Abstract
Free full text

Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome[S]
Abstract
PNPLA3 (adiponutrin, calcium-independent phospholipase A2 epsilon [iPLA2ϵ]) is an adipose-enriched, nutritionally regulated protein that belongs to the patatin-like phospholipase domain containing (PNPLA) family of lipid metabolizing proteins. Genetic variations in the human PNPLA3 gene (i.e., the rs738409 I148M allele) has been strongly and repeatedly associated with fatty liver disease. Although human PNPLA3 has triacylglycerol (TAG) hydrolase and transacylase activities in vitro, its in vivo function and physiological relevance remain controversial. The objective of this study was to determine the metabolic consequences of global targeted deletion of the Pnpla3 gene in mice. We found that Pnpla3 mRNA expression is altered in adipose tissue and liver in response to acute and chronic nutritional challenges. However, global targeted deletion of the Pnpla3 gene in mice did not affect TAG hydrolysis, nor did it influence energy/glucose/lipid homoeostasis or hepatic steatosis/injury. Experimental interventions designed to increase Pnpla3 expression (refeeding, high-sucrose diet, diet-induced obesity, and liver X receptor agonism) likewise failed to reveal differences in the above-mentioned metabolic phenotypes. Expression of the Pnpla3 paralog, Pnpla5, was increased in adipose tissue but not in liver of Pnpla3-deficient mice, but compensatory regulation of genes involved in TAG metabolism was not identified. Together these data argue against a role for Pnpla3 loss-of-function in fatty liver disease or metabolic syndrome in mice.
Obesity and the metabolic syndrome are major contributors to morbidity and mortality from a variety of diseases affecting virtually all organ systems (1). Obesity is essentially a disorder of lipid accumulation, primarily in the form of triacylglycerols (TAGs) in adipose tissue. TAGs serve as a critical reservoir for lipid metabolites involved not only in energy homeostasis but also other essential cellular processes including membrane synthesis and cell signaling. In the context of chronic energy excess and/or impaired lipid metabolism, TAGs accumulate in metabolically relevant nonadipose tissues such as liver, where they are associated with cellular and systemic metabolic dysfunction (2, 3). TAG accumulation in liver (hepatic steatosis) is the earliest hallmark of nonalcoholic fatty liver disease (NAFLD) and contributes to hepatocyte dysfunction, inflammation (steatohepatitis), fibrosis, cirrhosis, liver failure, and even hepatocellular carcinoma (4). NAFLD frequently occurs with other features of the metabolic syndrome (both in association with and independent of obesity) (4–6) and is the leading cause of liver disease in the Western world (4, 7, 8). However, the mechanisms underlying hepatocellular TAG accumulation and its relationship to lipid-induced toxic metabolic effects (lipotoxicity) remain unclear.
PNPLA3 (alternatively known as adiponutrin and calcium-independent phospholipase A2 epsilon [iPLA2ϵ]) belongs to a novel family of lipid metabolizing enzymes known as the patatin-like phospholipase domain-containing (PNPLA) family (9). Recently, genetic variation in the human PNPLA3 gene (i.e., the rs738409 I148M allele in particular) has been strongly and repeatedly associated with hepatic TAG content, hepatocellular injury, and progression of NAFLD in humans (10–24). PNPLA3 variants have also been associated with obesity and features of the metabolic syndrome in humans in several studies (16, 25, 26), although subsequent studies have not confirmed this association (10, 12, 18, 22). The mechanisms by which PNPLA3 genetic variation either predisposes to or protects against NAFLD and/or metabolic disease are not known but are obviously of considerable biomedical importance for the prevention and/or treatment of NAFLD and related metabolic disorders.
PNPLA3 was initially identified in 2001 as a membrane-associated, adipose-enriched protein that is induced during adipogenesis (27). PNPLA3 shares the greatest homology with PNPLA2 (alternatively known as adipose triglyceride lipase [ATGL] and calcium-independent phospholipase A2 zeta (iPLA2ζ]), which plays an extremely critical role in metabolism by mediating the rate-limiting step in TAG hydrolysis. Like Pnpla2/ATGL, Pnpla3 expression is altered in obesity-related dysmetabolic states (12, 25, 27–29). Also like Pnpla2/ATGL, Pnpla3 expression is strongly regulated by a variety of important nutritional (27–38), hormonal (25, 27, 35, 39–41), and pharmacological factors (36, 38, 42), although Pnpla2 and Pnpla3 are usually regulated in the opposite direction from each other. The regulation and subcellular localization of PNPLA3 are most consistent with a role in anabolic lipid metabolism (i.e., lipogenesis). Indeed, lipogenic stimuli dramatically increase Pnpla3 expression not only in adipose tissue (35) but also in liver (38). Interestingly, however, human PNPLA3 has TAG hydrolase (28, 43, 44) and acyl-CoA-independent transacylase activities (43) in cell-free systems in vitro. Furthermore, ectopic overexpression of wild-type (WT) human PNPLA3 in cultured cells (28, 44) or in murine livers (44) does not alter cellular TAG accumulation. Likewise, short interfering RNA-mediated knockdown of endogenous murine PNPLA3 in 3T3-L1 adipocytes does not affect measures of TAG hydrolysis (35). Thus, despite strong evidence implicating PNPLA3 in normal metabolism and disease, the in vivo function and physiological relevance of PNPLA3 remain controversial.
The goal of the present study was to better understand the function and physiological relevance of PNPLA3. To do so, we generated mice with global targeted deletion of Pnpla3. We then characterized these Pnpla3 knockout (Pnpla3-KO or KO) mice according to multiple aspects of metabolic phenotype under a variety of conditions, including those known to induce Pnpla3 expression. In doing so, we found that global Pnpla3 deletion in mice is not a major determinant of overall energy homeostasis, glucose homeostasis/insulin action, lipid homeostasis, or hepatic steatosis/injury. Assuming similar functions of PNPLA3 in mice and humans, these data suggest that PNPLA3 loss-of-function may not be the primary mechanism by which variation in PNPLA3 influences fatty liver disease or metabolic phenotypes.
METHODS
Generation of the Pnpla3 targeting construct and Pnpla3-KO mice
The Pnpla3 targeting construct was generated using bacterial artificial chromosome (BAC) recombineering techniques (45, 46). The BAC vector (pBACe3.6) containing Pnpla3 from the 129SvEV/AB2.2 mouse strain (BAC clone bMQ-381F07) was obtained from Wellcome Trust Sanger Institute. Nucleotides from −12 to +1927 (1,939 bp) of the Pnpla3 gene were replaced by a neomycin resistance cassette. The resulting targeting construct was electroporated into 129 embryonic stem (ES) cells, followed by antibiotic selection under standard conditions. Antibiotic-resistant ES cell clones were screened by Southern blotting to identify clones with homologous recombination. Appropriately targeted ES cells were then injected into C57BL/6 blastocysts and implanted into pseudopregnant females with the assistance of transgenic core facility staff at Beth Israel Deaconess Medical Center. Resulting chimeric mice were screened by PCR to identify mice with germline transmission of the targeted allele (WT allele: forward primer 5′ ggggcggggtacgtggattagaga, and reverse primer 5′ agccctgcccacaaccccaaaag [296 bp]; targeted allele: forward primer 5′ cggccgcttgggtggagag, and reverse primer 5′ caggtagccggatcaagcgtatgc [358 bp]). Mice carrying the targeted allele were then backcrossed to C57BL/6 mice for at least 4 generations (N4).
