Affymetrix (Santa Clara, CA) SNP 6.0 arrays according to recommended protocols and analyzed using the manufacturers' software. Where available, matched leukocyte DNA was used as a baseline for comparison of PAEC and PASMC array profiles. Data are deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/; accession numbers GSE20206 and GSE21097). Copy number abnormalities were identified within the SNP array data by genomic segmentation and confirmed by the observation of allele intensity ratio imbalances over the affected regions. No regions of loss of heterozygosity were detected. X-inactivation studies were conducted using a standard human androgen receptor assay (25) with the methylation sensitive enzyme HhaI and analyzed on an ABI 3730 fluorescent sequencer.
Fluorescence In Situ Hybridization
FISH analysis was conducted using the following probes: LSI 13/RB-1 (13q14) SpectrumOrange, LSI 13q34 SpectrumGreen, LSI LPL (8p22) SpectrumOrange, LSI MYC (8q24) SpectrumGreen, CEP8 (chromosome-8 centromere) SpectrumAqua, LSI Androgen Receptor (Xq12) SpectrumOrange, and CEP11 (chromosome-11 centromere) SpectrumGreen (Abbott Molecular, Des Plaines, IL). Formalin-fixed, paraffin-embedded, 4-μm lung sections were hybridized as previously described (26). The plexiform and concentric lesion areas were mapped on serial hematoxylin and eosin–stained sections. As controls, airway regions far away from the lesions and with nuclei of similar size and shape were selected. Fluorescence microscope analysis was performed using single band interference filters. Monochromatic images were captured for each specific filter using a CCD camera and merged using CytoVision (Applied Imaging Inc., Grand Rapids, MI). Interphase fluorescence in situ hybridization (FISH) analysis of thin tissue sections can be limited by nuclear truncation, whereby the entire nucleus of a cell may not be represented on the slide. To minimize this limitation, only nuclei with at least one signal from every probe in the set were scored. Hybridization for each probe was assumed to be independent and therefore tabulated separately, counting the number of nuclei with one, two, or more than two signals for each probe. The distribution of signals for each probe in control regions was compared using the chi-square test of association or Fisher's exact test, as appropriate for the number of cells scored, with the null hypothesis that normal cells show no significant variation between probes. As expected, each probe gave a similar distribution of one, two, or more than two signals in control cells, and there was no evidence of significant technical differences between the probes. This control distribution was then used to generate expected values for a chi-square goodness-of-fit test on the hybridization patterns observed in lesions as compared with nonvascular structures. Because all abnormalities predicted by the array data were chromosomal losses, we also performed a one-tailed z-score test to determine whether the proportion of nuclei in vascular lesions that showed a single signal was significantly greater than in control regions.
RESULTS
Two of the nine patients with PAH harbored multi-exon deletions of the BMPR2 gene (Table 1). Both mutations were present in leukocyte DNA, indicating that they are germline mutations. No SMAD9 mutations were identified. SNP microarray analysis detected mosaic chromosome abnormalities in PAEC from five patients (Table 1). Four of these were whole chromosome losses; the X chromosome was deleted in three female subjects, and chromosome-13 was lost in PAEC from subject PAH-1, who also harbors a germline BMPR2 mutation (Figure 1). The fifth abnormality was an interstitial deletion of the short arm of chromosome-8, 8p23.1-p12 (Figure 1). The estimated proportion of abnormal cells ranged from 12 to 46% (Table 1). To determine whether these changes were lung specific, peripheral blood mononuclear cells and/or other lung-derived cells were analyzed as available (Table 1). In four of the five cases with chromosome losses in PAEC, other cell types showed no detectable abnormalities, confirming that these are lineage-specific acquired somatic changes. PASMC from subject PAH-3 exhibited no detectable abnormality on the Affymetrix array, but Illumina analysis suggested a low frequency of X chromosome aneusomy. Significant X chromosome loss was detected in PASMC by FISH, as described below. As controls, SNP microarray analysis was performed on PAEC and PASMC cultured from the lungs of healthy individuals (n = 8) and subjects with other end-stage lung diseases (chronic obstructive pulmonary disease [n = 3] and cystic fibrosis [n = 2]). A total of 15 cultures were examined, and no chromosomal abnormalities were detected.

