Abstract
Free full text

Bottom-up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology
Abstract
DNA and protein have been extensively scrutinized for feasibility as parts in nanotechnology, but another natural building block, RNA, has been largely ignored. RNA can be manipulated to form versatile shapes, thus providing an element of adaptability to DNA nanotechnology, which is predominantly based upon a double-helical structure. The DNA-packaging motor of bacterial virus phi29 contains six DNA-packaging RNAs (pRNA), which together form a hexameric ring via loop/loop interaction. Here we report that this pRNA can be redesigned to form a variety of structures and shapes, including twins, tetramers, rods, triangles, and 3D arrays several microns in size via interaction of programmed helical regions and loops. Three dimensional RNA array formation required a defined nucleotide number for twisting of the interactive helix and a palindromic sequence. Such arrays are unusually stable and resistant to a wide range of temperatures, salt concentrations, and pH.
Introduction
DNA and proteins have been investigated extensively to further elucidate the mechanism underlying their bottom-up assembly and their possible applications in nanotechnology.1–6 Unlike DNA, RNA generally exhibits a single-stranded conformation in nature. The astonishing diversity evident in RNA function is attributable to the significant versatility present in RNA structure.7–11 The majority of the available models dealing with RNA functions such as mRNA transcription, maturation, translation, degradation, and the catalytic activity of ribozymes are based on specific RNA structure.12,13 However, the primary sequence of RNA governs its 3D structure, which exhibits helices, bulges, loops, stems, and hairpins.8,14 Phylogenetic analyses and complementary modification of RNA species have shown that covariation of bases, if in compliance with certain rules, can lead to the formation of a defined 3D structure.12,13,15 Stable RNA can be produced through the use of RNase-resistant nucleotide derivatives such as 2-F ribonucleotides or spiegelmer.16 There have been many 2D array constructions of biomacromolecules,2,17–21 but the construction of 3D arrays of RNA has not been reported.
The DNA-packaging motor of bacterial virus phi29 contains six copies of pRNA molecules that together form a hexameric ring as a crucial part of the motor.22–25 This ring is formed via hand-in-hand interaction by pairing of four nucleotides between the right loop (bases 45–48) and the left loop (bases 82–85).23,25–27 pRNA contains two functional domains (Figure 1). The procapsid binding domain is located at the central region,28 bases 23–97, while the DNA translocation domain is located at the 5′/3′ paired ends. This conclusion has been supported by several studies, including (a) base deletion and mutation;29–32 (b) ribonuclease probing;15,33,34 (c) oligo targeting;35 (d) competition assays to inhibit phage assembly;36–38 (e) UV-cross-linking to portal protein;39 (f) psoralen cross-linking;34 and (g) computer modeling.24 3D models of pRNA monomer and hexamer have also been constructed24 based upon constraints obtained from chemical modification,40 chemical modification interference, cross-linking, and cryo-AFM.28,40 pRNA dimers are the building blocks of hexamers.23,25,28 pRNA has many attributes that make it a novel choice for nanotechnological applications. Among these are its ATP-binding activity,41 its function in driving the DNA-packaging motor,42 its ability to form dimers, trimers, and hexamers,23,25,26,28,43 its tight and stable folding,24,30–32,44 its holding of two independent domains for structure and function,29,30,33,39 and its functional similarity to protein enzymes.22,42 Utilizing the novel properties of this pRNA, we have constructed pRNA dimers and trimers with a potential to serve as parts in nanotechnology.45 To make the description more clear, uppercase letters have been used in this phi29 pRNA system to describe the right loop of pRNA, and lowercase letters represent the left loop. The same letters in upper- and lowercase indicate complementary sequences, and different letters indicate noncomplementary loops. For example, pRNA (A–b′) contains a noncomplementary right loop A (5′-G45GAC) and left loop b′ (3′-U85GCG). pRNA (A–a′) contains a self-complementary right loop A (5′-G45GAC) and left loop a′ (3′-C85CUG). When two monomer RNAs with interlocking loops, such as (A–b′)/(B–a′), were mixed at equal molar concentrations in the presence of 5 mM MgCl2, RNAs formed dimers. When three monomer RNAs with interlocking loops, such as (A–b′)/(B–e′)/(E–a′), were mixed at equal molar concentrations, pRNA trimers were produced. This work suggests that RNA molecules can be used as versatile building blocks to construct patterned suprastructures in nanotechnology.