Animals
For the evaluation of adipose- and liver-specific Pnpla3 mRNA expression in response to high-fat (HF) feeding and leptin deficiency, tissues were collected from a) overnight-fasted 24 week-old male FVB mice (Taconic) fed chow diet (14 kcal% fat; Harlan Teklad RD8664) or HF diet (42 kcal% fat; Harlan Teklad TD88137) since weaning (47); and b) ad libitum-fed 10 week-old Lepob/Lepob mice (Jackson Laboratories) fed chow since weaning (35). For evaluation of the effects of global Pnpla3 deletion on metabolism, experimental mice were derived from breeding 129/C57BL/6(N4–N6) mice heterozygous for the targeted Pnpla3 allele. Resulting experimental WT (Pnpla3+/+) and KO (Pnpla3−/−) mice were fed one of four diets (composition in kcal%), as follows: standard (chow) diet (Prolab Isopro RMH 3000; 26% protein/60% complex carbohydrate in the form of starch/14% fat); low-fat low-sucrose (LFLS) diet (Research Diet D12328N; 16.4% protein/73% complex carbohydrate in the form of 61% starch and 12% maltodextrin/10.5% fat); low-fat high-sucrose (LFHS) diet (D12329N; 16.4% protein/73% carbohydrate in the form of sucrose/10.5% fat); or high-fat high-sucrose (HFHS) diet (D12331N; 16.4% protein/25% carbohydrate in the form of sucrose/58% fat) from weaning until 19 weeks of age. For evaluation of the metabolic response to pharmacological activation of liver X receptor (LXR), a separate cohort of 12 week-old male mice was fed chow diet containing 0.025% of the potent LXR agonist T0901317 (48, 49) for 6 days (equivalent to ~50 mg/kg of body weight/d) as described (38, 50). This dosing regimen was selected based on previous experiments demonstrating induction of hepatic Pnpla3 mRNA expression under these experimental conditions (38). Mice were housed at Beth Israel Deaconess Medical Center, University of Pittsburgh, or University of Texas-Southwestern Animal Facilities under standard conditions with access to water and food ad libitum. All investigations involving animals were conducted in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals outlined in the Institute for Laboratory Animal Research (ILAR) Guide for Care and Use of Laboratory Animals. Experimental procedures were approved by the institutional animal care and use committees at the institutions noted above.
Body composition, energy expenditure, and metabolic measurements
Body composition was determined by Echo magnetic resonance imaging (Echo Medical Systems). Oxygen consumption (Vo2), respiratory exchange ratio (RER), and physical activity were determined using a Comprehensive Laboratory Animal Monitoring System (Columbus Instruments). Plasma glucose was measured using a One-Touch FastTake glucometer (Lifescan). For glucose tolerance tests (GTTs), mice were injected intraperitoneally with 1.75 g/kg glucose following a 6 h fast. For insulin tolerance tests (ITTs), mice were injected intraperitoneally with regular human insulin (HumulinR; Lilly) at 0.80 units/kg following a 4 h fast. Serum corticosterone was determined by radioimmunoassay as described previously (47). Other serum parameters were determined using the following kits: insulin (Ultra Sensitive Mouse Insulin ELISA kit; Crystal Chem), TAG (Infinity Triglycerides Liquid Stable Reagent; Thermo Scientific), NEFA (HR series NEFA-HR[2] Reagents; Wako Diagnostics), cholesterol (Cholesterol E; Wako Diagnostics), alanine aminotransferase (ALT/SGPT test; Stanbio Laboratory), and aspartate aminotransferase (AST/SGOT test; Stanbio Laboratory).
TAG hydrolase activity
TAG hydrolase activity was determined as described previously (51). Briefly, tissues were homogenized in lysis buffer (0.25 M sucrose, 1 mM EDTA, 1 mM dithiothreitol, 20 μg/ml leupeptin, 2 μg/ml antipain, 1 μg/ml pepstatin, pH 7.0). Lysates were then centrifuged for 60 min at 100,000 g at 4°C to isolate lipid-free infranatants. Triolein substrate containing [9,10-3H]triolein as radioactive tracer was prepared by sonication as described previously (52). Cytosolic fractions (0.1 ml) with or without 500 ng/assay purified CGI-58 and/or 25 μM of the hormone sensitive lipase (HSL)-specific inhibitor 76-0079 (NNC 0076-0000-0079; Novo Nordisk) were incubated with 0.1 ml (167 nmol) of substrate at 37°C for 60 min. Reactions were terminated by adding 3.25 ml of methanol-chloroform-heptane (10:9:7) and 1 ml of 0.1 M potassium carbonate, 0.1 M boric acid, at pH 10.5. After centrifugation at 800 g for 20 min, the radioactivity in 1 ml of the upper phase was determined by liquid scintillation counting.
Ex vivo lipolysis in perigonadal white adipose tissue explants
Ex vivo tissue lipolysis was determined as described previously (53). Briefly, ~20 mg pieces of perigonadal white adipose tissue (WAT) were incubated in DMEM (Invitrogen) containing 2% fatty acid-free BSA (Sigma) with or without 10 μM isoproterenol (Sigma) at 37°C. NEFA and glycerol release were determined in aliquots of incubation buffer collected at the times indicated using NEFA-HR(2) reagents (Wako Diagnostics) and Infinity triglycerides Liquid Stable Reagent (Thermo Scientific), respectively. Results were normalized to tissue protein content as determined by the bicinchoninic acid method (Pierce).
Biochemical and histological determination of tissue TAG content
For biochemical determination of liver TAG content, liver tissue (~20–30 mg) was ground in liquid nitrogen and extracted by the method of Folch (54). Dried lipid extracts were redissolved in 60 µl of tert-butanol plus 40 µl of a 2:1 mixture of Triton X-114: methanol. TAG content was then determined using Infinity triglycerides Liquid Stable Reagent (Thermo Scientific). Liver TAG content was confirmed histologically by Oil Red O staining, using standard methods (55).
In vivo cholesterol and fatty acid synthesis
In vivo rates of cholesterol and fatty acid synthesis were measured in mouse tissues by using 3H-labeled water (ICN) as described (56). Mice were fasted for 2 h at the early phase of the light cycle and then injected intraperitoneally with [3H]H2O (50 mCi in 0.25 ml of isotonic saline). At 1 h after the injection, mice were sacrificed, and liver, perigonadal WAT, and intrascapular brown adipose tissue (BAT) were collected. Lipids were extracted from tissues and 3H-labeled fatty acids and digitonin-precipitable sterols (DPSs) were determined as described (57, 58). Rates of fatty acid and cholesterol synthesis were calculated as μmol of 3H radioactivity incorporated into fatty acids or DPSs per h per gram of tissue, respectively.
Mass spectrometry analysis of lipids
Quantitative analysis of TAG molecular species was performed as described (59, 60). Briefly, lipids were extracted from mouse tissues by using a modified method of Bligh and Dyer (61) in the presence of internal standards for electrospray ionization mass spectrometry (ESI-MS) analysis. ESI-MS analyses were performed using a TSQ Quantum Ultra Plus triple-quadrupole mass spectrometer (ThermoFisher Scientific) equipped with an automated nanospray apparatus (Nanomate HD; Advion Bioscience Ltd.) and Xcalibur system software.
RNA extraction, reverse transcription, and gene expression analysis
Total RNA was extracted from homogenized tissues using an RNeasy Lipid Tissue mini-kit with on-column DNase treatment (Qiagen). Reverse transcription of 1 μg of total RNA was performed using random decamers (RETROscript kit; Ambion, Inc.). Gene expression was determined by quantitative PCR (qPCR) (7300 real-time PCR system; ABI) using gene-specific primer-probe sets (TaqMan gene expression assays; Applied Biosystems) normalized to cyclophilin (forward primer 5′ ggtggagagcaccaagacaga; reverse primer 5′ gccggagtcgacaatgatg; and probe 5′ agccgggacaagccactgaaggat) or to 18S rRNA (Applied Biosystems) as reference genes, using the standard curve method as described (62). Appropriate analysis was performed to determine that expression of reference genes were unchanged under the experimental conditions described. Accuracy of RNA quantification was optimized by DNase treatment of samples, use of gene-specific primer probe sets that spanned intron-exon boundaries, and verification of the lack of amplification in no-RT and no-template controls. Experimental parameters conformed to MIQE guidelines (63).
Statistical analysis
Results are expressed as means ± SEM. Comparisons were made with unpaired two-tailed Student's t-test or factorial ANOVA, followed by determination of simple effects for pair-wise comparisons, if relevant (SPSS software). For repeated measurements, comparisons were made by two-way ANOVA with repeated measures. For all analyses, P values of <0.05 were considered statistically significant.