Chromosome abnormalities in pulmonary artery endothelial cells (PAEC) detected by single-nucleotide polymorphism (SNP) array analysis. Copy number changes can be detected on SNP arrays by a change in the locus-specific signal intensity relative to the genome-wide average and by the resulting distortion in B allele frequencies. This reduction in the chromosomal or regional copy number is presented as fractional monosomy. (A) Copy number profile of subject PAH-1 PAEC. The signal intensity for SNP probes on chromosome-13 is reduced relative to the genome-wide average, indicating a mosaic deletion. (B) Fractional monosomy in the regions of chromosome abnormality for cell lines from the 14 subjects in Table 1 (PAEC, n = 14; PASMC, n = 7; PBMC, n = 3). Statistical significance was determined using a one-sided t test, comparing the geometric mean of the candidate segment to the global mean. *Samples meeting the P value threshold of 10−7. (C) B-allele frequency distortion across the entire X chromosome in PAH-3. The value for heterozygous alleles is split into two levels, depending on the identity of the allele on the fractionally monosomic chromosome. The magnitude of this distortion is reduced in (D) PAH-4 and (E) PAH-2 in proportion to the fractional monosomy observed in these cases.
To validate the microarray results, FISH analysis was initially performed on cultured PAEC from subject PAH-1. Cells monosomic for chromosome-13 were detected by interphase and metaphase FISH (Figure 2). These abnormalities were detected in short-term (passage 5–6) primary cultures and are therefore unlikely to represent culture artifacts because abnormal cells arising in culture would be unlikely to proliferate to such high levels during this short culture period. To confirm that the abnormal cells were present in vivo at the time of lung explantation, FISH analysis was performed on formalin-fixed, paraffin-embedded tissue sections. Probes for the RB1 gene (13q14) and 13q34 were used in conjunction with CEP8 (chromosome-8) as a control. The background level of cells showing artifactual monosomic signals due to nuclear truncation was determined in histologically normal areas of the tissue; a similar frequency was observed for all probes (Table 2). We then compared FISH signals from the cells within vascular lesions to those in the normal areas of the same slide. Plexiform lesions exhibited a highly significant increase in the number of cells with a single chromosome-13 signal (RB1: χ2 = 50.46, P < 0.001; 13q34: χ2 = 22.30, P < 0.001) (Table 2). RB1 was apparently deleted more frequently than 13q34, suggesting the possibility of more complex rearrangements in some cells. This is further supported by identification of a translocation by metaphase FISH analysis (data not shown). In contrast, smooth muscle cells in concentric lesions showed no significant difference in hybridization pattern for either chromosome-13 probe (Table 2). Although CEP8 was intended as a control probe, monosomy 8 was more common in plexiform (z = 2.28; P = 0.01) and concentric lesions (z = 2.91; P = 0.002) than in control regions.
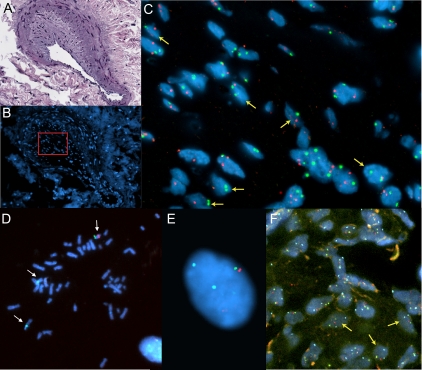
Fluorescent in situ hybridization analysis (FISH) of explant lung sections and cultured pulmonary artery endothelial cells (PAEC) demonstrate subject-specific chromosome monosomy in subjects PAH-3 (A, B, C) and PAH-1 (D, E, F). (A) Subject PAH-3 hematoxylin and eosin stained reference slide; (B) DAPI stained serial section with the region of intimal thickening shown in C (outlined in red); (C) FISH image of intimal region nuclei hybridized with an androgen receptor probe (Xq12, red) and CEP11 (green). Nuclei exhibiting X chromosome monosomy are indicated with yellow arrows. The proportion of cells showing a single Xq12 signal is significantly higher than those with a single chromosome-11 signal (P < 0.001; see Table 2). (D, E, F) Subject PAH-1 FISH images of cultured PAEC and lung sections hybridized with probes for RB1 (13q14.2, red), 13q34 (green), and CEP8 (aqua). PAEC metaphase (D) and interphase (E) nuclei exhibiting a single copy of chromosome-13 (yellow arrows) and two copies of chromosome-8 (white arrows). (F) Interphase FISH of a plexiform lesion within a lung section. Nuclei exhibiting chromosome 13 monosomy are indicated with yellow arrows. The proportion of cells with a single signal for both 13q14.2 and 13q34 probes is significantly higher that those with a single chromosome-8 signal (P < 0.005; see Table 2).
