Abstract
Free full text

Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway
Associated Data
Abstract
Systemic administration of synthetic small interfering RNAs (siRNAs) effectively silences hepatocyte gene expression in rodents and primates1–3. Whether or not in vivo gene silencing by synthetic siRNA can disrupt the endogenous microRNA (miRNA) pathway remains to be addressed. Here we show that effective target-gene silencing in the mouse and hamster liver can be achieved by systemic administration of synthetic siRNA without any demonstrable effect on miRNA levels or activity. Indeed, siRNA targeting two hepatocyte-specific genes (apolipo-protein B and factor VII) that achieved efficient (~80%) silencing of messenger RNA transcripts and a third irrelevant siRNA control were administered to mice without significant changes in the levels of three hepatocyte-expressed miRNAs (miR-122, miR-16 and let-7a) or an effect on miRNA activity. Moreover, multiple administrations of an siRNA targeting the hepatocyte-expressed gene Scap in hamsters achieved long-term mRNA silencing without significant changes in miR-122 levels. This study advances the use of siRNAs as safe and effective tools to silence gene transcripts in animal studies, and supports the continued advancement of RNA interference therapeutics using synthetic siRNA.
Recently, it was reported that adeno-associated viral (AAV)-expressed short hairpin RNA (shRNA), when administered to mice, can result in profound toxicity, presumably by saturation of the cellular miRNA pathway4. AAV expression of shRNA seemed to elicit toxicological effects by interference of cellular pathways for miRNA biogenesis, including transport of nuclear miRNA precursors to the cytoplasm by the nuclear karyphorin exportin-5. shRNA-mediated toxicological effects were evident by marked reductions in cytoplasmic levels of mature miRNA, and occurred in a manner independent of both ‘on-target’ silencing and shRNA sequence. The findings of ref. 4 can be explained by viral vector expression of shRNA at high levels, because lower-level shRNA expression seems to be tolerated4,5. However, the implications for cellular delivery of synthetic siRNA that acts downstream of miRNA biogenesis have remained undetermined.
We interrogated the biochemical and functional status of the miRNA pathway after in vivo administration of synthetic siRNA formulated in liposomal nanoparticles such as those recently described2,6. Formulated, synthetic siRNA targeting two hepatocyte-expressed gene transcripts (si-Apob targeting apolipoprotein B (Apob) and si-FVII targeting factor VII (F7)) and an irrelevant gene (si-Luc targeting luciferase) were administered by a single intravenous bolus injection at different dose levels in groups of three mice. At both 2 and 30 days, silencing of on-target gene transcripts in liver homogenates was measured by the branched DNA assay2, and levels of endogenous miRNA (miR-122, miR-16 and let-7a) were assessed by both northern blot and nuclease protection assays.
Administration of high-dose (5 mg kg−1) si-Apob and si-FVII resulted in marked silencing of the hepatocyte-expressed genes to 22 ± 3% and 17 ± 2% relative to placebo-treated mice at the 2-day time point, respectively (Supplementary Fig. 1). The siRNA effects were dose-dependent, with intermediate silencing effects observed in the animals treated with a low dose (2 mg kg−1) at the same time point. Consistent with previous findings1,2, the RNA interference (RNAi)-mediated silencing effect was selective, with no measurable effect on Apob mRNA levels with si-FVII or si-Luc treatment. Furthermore, there was no significant effect on F7 mRNA levels in mice treated with si-Apob or si-Luc siRNA. At the 30-day time point, Apob and F7 mRNA returned to near normal levels. Treatment with siRNA was not associated with any observed toxicities over the 30-day time period.
To determine whether either ~80% silencing of two different hepatocyte-expressed genes or hepatocyte delivery of three distinct siRNAs results in dysregulation of miRNA biogenesis, levels of the liver-specific miRNA, miR-122, and the broadly expressed miRNAs miR-16 and let-7a were measured in liver tissue samples at both time points for all treatment groups at all dose levels. As shown in Fig. 1, there was no significant reduction in miRNA levels for individual-siRNA-treated animals, as assessed by northern blot assay. To quantify more accurately miRNA levels, a nuclease protection assay was used to measure simultaneously each miRNA together with the small nuclear RNA U6. Inclusion of the U6 control allowed for normalization of the total amount of RNA in each assay. As summarized in Supplementary Table 1, no significant differences were measured in miRNA levels in siRNA-treated animals. Expression levels ranged from 80% to 110% of the levels measured in PBS-treated animals, and were not associated with any trend related to the siRNA dose level or degree of gene silencing. Certainly, no reduction of miRNA levels to match the >80% level previously reported in ref. 4 was observed with administration of synthetic siRNA.