RESULTS
Pnpla3 mRNA expression in adipose tissue and liver is altered in murine models of fatty liver and metabolic syndrome
Pnpla3 mRNA expression is highly regulated by acute nutritional and hormonal challenges in both adipose tissue and liver (25, 27–41). To determine the Pnpla3 response to chronic nutritional and hormonal challenges that predispose to fatty liver and the metabolic syndrome, we evaluated Pnpla3 mRNA expression in adipose tissue and liver of mice with obesity due to chronic high-fat diet feeding and leptin deficiency (Lep/Lepobob mice) (Fig. 1). In overnight-fasted diet-induced obese mice (Fig. 1A), Pnpla3 expression was increased in all adipose tissue depots. In addition, despite lower Pnpla3 expression in liver relative to adipose tissue, hepatic Pnpla3 expression was also increased (12-fold). Thus, prolonged high-fat feeding (sufficient to induce obesity [47]) increases Pnpla3 mRNA expression independent of acute nutritional effects. In ad libitum-fed genetically obese leptin-deficient mice (Fig. 1B), Pnpla3 expression was also altered in adipose tissue but in a depot-specific manner, consistent with prior results in leptin-resistant (Leprfa/Leprfa) rats (29). These data suggest that Pnpla3 may be regulated differently in leptin-deficient/resistant obesity than in diet-induced obesity. However, similar to diet-induced obesity, Pnpla3 expression was substantially increased in liver (22-fold) in mice with leptin deficiency. Thus, Pnpla3 mRNA expression is altered in adipose tissue and liver of two common models of fatty liver and metabolic syndrome, thereby supporting a potential role for PNPLA3 in the pathogenesis and/or physiological adaptation to these disorders.
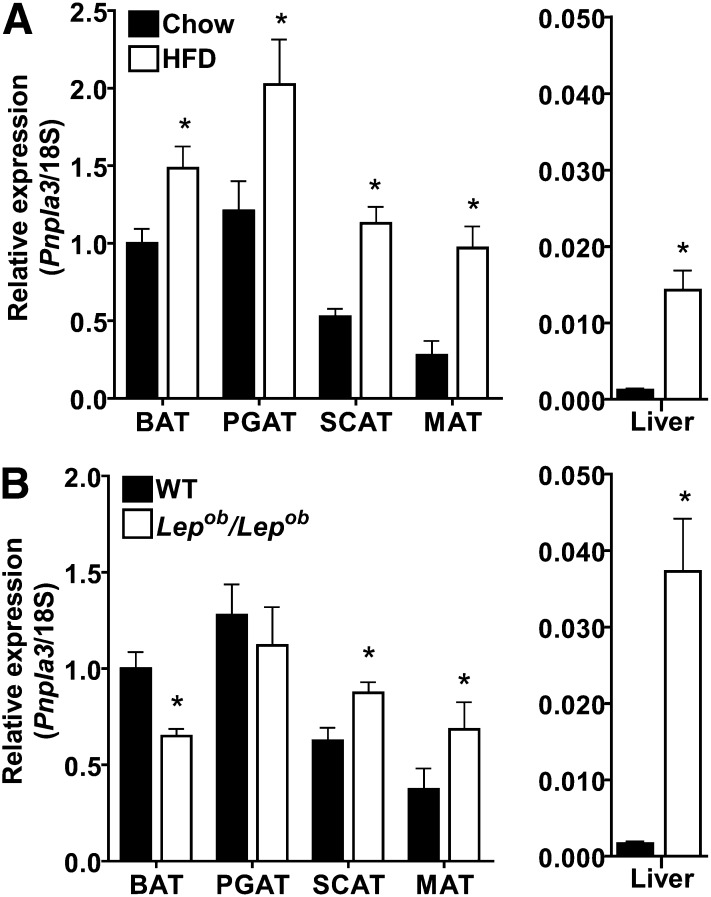
Pnpla3 mRNA expression in animal models of fatty liver and metabolic syndrome. (A) Pnpla3 mRNA expression in brown (BAT), perigonadal (PGAT), subcutaneous (SCAT), and mesenteric (MAT) adipose tissue, and liver of overnight-fasted male FVB mice fed chow (14 kcal% fat) or high-fat diet (HFD, 42 kcal% fat) from weaning until 24 weeks of age (n = 10 per group, male, 24 week-old, overnight-fasted). (B) Pnpla3 mRNA expression in the above adipose tissue depots and liver of WT or leptin-deficient (Lep/Lepobob) mice (n = 6 per group, male, 10 week-old, ad libitum-fed, chow diet). Pnpla3 expression is normalized to 18S rRNA and expressed relative to Pnpla3 expression in BAT of the chow-fed (A) or WT (B) control groups, respectively. * P < 0.05 by Student's t-test.
Mice with global targeted deletion of Pnpla3 lack Pnpla3 expression in metabolically relevant tissues
To better understand the physiological relevance of PNPLA3, including its contribution to fatty liver and metabolic syndrome, we generated a murine model with global targeted deletion of Pnpla3 (Pnpla3-KO mice) (Fig. 2A, B). As expected, Pnpla3 mRNA expression was decreased in a gene-dose-dependent manner in mice carrying the targeted allele and was completely absent in Pnpla3-KO mice in metabolically-relevant tissues where the Pnpla3 gene is normally expressed (i.e., in adipose tissue, adrenal glands, and liver) (Fig. 2C) (35). Similar mRNA expression results were obtained for primer-probe sets spanning exons 1–2 (proximal) (Fig. 2C), 4–5 (mid), and 7–8 (distal) of the Pnpla3 gene (data not shown). Gene expression analysis further confirmed that endogenous Pnpla3 expression was 100- to 1,000-fold higher in adipose tissue than in liver. Despite differences in adrenal Pnpla3 expression, no differences in adrenal histopathology (data not shown) or am serum corticosterone levels (25.3 ± 15.2 ng/ml for WT, vs. 27.1 ± 13.8 ng/ml for KO) were identified between genotypes. Finally, Pnpla3-KO mice were viable, fertile, and produced in the expected Mendelian ratios.
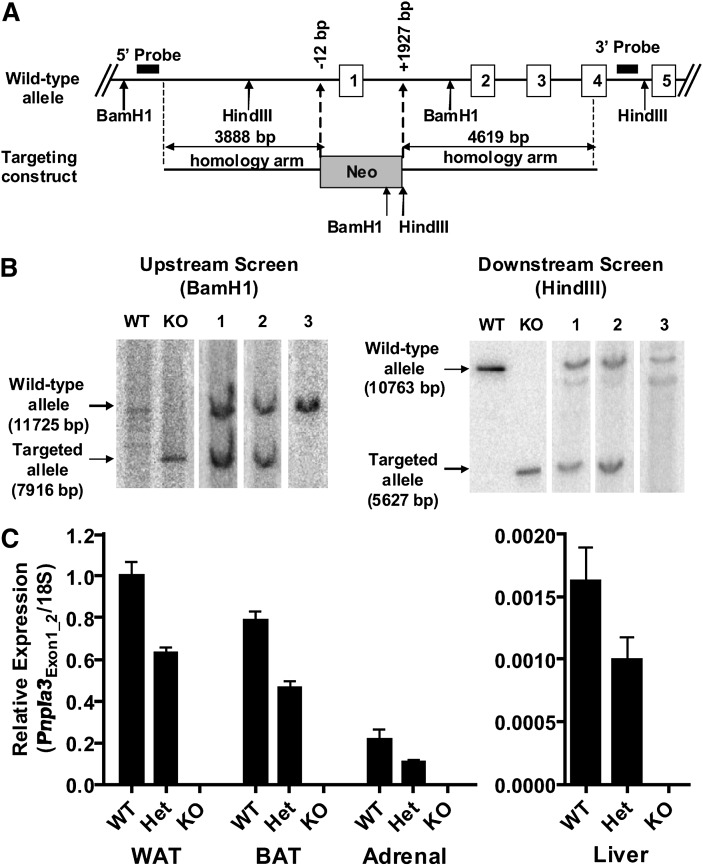
Generation of mice with global targeted deletion of Pnpla3 (Pnpla3-KO mice). (A) The native (wild-type, WT) Pnpla3 allele and targeting (knockout, KO) construct (not drawn to scale). Bacterial artificial chromosome (BAC) recombineering was used to replace 1939 bp of the native Pnpla3 gene from position -12 to +1927 including the ATG start signal and the entire exon 1 (which contains the GXSXG motif with the putative critical catalytic serine residue of the patatin domain) by a neomycin resistance cassette. Embryonic stem (ES) cell electroporation, selection, and screening were then performed using standard techniques. (B) Southern blots of ES cell clones confirming homologous recombination at upstream (BamH1) and downstream (HindIII) ends. BACs containing the WT and targeted (KO) allele are shown as controls. Lanes 1 and 2 demonstrate positive ES cell clones. Lane 3 demonstrates a negative ES cell clone. All gels were run under identical conditions. Images shown were run on separate gels. (C) Pnpla3 mRNA expression in perigonadal WAT, BAT, adrenal gland, and liver using a primer-probe set spanning exons 1–2 (Pnpla3Exon1_2) (n = 7–10/group, mixed gender, 7–8 week-old, ad libitum-fed, chow diet). Similar results were obtained for primer-probe sets spanning exons 4–5 and 7–8 (data not shown). Pnpla3 expression is normalized to 18S rRNA and expressed relative to Pnpla3 expression in WAT of the control group.