TABLE 2.
RESULTS OF FLUORESCENCE IN SITU HYBRIDIZATION ANALYSIS
| Cells Studied | RB 1 | 13q34 | CEP8 | Total Nuclei | Chi-Square Goodness of Fit to the Distribution in Control Cells | Increase in % of Monosomic Cells over Background | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject PAH-1 | |||||||||||||
 Signals per nucleus Signals per nucleus | 1 | 2 | 1 | 2 | 1 | 2 | RB 1 | 13q34 | CEP8 | RB1 | 13q34 | CEP8 | |
 Plexiform (EC) Plexiform (EC) | 51 | 73 | 31 | 93 | 31 | 93 | 124 | P < 0.001 | P < 0.001 | P < 0.001 | 24.3%; P < 0.001 | 13.8%;P = 0.04 | 11.9%; P = 0.01 |
| Concentric (SMC) | 6 | 28 | 7 | 27 | 12 | 22 | 34 | ns | nd | nd | ns | ns | 22.2%; P = 0.002 |
 Control areas Control areas | 18 | 89 | 12 | 95 | 14 | 93 | 107 | ns* | |||||
| Subject PAH-3 | AR | CEP11 | |||||||||||
 Signals per nucleus Signals per nucleus | 1 | 2 | > 2 | 1 | 2 | > 2 | AR† | CEP11† | AR | CEP11 | |||
 Plexiform (EC) Plexiform (EC) | 20 | 25 | 0 | 10 | 33 | 2 | 45 | P = 0.003 | ns | 20.0%; P = 0.009 | ns | ||
 Medical hypertrophy and intimal thickening (SMC) Medical hypertrophy and intimal thickening (SMC) | 61 | 41 | 2 | 27 | 71 | 6 | 104 | P < 0.001 | ns | 34.2%; P < 0.001 | ns | ||
 Control areas Control areas | 23 | 67 | 4 | 18 | 71 | 5 | 94 | ns* | |||||
Definition of abbreviations: EC = endothelial cells; nd = not done (one of the expected classes was < 5); ns = not significant (P > 0.05); SMC = smooth muscle cells;
FISH was also performed on lung sections from subject PAH-3 using a probe for the androgen receptor (Xq12), with CEP11 (chromosome-11) as a control. CEP11 results in lesional areas were not significantly different than in control lung regions, whereas endothelial cells and smooth muscle cells within areas of vascular remodeling showed a significant increase above background in the proportion of X-monosomy (Table 2; Figure 2), confirming the microarray data. In subject PAH-5, a similar FISH analysis with chromosome-8 probes failed to validate the interstitial 8p23.1-p12 deletion detected by SNP microarray. This discrepancy might reflect sampling differences between regions of the lung or the low level of abnormal cells present.
For female subjects with X chromosome loss, it was important to determine whether the deleted chromosome was the active or inactive copy because loss of the inactive X chromosome would likely have little or no phenotypic effect. Human androgen receptor analysis, which determines the methylation status of DNA immediately adjacent to the polymorphic CAG-repeat in exon 1 of the human androgen receptor gene, was informative in subjects PAH-3 and PAH-4. In both cases, this confirmed that the deleted allele was the active X chromosome. For subject PAH-2, the proportion of monosomic cells was too low to determine which allele was deleted. This analysis also showed that the background of non deleted cells in subject PAH-3 had 100% skewed X-inactivation, suggesting a clonal origin (Figure 3). This contrasts with DNA obtained from PASMC and bronchial smooth muscle cells from this patient, which were unequivocally polyclonal (Figure 3).