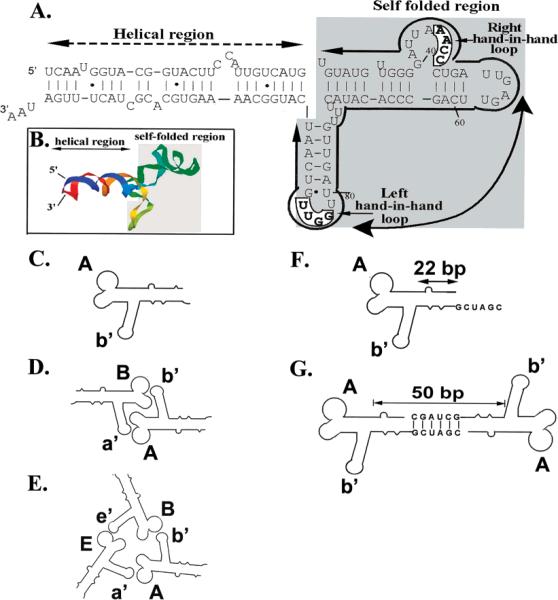
Sequence and structural elucidation of phi29 motor pRNA and related assemblages. (A) Primary and secondary structure of wild-type pRNA I–i′. The binding domain (shaded area) and the DNA translocation domain (the helical region) are marked with bold lines. The four bases in the right and left loops, which are responsible for inter-RNA interactions, are boxed. (B) Three-dimensional structure of wild-type pRNA I–i′ displayed as ribbon.24 (C) Diagrams depicting the pRNA monomer A–b′ with unpaired right/left loops. (D) pRNA dimers (A–b′)(B–a′). (E) pRNA trimers (A–b′)(B–e′)(E–a′). (F) pRNA monomer with unpaired right/left loops A–b′ and a 6-nucleotide palindromic sequence. (G) pRNA twin A–b′.
Results
1. Construction of a Variety of pRNA Building Blocks to Build RNA Arrays or Superstructures
Nanotechnology employs either the top-down or the bottom-up approach. Since the size of RNA ranges from the angstrom to the nanometer scale, the bottom-up approach could be reasonably applied to RNA in nanotechnological applications. Larger RNA complexes can be constructed from the following three building blocks: (a) Monomer with intramolecularly self-complementary left and right loops. (b) Monomer with noncomplementary left and right loops for intermolecular interaction. (c) Monomer with intermolecularly self-complementary left and right loops and palindromic 3′ ends. Building blocks a and b have been reported previously.45 The construction of building block c is depicted in Figure 1F,G.
2. Use of Monomeric Building Blocks to Construct RNA Twins, Tetramers, and Arrays
The use of monomers to construct dimers and trimers has been reported.45 The pRNA monomer derivatives with programmed left and right loops with the palindromic ends were used to build larger structures of RNA (Figures 1–3).