Global Pnpla3 deletion does not affect adipose tissue TAG hydrolysis
Because human PNPLA3 has TAG hydrolase activity in cell-free systems in vitro (43, 44), we next evaluated TAG hydrolysis in adipose tissue of Pnpla3-KO mice (Fig. 3). Pnpla3 mRNA expression is greatest in perigonadal WAT and BAT (35). However, in vitro TAG hydrolase activity in lysates of perigonadal WAT (Fig. 3A) and BAT (data not shown) did not differ between genotypes, either when assayed alone or in the presence of the PNPLA2/ATGL coactivator CGI-58 and/or the HSL inhibitor 76-0079. Likewise, basal and isoproterenol-stimulated glycerol (Fig. 3B) and NEFA (data not shown) release from perigonadal WAT explants did not differ between genotypes. Finally, there was no compensatory upregulation of Pnpla2/ATGL or HSL mRNA, the primary enzymes mediating TAG hydrolysis, in WAT (Fig. 3C, D, respectively) or BAT (data not shown). These data suggest that Pnpla3 deficiency does not significantly influence TAG hydrolysis in murine tissues.
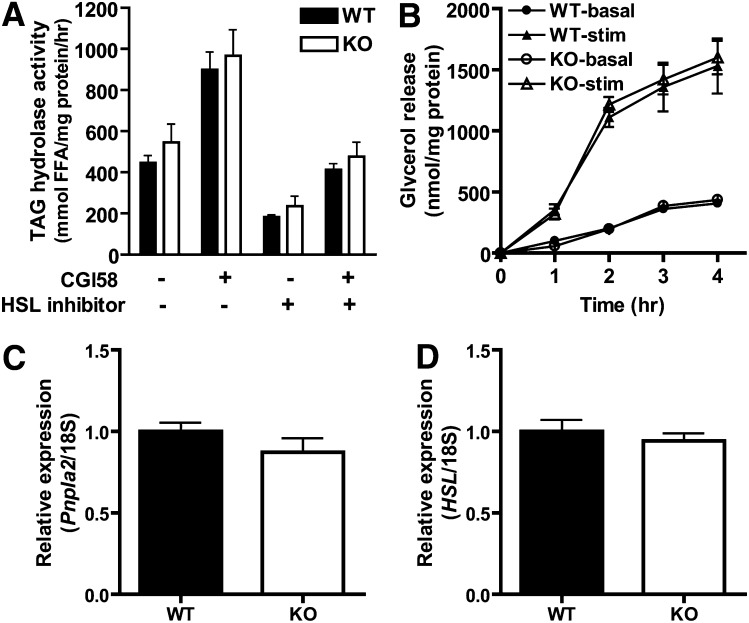
TAG hydrolysis in Pnpla3 knockout (KO) mice. (A) TAG hydrolase activity was determined in cytosolic fractions of perigonadal WAT lysates from WT and KO mice (n = 6/group, male, 16–17 week-old, ad libitum-fed, chow diet) in the presence or absence of the PNPLA2/ATGL coactivator CGI-58 and/or the HSL-specific inhibitor 76-0079. Similar results were obtained for BAT (data not shown). (B) Glycerol release over time from perigonadal WAT explants of WT and KO mice (n = 3/group, male, 8–10 week-old, ad libitum-fed, chow diet) in the absence (basal) or presence of 10 μM isoproterenol (stimulated, stim). Comparable results were obtained for NEFA release (data not shown). (C-D) mRNA expression of Pnpla2/ATGL (C) and HSL (D) in perigonadal WAT of WT and KO mice (n = 7–10/group, mixed gender, 7–8 week-old, ad libitum-fed, chow diet). Expression is normalized to 18S rRNA and expressed relative to gene expression in the WT control group. No significant differences were identified.
Global Pnpla3 deletion does not affect energy homeostasis
Because PNPLA3 could have activities other than TAG hydrolysis that could influence metabolism, we performed comprehensive metabolic phenotyping of Pnpla3-KO and WT mice in response to chronic nutritional challenges known to induce Pnpla3 expression (i.e., high-sucrose feeding and diet-induced obesity). Specifically, mice were fed a standard chow diet or one of three other diets with identical protein content but different sucrose and fat contents: a “control” low-fat low-sucrose (LFLS) diet, a low-fat high-sucrose (LFHS) diet, and a high-fat high-sucrose (HFHS) diet from weaning until 19 weeks of age. Importantly, gene expression analysis of terminal liver and perigonadal WAT confirmed substantially higher Pnpla3 mRNA expression in tissues from WT compared with Pnpla3-KO mice for all diet groups (see Fig. 7 and supplemental Table 1). Nevertheless, no differences were observed between genotypes for body weight (Fig. 4A–D), percent fat mass (Fig. 4E–H), percent lean body mass (chow diet: 70.28 ± 2.17% for WT vs. 69.11 ± 1.80% for KO; LFLS diet: 64.84 ± 1.64% for WT vs. 65.55 ± 1.09% for KO; LFHS diet: 62.45 ± 1.70 for WT vs. 62.06 ± 2.01 for KO; and HFHS diet: 57.04 ± 2.39 for WT vs. 50.15 ± 4.81% for KO); cumulative food intake (chow diet: 1,438 ± 35 for WT vs. 1,444 ± 55 kcal/mouse/19w for KO; LFLS diet: 1,515 ± 24 for WT vs. 1,466 ± 36 kcal/mouse/19w for KO; LFHS diet: 1,472 ± 48 for WT vs. 1,495 ± 77 kcal/mouse/19w for KO; HFHS diet: 1,738 ± 89 for WT vs. 1,764 ± 41 kcal/mouse/19w for KO), or fat pad/tissue weights (data not shown). Likewise, no differences were identified among genotypes for oxygen consumption, RER, or physical activity in the ad libitum-fed, fasted, or refed states (8 week-old males, n = 4–6/group; data not shown). Thus, Pnpla3 deficiency does not alter overall energy homeostasis in response to chronic nutritional challenges.
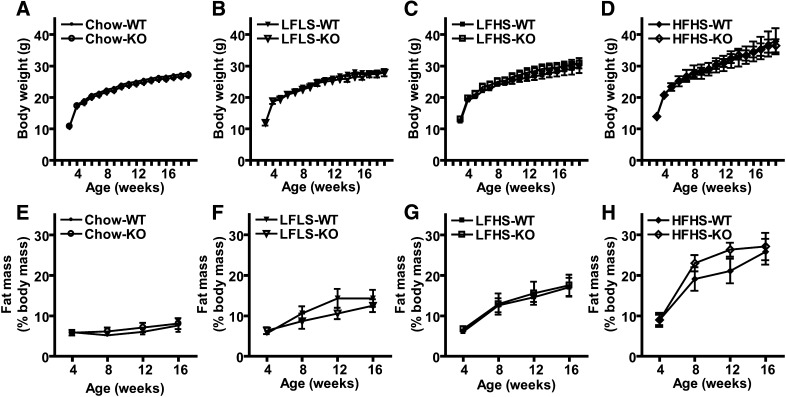
Energy homeostasis in Pnpla3 knockout (KO) mice. (A–D) Body weight in grams (g) of WT and KO mice fed chow (A), low-fat low-sucrose (LFLS) (B), low-fat high-sucrose (LFHS) (C), or high-fat high-sucrose (HFHS) (D) diets from weaning until 19 weeks of age. (E-H) Fat mass as a percent of total body mass (%) of WT and KO mice fed chow (E), LFLS (F), LFHS (G), or HFHS (H) diets from weaning until 19 weeks of age (n = 6–11/group, male). No significant differences were identified.