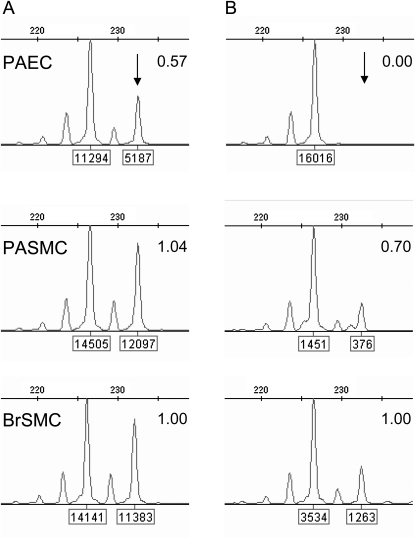
X-inactivation studies. Analysis of methylation at the androgen receptor locus in subject PAH-3. (A) Amplification of undigested DNA demonstrates a heterozygous pattern with alleles of 227bp and 233bp. Each allele is labeled with its peak height. The peak height ratio is shown, normalized to bronchial smooth muscle cells (BrSMC). Pulmonary artery smooth muscle cells (PASMC) exhibit the same peak height ratio as BrSMC, whereas in pulmonary artery endothelial cells (PAEC), the 233-bp allele (arrow) is markedly reduced in height, indicating that it is deleted in approximately 43% of cells, which is in good agreement with the microarray data (46%; see Table 1). (B) Alleles amplified from HhaI-digested DNA, where active chromatin is digested and only the inactive, methylated chromatin is PCR amplified. Peak height ratios were calculated as for panel A. PAEC show a complete absence of the 233bp allele, indicating that the 227-bp (nondeleted) allele is inactive in all cells.
DISCUSSION
The hypothesis that vascular remodeling in pulmonary hypertension is a quasi-neoplastic process has continued to find support (16, 17), yet until now there has been little genetic evidence beyond the initial reports of monoclonal endothelial cell proliferation and microsatellite instability (10–12). Our identification of chromosomal abnormalities in PAEC from more than half of the PAH cases examined therefore adds new weight to this hypothesis and pinpoints key somatic genetic events that may underlie the pathogenesis of the disease. These abnormalities were detected among patients with heritable, idiopathic, and associated forms of PAH, suggesting that somatic genetic changes may represent a shared feature across different types of the disease. Our data also provide the first evidence of a second (somatic) genetic hit in a patient with a germline BMPR2 mutation. PAEC from this patient (PAH-1) were deleted for chromosome-13, which contains the SMAD9 gene, as well as tumor suppressor genes RB1 and BRCA2. SMAD9 encodes the protein Smad-8, a downstream mediator of BMPR-II signaling, and mutations in this gene are known to predispose to PAH (27). Thus, somatic loss of chromosome-13 in this patient represents a second hit that likely further dysregulates the BMP pathway.
The pulmonary vascular cells that we studied are, by necessity, obtained at lung transplantation or post mortem and therefore represent the advanced stages of disease. Obtaining tissue by lung biopsy at earlier stages of pulmonary hypertension is contraindicated, and therfore it is not possible to determine how early in the disease process the chromosomal abnormalities arose or whether they represent an initiating event. However, their absence from blood or at least one other lung-derived cell type in all subjects strongly suggests that they are somatic events arising within the lung. Furthermore, no abnormalities were detected in PAEC from patients with cystic fibrosis or chronic obstructive pulmonary disease, suggesting that this not a generalized phenomenon in other end-stage lung diseases. Our analyses also demonstrate that cytogenetic aberrations detected in cells from the main pulmonary artery endothelium are present in plexiform lesions in vivo. Plexiform lesions consist of dysregulated endothelial and inflammatory cells (1). Little is known about how and when they arise, although their presence may correlate with advanced disease (28). Our FISH data indicate that the same chromosome abnormality is present in multiple discrete lesions disseminated within the lung, suggesting that chromosomally abnormal PAEC may predate the development of plexiform lesions, perhaps then migrating from the main vessels as the lesions develop. Indeed, we have previously shown that these PAEC are hyperproliferative, with increased migration, decreased apoptosis, and abnormal bioenergetics compared with control cells (14, 15).
Smooth muscle cell proliferation is also a key aspect of PAH pathogenesis, yet the incidence of chromosome abnormalities was lower than in endothelial cells from the same patients. This may reflect that endothelial cells are more susceptible to mitotic errors or that they are more likely to survive and proliferate once they become aneuploid. Endothelial cells within a tumor microenvironment can also acquire diverse chromosomal abnormalities that are distinct from those of the tumor itself (29). Although the molecular basis for this is unclear, it suggests that an abnormal microenvironment in cancer or PAH may support or even promote endothelial cell aneuploidy, which in turn contributes to the disease process through abnormal cell behavior and cell–cell interactions. In one subject, endothelial and smooth muscle cells within vascular lesions harbored the same chromosome abnormality. This suggests that the deletion may have occurred in a common vascular progenitor cell during embryonic development or early in the disease process. Alternatively, it may be evidence of endothelial-to-mesenchymal transition, which also has a putative role in vascular remodeling (30).