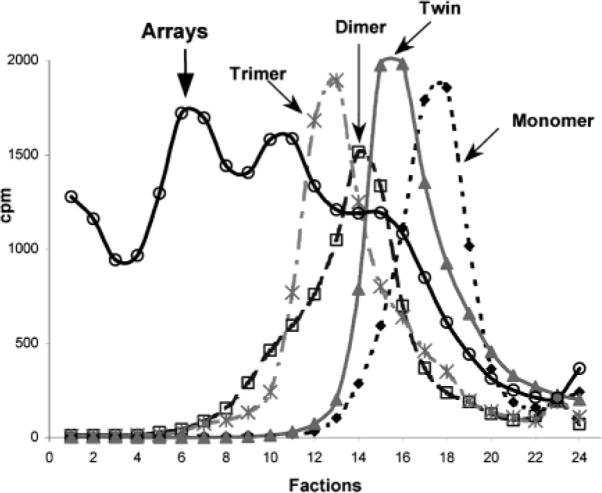
Separation of pRNA monomers, dimers, trimers, twins, and arrays by 5–20% sucrose gradient sedimentation. The [3H]-pRNA monomers, dimers, trimers, and twins were isolated from native polyacrylamide gel (see materials and methods). Arrays were prepared by mixing of equal molar amount of twin (A–b′), twin (B–e′) and twin (E–a′). All particles were loaded onto the top of the gradient and sedimented by ultracentrifugation. Sedimentation is from right to left.
When pRNA monomers with a noncomplementary right loop (e.g., pRNA A–b′) and palindromic ends were purified from denaturing gel and renatured by the addition of 5 mM MgCl2 into the solution, pRNA twins containing two identical monomers were formed. The formation of twins is highly efficient, approaching 100% even at concentrations as low as several ng/μL.
In this report, “dimer” (Figure 1D) refers to the complex composed of two different pRNA A–b′ and B–a′, while “twins” (Figure 1G) are composed of two identical pRNAs bridged by a palindromic sequence at the 3′ end of pRNA. For example, twin A–b′ is composed of two identical pRNA A–b′ linked by the palindromic sequence “3′CGAUCG”. When two twins, for example, twin A–b′ and twin B–a′, were mixed in the presence of 10 mM MgCl2, pRNA tetramers known as “double-twins” were produced (Figure 2). When three twins, such as twin A–b′, twin B–e′, and twin E–a′, were mixed, pRNA began to grow into a micron-sized array by serial addition of the twins A–b′, B–e′, or E–a′. The arrays displayed as bundles as revealed by AFM (Figure 4D) and grew to micron scale, suggesting that each bundle contains hundreds of pRNA molecules.
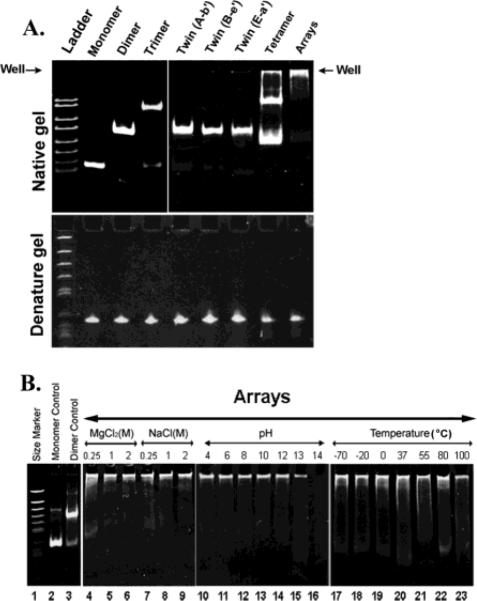
Polyacrylamide gel showing monomers, dimers, trimers, twins, tetramers, and arrays. (A) Native and denatured gel. (B) Test of the stability of pRNA dimers under different conditions.