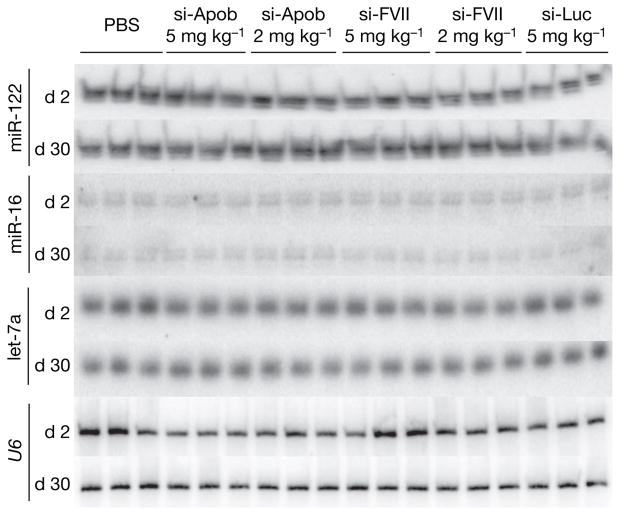
Northern blots of total RNA from mouse livers isolated two days (d 2) and 30 days (d 30) after intravenous injections of PBS, si-Apob (5 mg kg−1 and 2 mg kg−1), si-FVII (5 mg kg−1 and 2 mg kg−1) or si-Luc (5 mg kg−1) hybridized to radiolabelled probes specific for miR-122, miR-16 and let-7a (one animal per lane). Hybridization to a radiolabelled probe specific for U6 was used to confirm equal loading of RNA.
The northern blot and nuclease protection results established that synthetic siRNAs do not inhibit the synthesis or processing of cellular miRNAs. We extended these findings to investigate whether systemically administered siRNAs interfere with endogenous miRNA function. Although in vivo data on miRNA function are limited to only a few published reports7–9, it has been shown that inhibiting mouse miR-122 with antagomirs significantly increases the mRNA levels of a number of liver-expressed genes including Aldoa, Hfe2, Tmed3, Lass6, Slc35a4, Tmem50b and Gpx7 (ref. 9, see Supplementary Fig. 2). Accordingly, liver mRNA levels for these seven putative miR-122 targets were quantified using branched DNA assays (normalized to Gapdh) in mice treated with si-Apob, si-FVII and si-Luc (two days after injection), and compared to PBS-treated animals. Moreover, protein levels of total Aldolase and another miR-122 target, Iqgap1 (ref. 9), were assayed by western analysis of liver lysates. Consistent with the lack of effect on mature miR-122 levels, there was no increase in the mRNA or protein levels of any of these genes that could be correlated with the dose of siRNA administered or the degree of gene silencing observed (Fig. 2 and Supplementary Fig. 3).
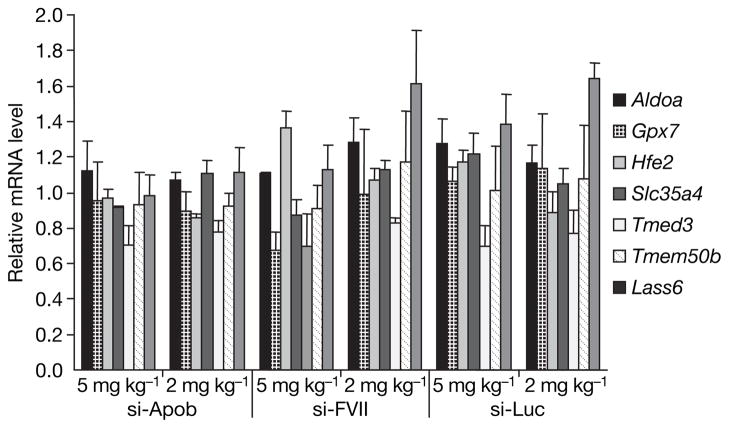
Mouse liver Aldoa, Gpx7, Hfe2, Slc35a4, Tmed3, Tmem50b and Lass6 mRNA levels normalized to Gapdh mRNA measured 2 days after intravenous injections of PBS or different doses (5 mg kg−1 and 2 mg kg−1) of formulated siRNA targeting Apob (si-Apob), F7 (si-FVII) or luciferase (si-Luc) (n = 3 per group). Data are shown as group mean ± s.d. normalized to the PBS group for each gene.
These studies in mice demonstrate that acute gene silencing of ~80% by single administration of siRNA is not associated with any changes in biosynthesis of miR-122, miR-16 or let-7a, nor with any increase in miR-122 target-gene expression. To explore whether prolonged gene silencing by multiple doses of siRNA would alter miRNA levels, and to extend our studies to another species, we targeted the hepatocyte-expressed gene SREBP cleavage-activating protein (Scap) in hamsters. To silence Scap, hamsters were given three, weekly injections of a formulated siRNA targeting Scap (si-Scap) or a mismatch control siRNA (si-Scapmm) at a dose of 2.5 mg kg−1. Repeated treatment with si-Scap resulted in significant (~75%) silencing of Scap mRNA, as well as a very significant reduction in Scap protein on day 21 relative to saline-treated animals (Supplementary Fig. 4). As shown in Fig. 3, silencing of Scap for three weeks by multiple doses of si-Scap had no effect on levels of miR-122, nor did repeat-dosing of the mismatch control, si-Scapmm.