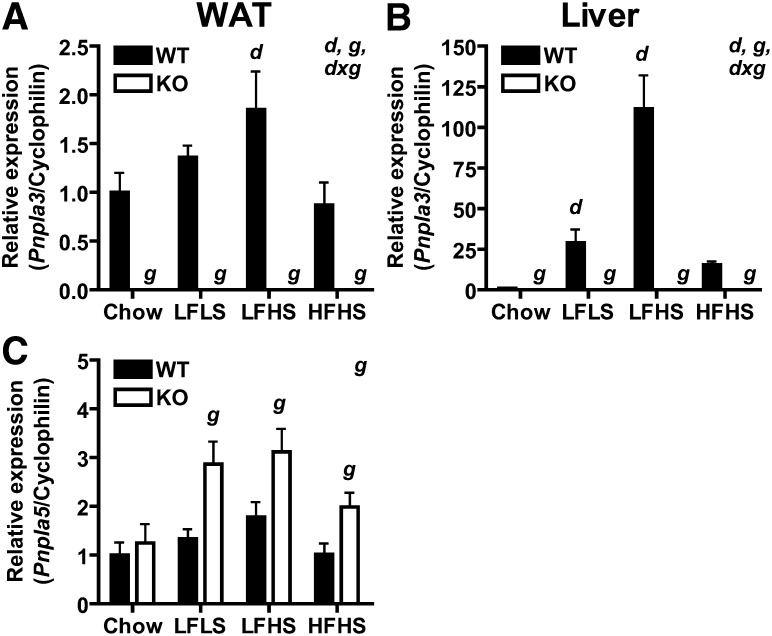
Gene expression in adipose tissue and liver of Pnpla3 knockout (KO) mice. Pnpla3 mRNA expression in (A) perigonadal WAT and (B) liver, and (C) Pnpla5 mRNA expression in WAT of ad libitum-fed 19 week-old male WT and KO mice fed chow, low-fat low-sucrose (LFLS), low-fat high sucrose (LFHS), or high-fat high sucrose (HFHS) diet since weaning (n = 6–11/group). Genes of interest are normalized to cyclophilin as a reference gene and expressed relative to gene expression in the WT chow-fed control for each gene/tissue. Significant main effects (P < 0.05) of diet (d) and genotype (g) as well as interactions between diet and genotype (d x g) are indicated in upper right corner. Simple effects for pair-wise comparisons are indicated over the appropriate bars (g for comparison of KO vs. WT within a given diet group; d for comparison of special diet vs. chow control within a given genotype group). Expression data for additional genes are found in supplemental Table I.
Global Pnpla3 deletion does not affect systemic glucose tolerance, insulin sensitivity, or serum lipid homeostasis
To determine the effect of Pnpla3 deficiency on systemic lipid/glucose homeostasis and insulin action, we next evaluated body weight and serum parameters in response to acute nutritional challenges (ad libitum feeding, fasting, refeeding) in mice fed LFLS, LFHS, and HFHS diets from weaning until 12 weeks of age (Table 1). As expected, both diet and feeding status had significant main effects on body weight and serum parameters, consistent with the appropriate metabolic responses. Further analysis revealed significant interactions between diet and genotype for NEFA (NEFA were lower in KO than WT mice fed LFHS diet but trended higher in KO than WT mice fed HFHS diet) and between feeding status and genotype for cholesterol (cholesterol was higher in KO than in WT refed mice). However, no main effects were identified for genotype on body weight, glucose, insulin, TAGs, NEFAs, or cholesterol. Furthermore, no consistent or reproducible differences between genotypes were identified for these parameters in terminal blood from ad libitum-fed 19 week-old mice from the above diet groups, in terminal blood from 11 week-old chow-fed mice fasted for intervals ranging from 4–16 h, or in terminal blood from 8–10 week-old chow-fed mice fasted for 12 h followed by 12 h of refeeding with the LFHS diet (data not shown). Finally, both genotypes from the above diet groups had similar serum glucose responses to intraperitoneal glucose (Fig. 5A–D) and insulin (Fig. 5E–H) challenges, indicating no differences in systemic glucose tolerance or insulin sensitivity, respectively. These data argue against a significant effect of genotype on these parameters but suggest that Pnpla3 deficiency may influence NEFA and/or cholesterol homeostasis under specific nutritional/metabolic conditions.
TABLE 1.
Body weight and serum parameters in fed, fasted, and refed WT and Pnpla3-KO mice fed LFLS, LFHS, and HFHS diets
| LFLS | LFHS | HFHS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Genotype | Fed | Fasted | Refed | Fed | Fasted | Refed | Fed | Fasted | Refed | P (<0.05) |
| Weight (g) | WT | 26.81 ± 0.63 | 24.09 ± 0.67 | 27.51 ± 0.42 | 27.14 ± 1.18 | 24.47 ± 1.16 | 26.90 ± 1.10 | 32.90 ± 2.84 | 30.27 ± 2.64 | 31.55 ± 2.69 | d, f |
| KO | 25.89 ± 0.94 | 23.19 ± 0.89 | 26.05 ± 0.89 | 28.86 ± 1.43 | 25.90 ± 1.40 | 28.69 ± 1.35 | 33.47 ± 1.48 | 30.86 ± 1.41 | 32.10 ± 1.39 | ||
| Glucose (mg/dl) | WT | 137 ± 14 | 96 ± 9 | 143 ± 9 | 116 ± 9 | 108 ± 8 | 121 ± 9 | 152 ± 5 | 124 ± 9 | 137 ± 13 | d, f |
| KO | 129 ± 6 | 109 ± 8 | 125 ± 10 | 139 ± 9 | 97 ± 8 | 134 ± 8 | 159 ± 10 | 123 ± 6 | 144 ± 7 | ||
| Insulin (ng/ml) | WT | 0.29 ± 0.08 | 0.12 ± 0.04 | 7.01 ± 3.09 | 0.64 ± 0.12 | 0.13 ± 0.04 | 5.64 ± 1.57 | 1.62 ± 0.50 | 0.22 ± 0.03 | 1.21 ± 0.18 | f, dxf |
| KO | 0.24 ± 0.10 | 0.16 ± 0.06 | 7.61 ± 3.01 | 0.94 ± 0.09 | 0.18 ± 0.04 | 9.05 ± 3.32 | 1.27 ± 0.23 | 0.32 ± 0.03 | 1.27 ± 0.17 | ||
| TAG (mg/dl) | WT | 75.70 ± 5.16 | 64.24 ± 2.94 | 134.14 ± 12.87 | 66.63 ± 2.85 | 68.94 ± 6.31 | 130.13 ± 7.92 | 71.65 ± 6.62 | 71.43 ± 8.53 | 85.15 ± 10.08 | d, f |
| KO | 71.18 ± 11.54 | 53.42 ± 2.53 | 145.37 ± 14.45 | 74.60 ± 11.29 | 62.11 ± 5.67 | 122.14 ± 15.73 | 55.045 ± 4.62 | 69.34 ± 5.38 | 109.55 ± 10.23 | ||
| NEFA (mEq/l) | WT | 0.55 ± 0.09 | 0.99 ± 0.09 | 0.57 ± 0.04 | 0.55 ± 0.08 | 1.17 ± 0.04 | 0.66 ± 0.06 | 0.51 ± 0.05 | 0.97 ± 0.07 | 0.72 ± 0.09 | d, f, dxg |
| KO | 0.56 ± 0.04 | 1.15 ± 0.03 | 0.53 ± 0.06 | 0.34 ± 0.03 | 1.04 ± 0.05 | 0.46 ± 0.04 | 0.53 ± 0.04 | 1.01 ± 0.04 | 0.93 ± 0.06 | ||
| Cholesterol (mg/dl) | WT | 98.97 ± 7.72 | 120.55 ± 11.40 | 122.12 ± 11.75 | 102.98 ± 4.86 | 134.25 ± 10.34 | 177.70 ± 11.51 | 131.54 ± 14.57 | 150.68 ± 15.58 | 157.44 ± 29.77 | d, f, fxg |
| KO | 92.69 ± 5.18 | 102.68 ± 10.50 | 155.11 ± 12.98 | 98.21 ± 3.18 | 108.05 ± 7.53 | 163.70 ± 13.49 | 143.16 ± 8.99 | 145.26 ± 10.13 | 203.47 ± 17.76 | ||
Twelve week-old male WT and Pnpla3 knockout (KO) mice fed low-fat low-sucrose (LFLS), low-fat high-sucrose (LFHS), or high-fat high-sucrose (HFHS) diet since weaning were weighed, and blood was collected in the morning following overnight ad libitum feeding (Fed), an overnight 12 h fast (Fasted), or a 24 h fast plus 12 h refeeding (Refed) (n = 6–11 mice/group). Significant main effects of diet (d), feeding status (f), and genotype (g) as well as interactions between these factors are indicated in the right-hand column. No main effects were identified for genotype for any of the above parameters. Since main effects of diet and feeding status were consistent with the appropriate metabolic responses, results of post hoc analysis for simple effects for these factors are omitted for clarity. However, results of post hoc analysis for interactions between these factors and genotype are as follows: d x g, significant interactions between diet and genotype were observed for NEFA, with NEFA tending to be higher in KO than in WT mice in the HFHS group (P = 0.061) but lower in the LFHS group; f x g, significant interactions between feeding status and genotype were observed for cholesterol with KO having higher cholesterol than WT mice in the refed group. P < 0.05, using factorial ANOVA followed by simple effects for pair-wise comparisons.