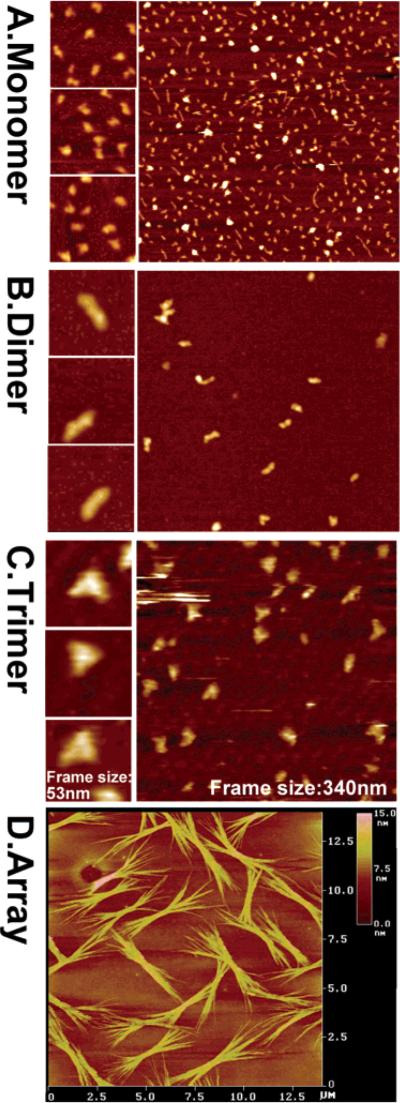
Atomic force microscopy (AFM) showing pRNA monomers (A), dimers (B), trimers (C), and arrays (D) of pRNA. The three insets at the left of each panel contain images with higher magnification, as indicated by the size of the frame. The pRNA monomers folded into a checkmark shape, dimers displayed a rod shape, trimer exhibited triangle shape, and arrays displayed as bundles. Formation of dimers requires Mg2+, while the sample on mica was briefly rinsed with water before freezing for cryo-AFM, which resulted in some dissociation of dimers or trimers even when the pRNA was already adsorbed to the activated mica surface. The color within each image reflects the thickness and height of the molecule. The brighter, or whiter, the color, the thicker or taller the molecule; the darker the image, the thinner the molecule.
When analyzed by 5% native polyacrylamide gel, pRNA monomers, dimers, trimers, twins, tetramers, and arrays exhibited different migration rates (Figure 2A). The array was too large to enter the gel and stayed trapped in the well (Figure 2). It did not run into the gel even after electrophoresis for 6 h in a 5% polyacrylamide gel.
When analyzed by sucrose gradient sedimentation, [3H] pRNA monomers, twins, dimers, and trimers sedimented to fractions 18, 16, 14, and 13, respectively (Figure 3). A plot of hypothetical molecular weight vs the log of migration distance (the fractional number) in longer sedimentation gradients revealed a linear relationship among monomers, dimers, and trimers (data not shown). The purified monomers, dimers, and trimers have been further confirmed by AFM imaging (Figure 4). The twins were sedimented to a position similar to the dimers. The array exhibited a rapid sedimentation rate that was close to that for a DNA with more than ten thousand base pairs.
3. The Effect of the Twisting Angle and the Length of the Interacting Helical Region on Array Formation
It is expected that arrays will grow by nucleation from one building block of pRNA and can grow in solution with three-dimensional extensions. Therefore, the twisting angle of the extending area between the two building blocks is of critical importance to proper array growth. The 5′/3′ paired helical junction region composed of nucleotides 1–28 and 92–117 (Figure 1) will be key in governing the extending direction of the next RNA building block. It would be ideal to restrict the joining of the two helical regions to an odd number of half-turns (180°). Since each helical turn of RNA is composed of eleven nucleotides, 50 nucleotides (Figure 1) will result in nine-half-turns (4.5 turns). When the 50 nucleotides were used as the initial design in array formation, array extension continued successfully (Figure 4D) and the formation of arrays was detected using this parameter. To test the effect of twisting angle and length of the interacting helical region in array formation, one nucleotide was eliminated from the helical region, resulting in a helical junction region with only 48 (27 for each monomer, with 6 bases to be paired) nucleotides. It was found that arrays would not form using this truncated pRNA, since 48 nucleotides generate only 8.7 half-turns.
4. Effect of the Sequence of the Interacting Left and Right Loops on Array Stability
It is expected that the interactive left and right loops play a critical role in the growth and extension of arrays. pRNA building blocks with different loop sequences were constructed and tested. The stability of arrays was tested by hour-long electrophoresis in polyacrylamide gel at elevated temperatures. It was found that arrays from the building blocks with the loop A–a′ were much more stable than pRNA with loop I–i′ (I, 5′AACC; i′, 3′UUGG), suggesting that such loops play a critical role in array stability.