The underlying cause of this aneuploidy is unknown. Chromosome aberrations are common in the bronchial epithelium of smokers and lung cancer patients but absent in control subjects who have never smoked (31). Smoking status was known for three of our five patients with abnormalities in PAEC; two have never smoked, and one quit three decades before transplant (Table 1). In contrast, three control subjects with no detectable chromosome abnormalities in PAEC had a smoking history of 20 to 60 pack-years, making tobacco use an unlikely explanation for our findings. Hypoxia and activation of hypoxia-inducible factor 1α are important contributors to pulmonary hypertension (13, 32, 33) and can play a role in triggering genomic instability in cancer cells (34, 35). Inflammation and reactive oxygen species are other possible causes (35–37). Although BMPR2 mutations are not known to play a direct role in genome stability, the BMP pathway is implicated in colon cancer; germline BMPR1A mutations cause juvenile polyposis and Cowden and Bannayan-Riley-Ruvalcaba syndromes (38), and somatic BMPR2 mutations were recently reported in sporadic colorectal cancers exhibiting microsatellite instability (39). Another possible explanation is that bone marrow–derived cells, which are dysregulated in PAH (24), may contribute to genome instability of the lung endothelium. However, a recent study found no evidence for cell fusion as a cause of genomic instability (26). The most fruitful areas for mechanistic studies are therefore likely to be hypoxia and inflammation, somatic mutations in genes important for mitotic control and genomic integrity (40), and a possible role for BMPR2.
The high incidence of X chromosome deletion is remarkable, particularly given that in the two cases where X-inactivation status could be determined, it was the active X that was deleted. X-monosomy has been reported in myelodysplastic syndrome and several cancers (41, 42) and in peripheral blood leukocytes of patients with autoimmune disorders (43, 44), although there the inactive X was lost. Indeed, it was assumed that loss of the active X would be cell lethal (44). The 100% skewed X-inactivation in PAEC from subject PAH-3 is also notable. Such extreme skewing is present in less than 2% of the adult female population (25), and, although there is generally good concordance between X-inactivation patterns in normal tissues from the same individual (45), PASMC and bronchial smooth muscle cells from this patient differed markedly. This suggests that the background of normal nondeleted PAEC may be clonal in origin, raising the possibility that another genetic mutation preceded the chromosome deletion. Further study is required to resolve the significance of these results and a potential role for the X chromosome in PAH.
FISH analysis in subject PAH-5 failed to validate the interstitial 8p23.1-p12 deletion detected by SNP arrays. This discrepancy might reflect genuine differences between the different regions of the lung sampled for these analyses, or the sensitivity of FISH analysis may be insufficient to validate a low level of abnormal cells. Conversely, FISH identified abnormalities that were not detected by microarray analysis, including apparent chromosome-8 loss in plexiform and concentric lesions of PAH-1 and a high frequency of X chromosome deletion in smooth muscle cells of PAH-3 that was barely detectable by array analysis. This highlights the limitations of current technologies in reliably detecting mosaic abnormalities. As our results show, limitations of the sensitivity of SNP microarrays to detect copy number changes can be partially circumvented by duplicate analysis on different platforms, but the overall threshold for detection is approximately 10% of cells abnormal. Technical considerations for interphase FISH include nuclear truncation and differential hybridization efficiencies of the probes. Our approaches to minimize their effects are described in Methods. Notwithstanding these limitations, our results raise the possibility that areas of vascular remodeling may harbor additional abnormalities that are not present in cells derived from the primary vessels. Future use of second-generation sequencing technologies to detect mosaic changes in PAH lung tissue at the DNA level and in allele-specific RNA expression offers a promising way to address these limitations and document the full extent of chromosomal changes in different parts of the lung (46, 47).
In summary, we have identified acquired chromosome abnormalities in PAEC from five of nine patients with PAH. These changes were also present in the plexiform lesions of primary lung tissue. These results provide the first molecular evidence of a second genetic hit in a patient with a germline BMPR2 mutation and provide new support for a neoplasia-like accumulation of somatic mutations in PAH lungs. In contrast to BMPR2 mutations, which are mainly confined to familial and idiopathic PAH, somatic chromosome abnormalities potentially represent a shared genetic event across idiopathic and associated forms of the disease.
Acknowledgments
The authors thank the patients and their families who consented to use of their tissue in this study; L. Mavrakis, M. Koo, and J. Sharp for cell and tissue acquisition, processing and cell culture; P. Harbor for extraction of DNA samples; M. Theodoro and the University of Colorado Cancer Center Cytogenetics Core for expert FISH analyses; and the Lerner Research Institute Genomics Core for sequencing and genotyping services.