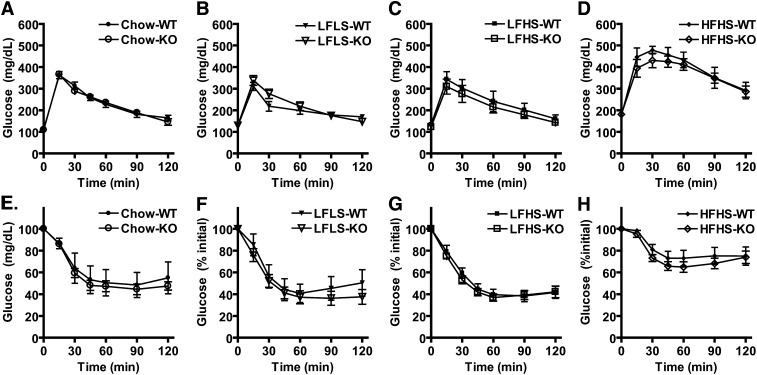
Glucose tolerance and insulin sensitivity in Pnpla3 knockout (KO) mice. (A–D) Glucose tolerance tests for WT and KO mice fed chow (A), low-fat low-sucrose (LFLS) (B), low-fat high-sucrose (LFHS) (C), or high-fat high-sucrose (HFHS) (D) diets (n = 6–11/group, male, 14 week-old). Mice were injected intraperitoneally with 1.75 g glucose per kg body weight following a 6 h fast, and plasma glucose was determined at the times indicated. (E–H) Insulin tolerance tests for WT and KO mice fed chow (E), LFLS (F), LFHS (G), or HFHS (H) diets (n-6–11/group, male, 16 week-old). Mice were injected intraperitoneally with regular human insulin at 0.80 units per kg of body weight following a 4 h fast, and plasma glucose was determined at the times indicated. No significant differences were identified.
Since Pnpla3 is positively regulated by the lipogenic nuclear transcription factor LXR (38), we additionally measured phenotypic parameters in a separate cohort of 12 week-old, 4 h-fasted, chow-fed male mice treated with the potent LXR agonist T0901317 for 6 days. This treatment regimen has previously been shown to induce hepatic Pnpla3 mRNA expression by 6- to 16-fold over the duration of treatment (38) and would therefore be expected to enhance phenotypic differences between genotype groups. As expected, T0901317 treatment increased serum cholesterol and hepatic TAG content to levels comparable to its previously reported effects under similar experimental conditions (50), thereby supporting the effectiveness of the treatment. However, again, no differences were observed between genotypes for body weight (27.27 ± 1.21 g for WT vs. 28.50 ± 0.47 g for KO), individual (data not shown) or total fat pad weight (3.80 ± 0.43% of body weight for WT vs. 3.27 ± 0.56% of body weight for KO), liver weight (7.70 ± 0.49% of body weight for WT, vs. 7.51 ± 0.32% of body weight for KO), hepatic TAG content (85.07 ± 22.39 mg/g tissue for WT vs. 89.94 ± 19.61 mg/g tissue for KO), hepatic cholesterol content (1.86 ± 0.17 mg/g tissue for WT vs. 1.85 ± 0.33 mg/g tissue for KO), blood glucose (367 ± 13 mg/dl for WT vs. 380 ± 10 mg/dl for KO), serum TAG (75.33 ± 15.33 mg/dl for WT vs. 72.00 ± 7.23 mg/dl for KO), serum NEFA (0.81 ± 0.04 mEQ/l for WT vs. 0.82 ± 0.06 mEq/l for KO), or serum cholesterol (188.79 ± 20.79 mg/dl for WT vs. 186.0 ± 15.89 mg/dl for KO). Thus, Pnpla3 deficiency does not alter systemic glucose or lipid homeostasis in response to pharmacological induction by an LXR agonist.
Global Pnpla3 deletion does not contribute to hepatic steatosis or injury
Recently, genetic variation in the human PNPLA3 gene has been strongly and repeatedly associated with fatty liver disease in humans. We therefore assessed the effect of Pnpla3 deficiency on hepatic lipid content and injury (Fig. 6). Although the expected increases in hepatic TAG content were observed for the LFHS and HFHS diet groups compared to the LFLS control group, no differences were identified for liver weight (data not shown) or liver TAG content between genotypes for any of the diet groups, whether measured biochemically (Fig. 6A) or histologically with Oil Red O staining (data not shown). Since transacylase activity of PNPLA3 could result in physiologically significant changes in TAG molecular species without a net overall change in TAG mass, we further characterized TAG molecular species using ESI-MS/MS (59, 60). However, individual TAG molecular species also did not differ between genotypes for liver (Fig. 6B) or WAT (data not shown). Serum AST (Fig. 6C) and ALT (Fig. 6D), markers of hepatic injury, also did not differ between genotypes. Given the above interactive effects on serum NEFA and cholesterol, we additionally evaluated in vivo cholesterol and fatty acid metabolism in a separate cohort of 8–9 week-old chow-fed mice using tritiated water. However, no differences in hepatic incorporation of [3H]H2O into digitonin precipitable sterols (i.e., cholesterol) (Fig. 6E) or NEFAs (Fig. 6F) were identified. Similar results were obtained for NEFA biosynthesis in WAT and BAT (Fig. 6F). These data suggest that Pnpla3 deficiency does not contribute to hepatic lipid metabolism, steatosis, or injury.
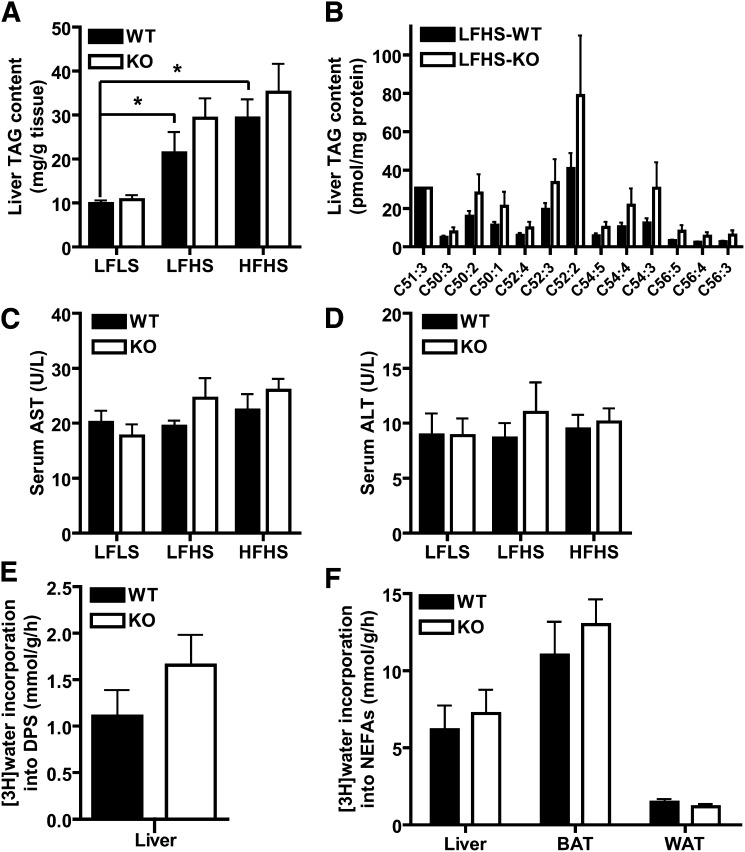
Lipid homeostasis in Pnpla3 knockout (KO) mice. (A) Liver TAG content normalized to tissue weight in WT and KO mice fed low-fat low-sucrose (LFLS), low-fat high-sucrose (LFHS), or high-fat high-sucrose (HFHS) diets (n = 6–11/group, male, 19 week-old, ad libitum-fed). (B) TAG molecular species normalized to protein content in liver of WT and KO mice fed LFHS diet (n = 7–10/group, male, 19 week-old, ad libitum-fed). TAG molecular species in liver lipid extracts were determined by electrospray ionization mass spectrometric analysis using C51:3 as an internal standard (tri 17:1). TAG species listed on the x axis are identified by the total number of carbons in the fatty acid moieties and the total number of double bonds in those fatty acids moieties (i.e., C51:3 represents a glycerol backbone esterified to three fatty acids with 17 carbons and 1 double bond each). (C–D) Serum aspartate aminotransferase (AST) (C) and alanine aminotransferase (ALT) (D) in WT and KO mice fed LFLS, LFHS, or HFHS diets (n = 6–11/group, male, 19 week-old, ad libitum-fed). (E–F) In vivo cholesterol (E) and fatty acid (F) biosynthesis in liver, BAT, and perigonadal WAT of WT and KO mice (n = 5/group, male, 8–9 week-old, chow diet). Fatty acid and cholesterol biosynthesis rates were calculated as micromoles of 3H-radioactivity from [3H]H2O incorporated into fatty acids or digitonin-precipitable sterols (DPSs), respectively, per gram of tissue per h as described in methods. * P < 0.05 by Student's t-test (for effect of diet, only comparisons in the WT group are shown).