5. Determination of the Effect of Salt, pH and Temperature on pRNA Complex Formation and Stability
The minimum ion concentration requirement for pRNA array formation was determined by both polyacrylamide gel shift assay and sucrose gradient sedimentation. It was found that although 5 mM of divalent ions such as MgCl2, CaCl2, and MnCl2 were needed, a minimum of 1 M of monovalent ions such as NaCl alone was needed for the formation of pRNA arrays. The arrays were resistant to pH values as low as 4 and as high as 12, temperatures as low as −70 °C and as high as 100 °C, and high salt concentrations of 2 M NaCl and 2 M MgCl2. At pH 13, only a portion of the arrays were degraded (Figure 2, lane 15). This indicates that pRNA arrays are much more stable than regular 120-nucleotide RNA, which has an unfolding TM between 50 and 70° and is sensitive to pH values higher than 11. Such stability is credited to the tightly folded pRNA structure and to the intertwining of the RNA molecule in the arrays.
Discussion
The emergent field of nanotechnology generally involves the characterization, manipulation, modification and/or assembly of organized materials on the nanoscale level.46,47 These materials can then be used as building blocks for the construction of larger devices and systems, thereby helping to form supramolecular structures.48–50 Since biological macromolecules intrinsically have defined features at the nanometer scale, they have the potential to serve as powerful building blocks for the bottom-up fabrication of nanostructures and nanodevices. RNA is a particularly interesting candidate for such applications, since RNA molecules can be designed and manipulated with the simplicity that is typical for DNA nanotechnology,51,52 yet they exhibit a versatility and flexibility in structure and function similar to that of proteins. One of the major challenges in the design of functional nanostructures is to successfully integrate bottom-up assembly approaches with top-down fabrication techniques. Using circular permutation,32,53 we have been able to incorporate varieties of chemical groups into pRNA for linkage and bridging for further applications. In this report, we show that self-assembled RNA nanostructures can be formed with dimensions that are well within the range of state-of-the-art microfabrication technology.
Of considerable interest in current nanotechnology is the synthesis of patterned arrays2,54 for technological applications. Arrays can be created and applied in high-throughput proteomics, as chips in the diagnosis of diseases, or as ultrahigh-density data storage systems.55 Ordered nanocrystals have already been assembled into superlattices by techniques such as colloidal crystallization,56,57 macromolecule self-assembly, complementary interaction,58,59 and patterned etch pits. Ordered biological structural arrays can serve as templates for the further construction of superlattices.
Potentially, RNA arrays can be constructed through the use of pRNA twins, dimers, trimers, or hexamers as building blocks (Figure 4). Palindromic sequences have been introduced into the 3′ end of the pRNA, and can serve as links for bridging and intermolecular interaction. The left and right loops can be used to aid array growth by continuous extension via loop/loop intermolecular interaction. Gel electrophoresis and AFM images have revealed that interaction of the palindromic sequences with the right and left loops causes the formation of pRNA arrays composed of a huge number of twins (Figure 4D).
After mixing two different twins with intermolecularly compatible loop sequences, we observed two alternate assembly pathways, which we attribute to the structural flexibility inherent to pRNA. The formation of tetramers indicates that the two twins were able to form a closed circular structure (Figure 5), and since the four domains for intermolecular interaction were partnered, assembly ceased. This pathway competed with the formation of chains (Figure 5) or possibly larger circles of twins, which assembled to a size above the polyacrylamide gel separation limit (Figure 2, the tetramer lane). We have reported previously that the UUU bulge at the three-way junction served as a hinge to provide for the flexibility of pRNA, which enables the dimerization of twins and is also evident in the assembly into hexamers28 from dimers via hand-in-hand interactions.13,23 In dimers, each pRNA monomer subunit only holds the hands of one additional pRNA. In hexamers, however, each pRNA monomer subunit holds the hands of two additional pRNAs (Figure 15 in ref 24). This may at first seem paradoxical given the hand interactions of dimers and hexamers, but such interaction can be accounted for if the conformational change of pRNA and the presence of a hinge at the three-way junction are considered. The flexibility displayed by pRNA on the dimer-to-hexamer assembly pathway may also be an essential intrinsic feature of pRNA, enabling its function in DNA translocation.24 In dimer-to-hexamer assembly, connector binding of a closed dimer is associated with breaking up one of the two hand-in-hand interactions and a dramatic change in the relative orientation of the two pRNA monomers, which requires a reorientation of the binding loops. The dimer of twin formation reported here may be enabled by a similar structural transition involving a loop hinge.
