Abstract
Free full text

Distinct genetic regions modify specific muscle groups in muscular dystrophy
Abstract
Phenotypic expression in the muscular dystrophies is variable, even with the identical mutation, providing strong evidence that genetic modifiers influence outcome. To identify genetic modifier loci, we used quantitative trait locus mapping in two differentially affected mouse strains with muscular dystrophy. Using the Sgcg model of limb girdle muscular dystrophy that lacks the dystrophin-associated protein γ-sarcoglycan, we evaluated chromosomal regions that segregated with two distinct quantifiable characteristics of muscular dystrophy, membrane permeability and fibrosis. We previously identified a single major locus on murine chromosome 7 that influences both traits of membrane permeability and fibrosis in the quadriceps muscle. Using a larger cohort, we now found that this same interval strongly associated with both traits in all limb skeletal muscle groups studied, including the gastrocnemius/soleus, gluteus/hamstring, and triceps muscles. In contrast, the muscles of the trunk were modified by distinct genetic loci, possibly reflecting the embryological origins and physiological stressors unique to these muscle groups. A locus on chromosome 18 was identified that modified membrane permeability of the abdominal muscles, and a locus on chromosome 3 was found that regulated diaphragm and abdominal muscle fibrosis. Fibrosis in the heart associated with a region on chromosome 9 and likely reflects differential function between cardiac and skeletal muscle. These data underscore the complexity of inheritance and penetrance of single-gene disorders.
the muscular dystrophies are genetically heterogeneous. Mutations in the genes encoding dystrophin and the sarcoglycan genes disrupt the dystrophin protein complex and lead to muscular dystrophy and cardiomyopathy. Dystrophin, together with its associated proteins, underlies the membrane of all muscles, providing mechanical support during muscle contraction (4, 16, 21). The complex includes dystrophin, the sarcoglycans, including α-, β-, γ-, and δ-, as well as a number of other transmembrane and cytoplasmic components (28). Mutations in the dystrophin gene lead to Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD), while mutations in the genes encoding the sarcoglycan subcomplex result in the recessively inherited limb girdle muscular dystrophies (LGMD). The dystrophin complex provides mechanical support to the plasma membrane of contracting muscle. Mutations that disrupt the complex result in instability of the muscle fibers. Myofiber damage leads to necrosis and subsequent muscle degeneration. Eventually, skeletal and cardiac muscle fibers are replaced by adipose and connective tissue. This leads to muscle weakness, a hallmark of muscular dystrophy, as well as cardiomyopathy and heart failure.
The muscular dystrophies are characterized by phenotypic variability. Disease factors such as age of onset, disease progression, muscle group involvement, and accompanying cardiac and central nervous system impairment, as well as the clinical marker serum creatine kinase (CK), cannot be predicted by the specific gene mutation. Although allelic heterogeneity accounts for some of the phenotypic range, the identical mutation in different individuals can manifest with a broad array and timing of symptoms. That family members with the same mutation may display phenotypic variability suggests that additional environmental and genetic factors interact with the primary mutation to determine outcome (26).
In this study, we used the Sgcg mouse model to map genetic modifiers. We selected this model because the most common Sgcg mutation in humans, c.525delT, is a null mutation that displays a phenotypic range from mild to severe (13, 18). Environmental variation is not thought to account for all of the variability seen between or within families (13). The Sgcg mouse model was engineered by eliminating exon 2, which encodes the initiator methionine and the transmembrane domain. This mutation results in a complete absence of γ-sarcoglycan protein at the plasma membrane, as well as secondary reduction of the remaining sarcoglycan subunits (7). Sgcg mice display a reduced life span and develop cardiomyopathy (7). A second independent model using the same gene targeting strategy for the γ-sarcoglycan gene results in mice with less lethality and more hypertrophy of the muscle (22). This finding indicates that additional genetic loci influence outcome.