Global Pnpla3 deletion promotes compensatory regulation of Pnpla5 mRNA but not of other genes/proteins involved in TAG metabolism
To identify genes and/or pathways that might be compensating for Pnpla3 deficiency, we evaluated mRNA expression of genes involved in TAG metabolism in liver and perigonadal WAT of ad libitum-fed WT and Pnpla3-KO mice fed chow or an LFLS, LFHS, or HFHS diet from weaning until 19 weeks of age (Fig. 7 and supplemental Table I). As expected, both diet and genotype had significant main effects on Pnpla3 mRNA expression in both WAT (Fig. 7A) and liver (Fig. 7B). The diet-induced effect was particularly dramatic in liver, where Pnpla3 mRNA expression was induced over 100-fold in LFHS-fed mice. It is worth noting that LFLS diet (61% starch, 12% maltodextrin) also induced hepatic Pnpla3 mRNA expression by 29-fold compared with chow (60% starch) and 2-fold compared with HFHS diet (25% sucrose), suggesting a positive regulatory effect of the more palatable/digestible maltodextrin carbohydrate component in the former and/or a suppressive regulatory effect of dietary fat in the latter. As expected, significant main effects of diet on mRNA expression of numerous genes involved in TAG metabolism were identified in both liver and WAT (supplemental Table 1). However, no main effects of genotype on mRNA expression of these genes were identified for either tissue. In contrast, Pnpla5, a Pnpla3 paralog with unclear function, was significantly increased in WAT of Pnpla3-KO mice compared with WT mice (Fig. 7C). Pnpla5 mRNA expression could not be detected in liver of WT or KO mice even after 60 cycles using undiluted cDNA prepared from 1 μg of RNA. Thus, Pnpla3 deficiency promotes compensatory upregulation of Pnpla5 but not of other genes involved in TAG metabolism.
DISCUSSION
Substantial data support an important role for PNPLA3 in normal metabolism and disease including the following: i) PNPLA3 shares significant homology with proteins known to play critical roles in metabolism (9, 64, 65); ii) PNPLA3 is highly regulated by important nutritional/metabolic factors (25, 27–42); iii) PNPLA3 expression is altered in obese/dysmetabolic states (25, 27–29); iv) PNPLA3 has lipid hydrolase and transacylase activities in vitro (28, 43, 44); and v) Genetic variation in PNPLA3 is associated with NAFLD in humans (10–26). Despite this strong evidence, the in vivo function and physiological relevance of PNPLA3 remain unclear. In this study, we determined the metabolic consequences of global Pnpla3 deletion in mice. We confirmed that Pnpla3 mRNA expression is indeed altered in adipose tissue and liver in response to acute and chronic nutritional challenges. Importantly, however, global Pnpla3 deletion did not affect TAG hydrolysis or influence metabolic phenotypes, including energy homeostasis, glucose homeostasis/insulin action, lipid homeostasis, or hepatic steatosis/injury. Furthermore, experimental interventions designed to increase Pnpla3 expression and enhance differences between genotypes (e.g., refeeding, high-sucrose diet, diet-induced obesity, and pharmacological activation by LXR) likewise failed to reveal any differences in the above metabolic phenotypes. It is worth noting that while this manuscript was in preparation, another group published similar data from an independently generated model of global Pnpla3 deletion and additionally failed to identify any metabolic consequences of Pnpla3 deficiency in mice challenged with a methacholine-deficient diet or leptin deficiency (66). Together these data argue against a role for Pnpla3 loss-of-function in NAFLD and the metabolic syndrome, at least in mice.
Although genotype had no main effects on phenotype overall, two noteworthy interactions were identified in our study: i) serum NEFAs were lower in LFHS-fed Pnpla3-KO mice (but tended to be higher in HFHS-fed mice); and ii) serum cholesterol was higher in refed Pnpla3-KO mice. These data suggest that Pnpla3 deficiency may influence NEFA and/or cholesterol homeostasis under certain metabolic conditions (e.g., chronic high-carbohydrate feeding and/or the postprandial state), perhaps in combination with other factors (e.g., stress/glucocorticoids, concurrent genetic/environmental influences). These findings may be particularly relevant to humans who consume increasing quantities of simple carbohydrates (particularly fructose) and who spend a significant amount of time in the postprandial state (67). Interestingly, Kollerits et al. (16) recently identified an association between the PNPLA3 rs738409 (I148M) variant and serum apolipoprotein B-containing lipoprotein concentrations in humans (16). However, numerous other studies have failed to identify an association between PNPLA3 variants and cholesterol/lipoprotein concentrations (10, 22, 23). Furthermore, in the present study, we were not able to identify differences between genotypes for NEFA or cholesterol metabolism using in vivo tritiated water methods. The lack of consistency of these findings across outcomes stresses the need for further exploration of these results. Finally, it is also worth noting that hepatic TAG content, although not statistically different, was consistently higher in Pnpla3-KO mice in our studies (Fig. 6A, B, and unpublished observations). Thus, it is possible that alternative metabolic conditions, enhanced statistical power, and/or a more homogeneous background strain could unmask previously unrecognized phenotypes in Pnpla3-KO mice.
Thus, the question remains - what is the function of PNPLA3? The regulation and subcellular localization of PNPLA3 are most consistent with a role for PNPLA3 in anabolic lipid metabolism (i.e., lipogenesis), and yet, in vitro functional data support a role in TAG hydrolysis (and/or transacylation) (43, 44). The latter could account for a metabolically neutral phenotype, but the physiological significance of such an activity is unclear. Thus far, overexpression and/or knockdown of PNPLA3 in cells (28, 35, 44) and now also global targeted deletion in mice (66) have failed to identify significant effects of PNPLA3 on metabolic phenotypes including TAG metabolism. Furthermore, no clear compensatory regulation of key enzymes involved in TAG synthesis or hydrolysis was identified in Pnpla3-KO mice. Interestingly, Pnpla5 mRNA expression was increased in WAT of Pnpla3-KO mice, thereby corroborating findings by Chen et al. (66). Pnpla5 (also known as GS2-like) is a paralog of Pnpla3. Like PNPLA3, PNPLA5 has been shown to have TAG hydrolase activity, and ectopic expression reduces incorporation of radiolabeled FFAs into TAG in HEK293 cells (28). However, another study failed to identify any TAG hydrolase activity for PNPLA5 (68). Furthermore, it is not clear that PNPLA5 is expressed at physiologically significant levels, at least in liver, of humans (65) or mice (28). Indeed, we were unable to detect Pnpla5 expression in murine liver. Likewise, the hepatic Pnpla5 expression reported by Chen et al. was also extremely low (66). Thus, it is possible that PNPLA5 may compensate for loss of PNPLA3 in WAT but not in liver in this model. Understanding the function of PNPLA5 may provide clues to the function of PNPLA3 and vice versa.