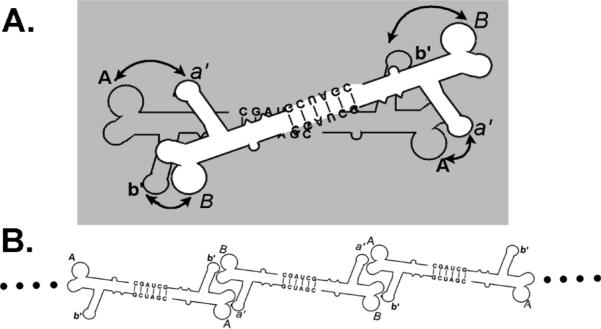
A mixture of two complementary twins, A–b′ and B–a′, assembled into two distinct supramolecular structures. (A) Two complementary twins were able to form a stable tetramer (double-twins) by assembling into a circular structure. (B) Concatemers of alternating twins formed when a twin interacted with two rather than one complementary twin.
The rate of sedimentation is generally dependent not only on molecular weight but also on the shape of the molecule. Dimers are more compact than twins, which explains why dimers migrate more quickly in sucrose gradient. At any movement, the extension of arrays can terminate and lead to the abortive smaller structure. This might explain the broad peak and multiple curves in sucrose gradient sedimentation. Such a broad peak and multiple curves could not be separated by polyacrylamide gel since molecules of more than 1000 nucleotides are beyond the resolution limit of polyacrylamide gel.
As expected, the twisting angle between the two loop regions in a twin had a major effect on array formation. Deletion of two bases from the stem of the twin is expected to change the angle between the two loop regions by about 65.5°. We speculate that in the twins that gave rise to extended arrays, the two loop regions were roughly in a planar alignment.
Materials and Methods
Synthesis of RNAs
The construction of pRNA and the synthesis, purification, and nomenclature of pRNA have been reported previously.53
Native or Denatured Polyacrylamide Gel for RNA Purification and the Detection of RNA Complexes and Arrays
After transcription, RNA was first purified from 8% denaturing polyacrylamide gel in the presence of 8 M urea. The pRNA monomer, twin, dimer, and trimer bands were excised from the gels and eluted overnight using elution buffer (0.5 MNH4OAC, 0.1 mM EDTA, 0.1% SDS, and 0.5 mM MgCl2 at 37 °C). The purified RNAs were used to construct dimers, trimers, or arrays which were analyzed by 5% to 8% native polyacrylamide prepared in TBM buffer (Tris 89 mM, boric acid 200 mM, MgCl2 5 mM, pH 7.6). The RNA was visualized by ethidium bromide staining. Images were captured by an Eagle Eye II system (Stratagene). These complexes were then either kept in TBM buffer at 4 °C for further use or frozen at −20 °C.
Separation of pRNA Complexes by Sucrose Gradient Sedimentation
Linear 5–20% sucrose gradients were prepared in TBM buffer. The RNA of multimers was loaded onto the top of the gradient. Samples were spun in an SW55 rotor at 40,000 rpm for 12 h at 4 °C. After sedimentation, 12-drop fractions were collected and subjected to scintillation counting.