Notes
Supported in part by NIH grants R37HL060917, R21HL094927, and CTSA UL1RR024989 from the National Center for Research Resources, the Cardiovascular Medical Research and Education Fund, and start-up funding from Strategic Investment Funds to the Genomic Medicine Institute, Cleveland Clinic.
Originally Published in Press as DOI: 10.1164/rccm.201003-0491OC on June 25, 2010
Author Disclosure: M.A.A. received more than $100,001 from the NIH/NHLBI and $50,001 to $100,000 from the AHA in sponsored grants. S.A.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.V-G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.A. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. W.X. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.P.N. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.A.T. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.M.T. received up to $1,000 from Novartis in consultancy fees and more than $100,001 from the NIH in sponsored grants. S.C.E. received more than $100,001 from Asthmatx as an investigator in the industry-sponsored grant. M.W.G.'s institution received more than $100,001 from the NIH in sponsored grants. C.D.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
Articles from American Journal of Respiratory and Critical Care Medicine are provided here courtesy of American Thoracic Society
Full text links
Read article at publisher's site: https://doi.org/10.1164/rccm.201003-0491oc
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3001257?pdf=render
Free to read at ajrccm.atsjournals.org
http://ajrccm.atsjournals.org/cgi/content/abstract/182/9/1153
Free after 12 months at ajrccm.atsjournals.org
http://ajrccm.atsjournals.org/cgi/content/full/182/9/1153
Free after 12 months at ajrccm.atsjournals.org
http://ajrccm.atsjournals.org/cgi/reprint/182/9/1153.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Identification of TFRC as a biomarker for pulmonary arterial hypertension based on bioinformatics and experimental verification.
Respir Res, 25(1):296, 03 Aug 2024
Cited by: 1 article | PMID: 39097701 | PMCID: PMC11298087
Pulmonary Hypertension and Hyperglycemia-Not a Sweet Combination.
Diagnostics (Basel), 14(11):1119, 28 May 2024
Cited by: 0 articles | PMID: 38893645 | PMCID: PMC11171670
Review Free full text in Europe PMC
Reduced FOXF1 links unrepaired DNA damage to pulmonary arterial hypertension.
Nat Commun, 14(1):7578, 21 Nov 2023
Cited by: 0 articles | PMID: 37989727 | PMCID: PMC10663616
Emerging role of exosomes in vascular diseases.
Front Cardiovasc Med, 10:1090909, 02 Mar 2023
Cited by: 3 articles | PMID: 36937921 | PMCID: PMC10017462
Review Free full text in Europe PMC
Tensions in Taxonomies: Current Understanding and Future Directions in the Pathobiologic Basis and Treatment of Group 1 and Group 3 Pulmonary Hypertension.
Compr Physiol, 13(1):4295-4319, 30 Jan 2023
Cited by: 4 articles | PMID: 36715285 | PMCID: PMC10392122
Review Free full text in Europe PMC
Go to all (103) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE21097
- (1 citation) GEO - GSE20206
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genetic ablation of the BMPR2 gene in pulmonary endothelium is sufficient to predispose to pulmonary arterial hypertension.
Circulation, 118(7):722-730, 28 Jul 2008
Cited by: 149 articles | PMID: 18663089 | PMCID: PMC3920834
Receptor for activated C-kinase 1, a novel interaction partner of type II bone morphogenetic protein receptor, regulates smooth muscle cell proliferation in pulmonary arterial hypertension.
Circulation, 115(23):2957-2968, 21 May 2007
Cited by: 33 articles | PMID: 17515463
CCL5 deficiency rescues pulmonary vascular dysfunction, and reverses pulmonary hypertension via caveolin-1-dependent BMPR2 activation.
J Mol Cell Cardiol, 116:41-56, 31 Jan 2018
Cited by: 19 articles | PMID: 29374556
BMP type II receptor as a therapeutic target in pulmonary arterial hypertension.
Cell Mol Life Sci, 74(16):2979-2995, 26 Apr 2017
Cited by: 65 articles | PMID: 28447104 | PMCID: PMC5501910
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: UL1RR024989
NHLBI NIH HHS (7)
Grant ID: R37 HL060917
Grant ID: R21 HL094927-01A1
Grant ID: R21HL094927
Grant ID: R21 HL094927-02
Grant ID: R01 HL089508
Grant ID: R21 HL094927
Grant ID: R37HL060917