The Sgcg mutation was previously bred into four distinct inbred strains, resulting in a suppressed or enhanced phenotype (10). In the 129T2/SvEmsJ (129) background, Sgcg displayed a milder phenotype than was seen in the DBA/2J (D2) background (10). With the use of the mild and severely affected strains, an F2 D2/129 Sgcg (SgcgD2/129) cohort was used in a genomewide scan of disease severity in an effort to identify genetic modifiers. Phenotypic severity was determined with two quantifiable methods. The first marker of phenotypic severity in muscular dystrophy is disruption of muscle membrane integrity that is monitored with the vital tracer Evans blue dye. Membrane damage is an early marker of disease progression, and normal muscle tissue is not permeable to the dye. Dye uptake can readily be detected and quantified in Sgcg mice, and the level of dye uptake reflects the degree of membrane fragility that arises from disruption of the dystrophin complex in muscle. The second phenotype, fibrosis, is also a marker of disease progression and is determined by the degree of hydroxyproline (HOP) deposition in muscle. HOP is a modified amino acid found in collagen, a major component of fibrosis, and more severely affected mice display more significant fibrosis.
A genomewide quantitative trait locus (QTL) analysis of these traits previously identified a single major locus on chromosome 7 (9). This analysis was conducted in F2 SgcgD2/129 quadriceps muscle, a representative limb-based skeletal muscle, and the single locus on chromosome 7 modified both traits of dye uptake and fibrosis (9). This result was unanticipated since it was expected that membrane leak and fibrosis employed distinct biological processes and therefore would be altered by distinct genetic loci. This prior analysis was conducted with a relatively small cohort of 80 animals and focused only on the quadriceps muscle. Further analysis of the chromosome 7 region, referred to as DMOD1 (dystrophic modifier), uncovered an insertion/deletion polymorphism in the Ltbp4 gene (9).
Limb-based skeletal muscles, respiratory muscle, and cardiac muscle are all significantly diseased in muscular dystrophy mediated by dystrophin and sarcoglycan gene mutations. We now extended our previous study by conducting a genomewide analysis using a larger cohort of F2 SgcgD2/129 mice. We found that the chromosome 7 interval that harbors Ltbp4 accounted for a significant degree of the variation in all limb-based skeletal muscles examined. However, when trunk musculature was evaluated, such as the diaphragm and abdominal muscles, other genomic intervals emerged that regulated the severity of fibrosis and membrane leak. Cardiac fibrosis harbors its own modifier genetic loci. The regulation of distinct muscle groups in muscular dystrophy by specific genetic regions indicates that unique pathways may be differentially active in these muscles.
METHODS
Animals.
The mice used in this study were described previously (7, 9, 10). Exon 2 of the Sgcg gene was deleted in the Sgcg mice, and these mice were backcrossed to wild-type DBA 2J JAX (D2) or 129/SVEMS+/J JAX (129) mice over 10 generations. Heterozygous Sgcg+/− mice were used for breeding, and after 10 generations of backcrossing heterozygotes were interbred to create homozygous Sgcg mice on the D2 background or on the 129 background. SgcgD2 mice and Sgcg129 mice were interbred to create SgcgD2/129. The animals used for genotyping were from the F2 generation. Both males and females were used, since we previously have not detected differences for these traits between male and female mice (10, 11). All animals were housed uniformly in a single pathogen-free barrier facility and treated under the approval of the University of Chicago Animal Care and Use Committee. All animals were studied at 8 wk of age.
Quantitative dye uptake.
The Evans blue dye uptake assay was performed as described previously (10, 25). Mice were injected intraperitoneally with 5 μl/g body mass Evans blue dye (Sigma, E-2129) in phosphate-buffered saline at 10 mg/ml 40 h before death. Tissues were dissected, minced, weighed, and incubated in 1 ml of formamide for 2 h at 55°C. The dye uptake assay was performed on half the abdominal muscle group and half of each quadriceps muscle. The other halves of these muscles were subjected to HOP measurements (see below). The entire gastrocnemius/soleus, gluteus/hamstrings, and triceps were used in this assay because of their smaller size and the inability to bisect these muscle groups for both assays. Absorbance was measured at 620 nm, and results are reported as the Z score of square root-transformed absorbance per milligram of tissue. Square root transformation was used to normalize the phenotype distributions for QTL analysis.
Hydroxyproline measurement.
The HOP assay was performed as described previously (5, 10). Muscle was dissected, minced, weighed, and hydrolyzed in 2 ml of 6 M hydrochloric acid at 110°C overnight. After addition of Ehrlich's reagent, absorbance of the supernatant was measured at 558 nm. The cardiac ventricles were assayed after removal of the atrial and valvular structures. Results for the quadriceps, abdominal, and diaphragm muscles were reported as the Z score of log-transformed nanomoles per liter of HOP per milligram of tissue. Heart HOP was reported as the Z score of nanomoles per liter of HOP per milligram of tissue. Log transformation was used to normalize the phenotype distributions for QTL analysis. Neither square root nor log transformation normalized the distribution of heart HOP data. Tissues were harvested from the same animals used for the dye uptake assay and frozen at −80°C until use. The HOP assay was performed on a portion of the abdominal and quadriceps muscles and on the entire diaphragm muscle and cardiac ventricles, both right and left.