Cryoatomic Force Microscopy (cryo-AFM) of pRNA Oligomers
Oligomeric pRNA was purified from native polyacrylamide gels or sucrose gradient. To prepare the sample for cryo-AFM imaging of monomers, dimers, and trimers, a piece of mica was freshly cleaved and soaked with spermidine. Excess spermidine was removed by repeated rinsing with deionized water. The pRNA sample (10 μg/mL) was applied to the mica, which had been preincubated with TBM buffer. After 30 s, the unbound pRNA was removed by rinsing with the same buffer. Before the sample was transferred to cryo-AFM for imaging, it was quickly rinsed with deionized water (<1 s) and the solution was removed with dry nitrogen within seconds. All cryo-AFM images were collected at 80 K. Scan lines were removed by an offline matching of the basal line. Calibration of the scanner was performed with mica and 1 μm dot matrix. To prepare the arrays, a 5 μL sample drop was spotted on freshly cleaved mica (Ted Pella, Inc.) and left to adsorb to the surface for 2 min. To remove buffer salts, 5–10 drops of doubly distilled water were placed on the mica, the drops were shaken off, and the sample was dried with compressed air. Imaging was performed under 2-propanol in a fluid cell on a NanoScope IIIa, using an NP–S oxide-sharpened silicon nitride probe (Veeco Probes).
Acknowledgment
We thank Songchuan Guo and Jeremy Hall for their assistance in the preparation of this manuscript. Dr. Shitong Sheng in Dr. Zhifeng Shao's lab prepared the cryo-AFM images. This work was supported by NIH grant GM59944.
References
Full text links
Read article at publisher's site: https://doi.org/10.1021/nl0494497
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3010238?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1021/nl0494497
Article citations
RNA four-way junction (4WJ) for spontaneous cancer-targeting, effective tumor-regression, metastasis suppression, fast renal excretion and undetectable toxicity.
Biomaterials, 305:122432, 21 Dec 2023
Cited by: 3 articles | PMID: 38176263
Nucleic acid nanostructures for in vivo applications: The influence of morphology on biological fate.
Appl Phys Rev, 10(1):011304, 01 Mar 2023
Cited by: 3 articles | PMID: 36874908 | PMCID: PMC9869343
Review Free full text in Europe PMC
RNA origami: design, simulation and application.
RNA Biol, 20(1):510-524, 01 Jan 2023
Cited by: 9 articles | PMID: 37498217 | PMCID: PMC10376919
Review Free full text in Europe PMC
Combining Experimental Restraints and RNA 3D Structure Prediction in RNA Nanotechnology.
Methods Mol Biol, 2709:51-64, 01 Jan 2023
Cited by: 0 articles | PMID: 37572272 | PMCID: PMC10680996
Nucleic acid nanoassembly-enhanced RNA therapeutics and diagnosis.
Acta Pharm Sin B, 13(3):916-941, 27 Oct 2022
Cited by: 26 articles | PMID: 36970219 | PMCID: PMC10031267
Review Free full text in Europe PMC
Go to all (104) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy.
J Nanosci Nanotechnol, 5(12):1964-1982, 01 Dec 2005
Cited by: 83 articles | PMID: 16430131 | PMCID: PMC2842999
Review Free full text in Europe PMC
Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices.
J Nanosci Nanotechnol, 3(4):295-302, 01 Aug 2003
Cited by: 56 articles | PMID: 14598442
Construction and 3-D computer modeling of connector arrays with tetragonal to decagonal transition induced by pRNA of phi29 DNA-packaging motor.
J Nanosci Nanotechnol, 5(6):856-863, 01 Jun 2005
Cited by: 26 articles | PMID: 16060143
Construction of bacteriophage phi29 DNA packaging motor and its applications in nanotechnology and therapy.
Ann Biomed Eng, 37(10):2064-2081, 04 Jun 2009
Cited by: 27 articles | PMID: 19495981 | PMCID: PMC2855900
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: R01 GM059944-04
Grant ID: R01 GM059944