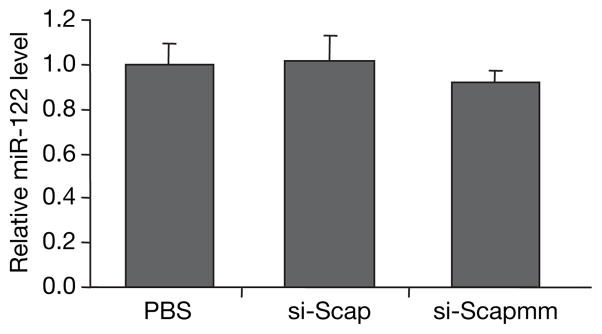
Hamster liver miR-122 levels measured by nuclease protection (normalized to U6 RNA levels) 7 days after three, weekly intravenous injections of PBS or 2.5 mg kg−1 formulated siRNA targeting Scap (si-Scap) or a mismatch control siRNA (si-Scapmm). Data are shown as mean ±s.d.
Our findings demonstrate that robust gene silencing, both acute and longer-term, can be achieved in the liver without any detectable alteration of cellular miRNA biogenesis or function. Certainly, additional studies are warranted such as those with longer ‘dicer substrate’ siRNAs (>21 nucleotides in length) that act upstream of 21-nucleotide siRNA and could interfere with miRNA biogenesis10. Meanwhile, effective gene silencing with siRNA, in which RNA interference is mediated downstream of miRNA biogenesis, can be dissociated from toxicities that may be intrinsic to DNA-expressed shRNA.
METHODS SUMMARY
Antagomirs and siRNAs were synthesized as described1,9. Formulation of siRNAs into liposomes was performed by extrusion. Antagomirs and formulated siRNAs were administered to mice and hamsters by intravenous injection. Expression levels of miRNAs were determined by northern blotting and nuclease protection assay of total RNA isolated from livers of treated animals. Levels of miR-122 target mRNAs were measured in liver lysates using the QuantiGene assay (Genospectra). Western blot analysis was used to measure Scap protein levels in hamster liver lysates and miR-122 target (Aldoa, Iqgap1) protein levels in mouse liver lysates.
Acknowledgments
We thank A. Wetzel, M. Jayaraman, K. N. Jayaprakash, K. G. Rajeev, L. Nechev and G. Wang for assistance in chemistry; R. Dorkin for liposome preparation; M. Duckman for assistance with the figures; and J. Maraganore for critical advice on the manuscript. M. Spranger is supported by CC-SPMD/ETH. M.G., R.L. and D.G.A. are supported by an NIH grant.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Experimental work was performed by M.J., R.C., A.A., M.G., Y.-A.M. and M. Spranger. Experimental design and critical advice was provided by M.J., R.C., A.A., P.H., J.S., H.-P.V., M.M., M. Stoffel, R.L., D.G.A., J.D.H., V.K. and D.B. The paper was written by D.B.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature06179
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3019095
Citations & impact
Impact metrics
Article citations
AMD Genomics: Non-Coding RNAs as Biomarkers and Therapeutic Targets.
J Clin Med, 11(6):1484, 09 Mar 2022
Cited by: 7 articles | PMID: 35329812 | PMCID: PMC8954267
Review Free full text in Europe PMC
Liver-Specific siRNA-Mediated Stat3 or C3 Knockdown Improves the Outcome of Experimental Autoimmune Myocarditis.
Mol Ther Methods Clin Dev, 18:62-72, 22 May 2020
Cited by: 3 articles | PMID: 32577433 | PMCID: PMC7301178
Safety Considerations for Humans and Other Vertebrates Regarding Agricultural Uses of Externally Applied RNA Molecules.
Front Plant Sci, 11:407, 23 Apr 2020
Cited by: 16 articles | PMID: 32391029 | PMCID: PMC7191066
Review Free full text in Europe PMC
A Nuclease Protection ELISA Assay for Colorimetric and Electrochemical Detection of Nucleic Acids.
Anal Methods, 11(8):1027-1034, 24 Jan 2019
Cited by: 4 articles | PMID: 31656535 | PMCID: PMC6814143
Biodegradable hybrid mesoporous silica nanoparticles for gene/chemo-synergetic therapy of breast cancer.
J Biomater Appl, 33(10):1382-1393, 16 Mar 2019
Cited by: 6 articles | PMID: 30880565
Go to all (84) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
RNAi-mediated gene silencing in non-human primates.
Nature, 441(7089):111-114, 26 Mar 2006
Cited by: 770 articles | PMID: 16565705
Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs.
Nature, 432(7014):173-178, 01 Nov 2004
Cited by: 1230 articles | PMID: 15538359
MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells.
Cancer Res, 67(20):9762-9770, 01 Oct 2007
Cited by: 512 articles | PMID: 17942906
Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting.
RNA, 13(4):431-456, 28 Feb 2007
Cited by: 123 articles | PMID: 17329355 | PMCID: PMC1831859
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIBIB NIH HHS (3)
Grant ID: R01 EB000244-27
Grant ID: R01 EB000244
Grant ID: R37 EB000244