Genetic and statistical analysis.
Autosomal single nucleotide polymorphisms (SNPs) (n = 330) informative between the parental D2 and 129 strains were genotyped in 282 F2-SgcgD2/129 animals on the Illumina GoldenGate platform. J/qtl 1.3.0 software (http://churchill.jax.org/software/jqtl.shtml) was used to perform whole genome QTL mapping for each of the membrane permeability and fibrosis phenotypes. Each phenotype was transformed to normalize the distribution of the data. Neither square root nor log transformation normalized the heart HOP data, so nonparametric statistics were used instead. For each scan of the transformed data values, a normal model and the EM algorithm were used. Significance thresholds were determined by 1,000 permutation tests. A nonparametric model, with default settings, was used for heart HOP. The 95% Bayes credible interval was calculated in J/qtl. The percent variance explained was calculated with the fit.qtl command in J/qtl. Tests of normality and other statistics were calculated in Prism. Build 37.1 was used for genomic analysis.
RESULTS
Modifiers of muscle membrane permeability.
Evans blue dye is a small vital tracer that complexes with albumin. Normal cells, including muscle cells, are impermeable to the vital tracer Evans blue dye unless injury is present to disrupt the plasma membrane barrier. In muscular dystrophy, as in other injury states, the dye couples to albumin and is retained by myofibers and cardiomyocytes (24). Typically, in muscular dystrophy, dye uptake is seen as clusters of dye-positive fibers within the muscle or myocardium. Because of this patchy, nonuniform pattern, an accurate measurement of dye uptake should assess the entire muscle. Sgcg mice are a model of muscular dystrophy, and Sgcg mice on a mixed DBA/2J and 129/SVEMS+/J background were used in this study. As with dystrophin or sarcoglycan gene mutations, dye uptake can be detected readily in Sgcg mice and reflects the degree of muscle membrane leakiness or fragility. A second assay measured HOP content as an indicator of fibrosis or scarring. Given the requirements of these assays, both dye uptake and HOP measurements could not be performed on the same muscle group. Thus we collected half of the quadriceps muscles and the entire gastrocnemius/soleus, gluteus/hamstrings, and triceps for measurement of dye uptake. The first three muscle groups are found in the hindlimb and are subject to heavy use with daily activities, even in a cage-bound animal. As with most muscle groups in the mouse, these muscles are primarily composed of fast fibers, with the exception of the soleus muscle. The amount of dye taken up into these muscles was quantified with a spectrophotometric assay that allows assessment of uptake into the entire muscle and avoids the bias that can be associated with histological examination.
Whole genome SNP analysis was used to map loci that segregate with mild or severe membrane permeability. The analysis was performed on 281 F2-Sgcg animals and utilized 330 SNPs informative between the parental D2 and 129 strains. A single major modifier locus influencing membrane permeability was identified on chromosome 7 (Fig. 1A). For quadriceps muscle, this locus accounted for >37% of the phenotypic variability of dye uptake. We previously reported this locus, using a smaller cohort of 80 animals, and found an insertion/deletion in the Ltbp4 gene within this interval (9). Using this cohort plus additional animals, we now found that the same locus influences membrane permeability in all limb-based skeletal muscles examined, including the gastrocnemius/soleus, gluteus/hamstring, and triceps muscle groups (Fig. 1). The approximate 95% confidence intervals for these overlapping loci are as follows: 27.10065–29.10065 Mb in quadriceps, 21.10065–33.10066 Mb in gastrocnemius/soleus, 15.10065–28.01074 Mb in gluteus/hamstrings, and 19.10065–35.10066 Mb in triceps. In quadriceps muscle, the peak logarithm of odds (LOD) score for this locus was 25.94; it was 13.52 as reported in the previous study using the smaller cohort (9). Peak LOD scores for the gastrocnemius/soleus, gluteus/hamstrings, and triceps were 10.24, 8.60, and 6.12, respectively. We previously referred to this interval as DMOD1 (dystrophic modifier). No other peaks reached significance in these four muscles.