Nevertheless, numerous studies have demonstrated a strong association between genetic variation in PNPLA3 and hepatic steatosis, hepatic injury, and progression of NAFLD (10–24). The human rs738409 variant is of particular interest because it contains a C → G nucleotide change that encodes a methionine in place of isoleucine at position 148 (the I148M mutation). Computer modeling suggests that this mutation might alter substrate access to the putative catalytic serine residue in the conserved GXSXG motif (44). Indeed, both the human I148M mutation as well as a mutant in which the catalytic serine has been replaced by an alanine (S47A) lack in vitro TAG hydrolase activity (28, 44), suggesting that the I148M allele contributes to disease via loss of TAG hydrolytic function. Interestingly, adenoviral overexpression of these mutants (I148M and S47A) in murine liver enhances hepatic TAG accumulation, even in the presence of normal endogenous expression of the WT murine allele, which is not consistent with simple loss-of-function. Thus, the I148M and S47A mutations may also affect TAG metabolism by mechanisms distinct from loss of TAG hydrolytic function. For example, PNPLA3 may have additional substrates and/or functions that directly or indirectly promote TAG synthesis and that are enhanced by the above mutations. Certainly, a predominant role for PNPLA3 in lipid synthesis would be more consistent with its regulation. It remains possible that species-specific differences exist in function, expression, and/or regulation of PNPLA3. Indeed, human and rodent PNPLA3 proteins differ considerably at their C termini and exhibit different expression patterns (e.g., human PNPLA3 mRNA is more highly expressed in liver) (38). However, the impact of these differences on function and physiology remains to be determined.
In summary, our data provide evidence against a critical role for PNPLA3 in murine TAG hydrolysis and also provide further important corroborative evidence against a role for Pnpla3 deficieny/loss-of-function in the pathogenesis of NAFLD and the metabolic syndrome in mice. It is important to note, however, that our data do not rule out a potential contribution of PNPLA3 gain-of-function or of specific genetic PNPLA3 variants (i.e., the rs738409 I148M mutant) to lipid metabolism or metabolic disease in humans. Data connecting genetic variation in PNPLA3 to human disease are extremely compelling and clearly warrant continued efforts to understand the precise function and physiological relevance of this gene, which, unfortunately, remain elusive nearly a decade after its initial identification. Purification and systematic analysis of its enzymatic activities and substrate specificity in vitro are necessary to more clearly characterize its function but are challenging given the numerous potential substrates/activities and inherent differences between in vitro and in vivo systems. Thus, these studies should also be complemented by further in vivo metabolic characterization of mice with targeted genetic “knock-in” of the I148M allele as well as overexpression of WT and/or PNPLA3 mutant alleles in metabolically-relevant tissues. Such studies will be crucial for determining whether therapeutic manipulation of PNPLA3 might be useful in the prevention and/or treatment of NAFLD and related metabolic disorders.
Acknowledgments
The authors thank Jeffrey S. Flier, in whose laboratory the project was initiated.
Footnotes
Abbreviations:
- ALT
- alanine aminotransferase
- AST
- aspartate aminotransferase
- BAC
- bacterial artificial chromosome
- BAT
- brown adipose tissue
- DPSs
- digitonin-precipitable sterols
- ES cell
- embryonic stem cell
- GTT
- glucose tolerance test
- HF
- high-fat
- HFHS
- high-fat high-sucrose diet
- ITT
- insulin tolerance test
- HSL
- hormone sensitive lipase
- iPLA2
- calcium-independent phospholipase A2
- KO
- knockout
- LFHS
- low-fat high-sucrose diet
- LFLS
- low-fat low-sucrose diet
- LXR
- liver X receptor
- RER
- respiratory exchange ratio
- MAT
- mesenteric adipose tissue
- NAFLD
- nonalcoholic fatty liver disease
- Pgat
- perigonadal adipose tissue
- Pnpla3 (PNPLA3, Pnpla3)
- patatin-like phospholipase domain containing 3
- SCAT
- subcutaneous adipose tissue
- TAG
- triacylglycerol
- WAT
- white adipose tissue
- WT
- wild-type
This work was supported by National Institutes of Health Grant P30 DK-036836 Joslin Diabetes and Endocrinology Research Center Pilot and Feasibility Award, Howard Hughes Medical Institute Physician-Scientist Early Career Award, and University of Pittsburgh Department of Medicine Junior Scholar Award (E.E.K.); National Institutes of Health Grants P01 HL-020948; and RL1 HL-092550 (J.Z.L.); and National Institutes of Health Grant P01 HL-057278 (R. G.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. Work was also supported by an Austrian Ministry of Science and Research GEN-AU GOLD (Genomics of Lipid-associated Disorders) Grant, an F30 (SFB LIPOTOX) Grant, and Wittgenstein Award Z136 (R. Z.).
[S]The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table.
REFERENCES
Articles from Journal of Lipid Research are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1194/jlr.m011205
Read article for free, from open access legal sources, via Unpaywall:
http://www.jlr.org/article/S0022227520405279/pdf
Open access at www.jlr.org
http://www.jlr.org/cgi/content/abstract/52/2/318
Open access at www.jlr.org
http://www.jlr.org/cgi/reprint/52/2/318.pdf
Open access at www.jlr.org
http://www.jlr.org/cgi/content/full/52/2/318
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1194/jlr.m011205
Article citations
Team players in the pathogenesis of metabolic dysfunctions-associated steatotic liver disease: The basis of development of pharmacotherapy.
World J Gastrointest Pathophysiol, 15(4):93606, 01 Aug 2024
Cited by: 0 articles | PMID: 39220834 | PMCID: PMC11362842
Review Free full text in Europe PMC
Gut microbes, diet, and genetics as drivers of metabolic liver disease: a narrative review outlining implications for precision medicine.
J Nutr Biochem, 133:109704, 17 Jul 2024
Cited by: 1 article | PMID: 39029595
Review
A PNPLA3-Deficient iPSC-Derived Hepatocyte Screen Identifies Pathways to Potentially Reduce Steatosis in Metabolic Dysfunction-Associated Fatty Liver Disease.
Int J Mol Sci, 25(13):7277, 02 Jul 2024
Cited by: 0 articles | PMID: 39000384
PNPLA3 is a triglyceride lipase that mobilizes polyunsaturated fatty acids to facilitate hepatic secretion of large-sized very low-density lipoprotein.
Nat Commun, 15(1):4847, 06 Jun 2024
Cited by: 1 article | PMID: 38844467
The Role of PNPLA3_rs738409 Gene Variant, Lifestyle Factors, and Bioactive Compounds in Nonalcoholic Fatty Liver Disease: A Population-Based and Molecular Approach towards Healthy Nutrition.
Nutrients, 16(8):1239, 21 Apr 2024
Cited by: 2 articles | PMID: 38674929 | PMCID: PMC11054963
Go to all (155) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs
- (1 citation) dbSNP - rs738409
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease.
Hepatology, 52(3):1134-1142, 01 Sep 2010
Cited by: 153 articles | PMID: 20648554 | PMCID: PMC2932863
Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis.
J Clin Invest, 122(11):4130-4144, 01 Nov 2012
Cited by: 166 articles | PMID: 23023705 | PMCID: PMC3484461
Shifts in dietary carbohydrate-lipid exposure regulate expression of the non-alcoholic fatty liver disease-associated gene PNPLA3/adiponutrin in mouse liver and HepG2 human liver cells.
Metabolism, 63(10):1352-1362, 21 Jun 2014
Cited by: 20 articles | PMID: 25060692 | PMCID: PMC4175036
Unravelling the pathogenesis of fatty liver disease: patatin-like phospholipase domain-containing 3 protein.
Curr Opin Lipidol, 21(3):247-252, 01 Jun 2010
Cited by: 55 articles | PMID: 20480550
Review
Funding
Funders who supported this work.
Austrian Science Fund FWF (4)
Lipotoxicity: Lipid-induced Cell Dysfunction and Cell Death
Prof Rudolf Zechner, University of Graz
Grant ID: F 3001
Lipases and lipotoxicity
Prof Rudolf Zechner, University of Graz
Grant ID: F 3002
Molecular Enzymology: Stucture, Function and Biotechnological Exploitation of Enzymes
Univ.Prof. Dr. Ellen ZECHNER, University of Graz
Grant ID: W 901
Metabolic lipases in lipid and energy metabolism
Prof Rudolf Zechner, University of Graz
Grant ID: Z 136
Howard Hughes Medical Institute
NHLBI NIH HHS (6)
Grant ID: P01 HL-020948
Grant ID: P01 HL-057278
Grant ID: P01 HL057278
Grant ID: RL1 HL-092550
Grant ID: P01 HL020948
Grant ID: RL1 HL092550
NIDDK NIH HHS (2)
Grant ID: P30 DK036836
Grant ID: P30-DK-036836