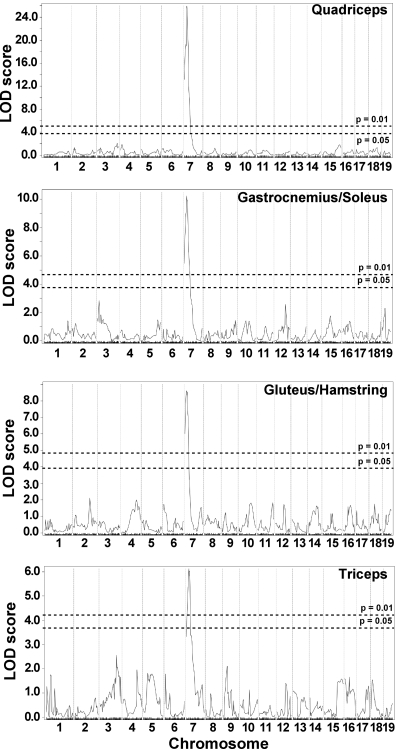
Membrane permeability in limb-based skeletal muscle is modified by a single major locus on chromosome 7. Genomewide association scans were performed correlating membrane permeability in Sgcg mice, a model of muscular dystrophy. Sgcg mice on a mixed genetic background of DBA/2J and 129/SVEMS+/J were used for mapping. Membrane permeability was measured as Evans blue dye uptake into individual muscle groups. A single locus on chromosome 7, DMOD1, regulates dye uptake in quadriceps, gastrocnemius/soleus, gluteus/hamstring, and triceps muscles. Autosomal chromosomes and the 330 informative single nucleotide polymorphism (SNP) markers used are plotted on the x-axis; logarithm of odds (LOD) scores are represented on the y-axis. The same region on chromosome 7 was tightly associated with membrane permeability in all 4 muscles, with peak LOD scores of 25.94 in quadriceps, 10.24 in gastrocnemius/soleus, 8.60 in gluteus/hamstring, and 6.12 in triceps. No other regions of the genome reached significance.
In contrast to the limb-based skeletal muscles, the abdominal muscle group did not show significant evidence for being modified by the chromosome 7 DMOD1 interval (Fig. 2). Instead, dye uptake in the abdominal muscles was associated with a locus on chromosome 18 with a peak LOD score of 5.10 and a 95% confidence interval from 6.602274 to 17.516539 Mb. This region, termed DMOD2, was not identified in the limb-based skeletal muscle and may reflect the unique function and/or distinct developmental origins of the abdominal muscles compared with the limb-based skeletal muscles of the fore- and hindlimbs.
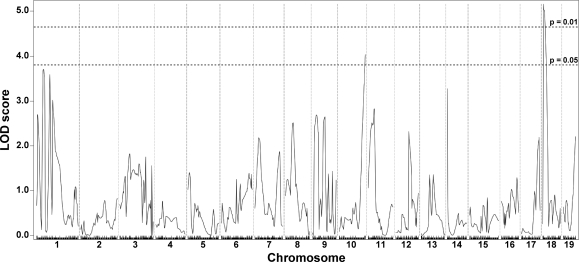
Membrane permeability in the abdominal muscles is modified by a locus on chromosome 18, DMOD2. A genomewide association scan for abdominal dye uptake was performed with data from 281 SgcgD2/129 F2 progeny. Autosomal chromosomes and the 330 informative SNP markers used are plotted on the x-axis; LOD scores are represented on the y-axis. A single region on chromosome 18 showed significant association with Evans blue dye content in abdominal muscle, with a peak LOD score of 5.16.
Modifiers of fibrosis in quadriceps, diaphragm, and abdominal muscle.
The second quantifiable trait of muscular dystrophy that we measured was fibrosis. Fibrosis is an increase in extracellular matrix deposition and is reflected by an increase in collagen content. HOP is a measure of collagen content, and by performing this assay on entire muscles we avoided the sampling error that can arise from evaluating histopathology sections of muscle. The HOP assay was performed with half the quadriceps and abdominal muscles, representative of limb-based and trunk-based skeletal muscle, respectively, since these muscle groups could be bisected in an unbiased manner (n = 270). Given the small size of the trunk-based diaphragm muscle, the entire diaphragm muscle was assayed for HOP content (n = 272). For the quadriceps muscle, we found evidence supporting the previously reported DMOD1 locus on chromosome 7 that contains Ltbp4 (9) (Fig. 3). In contrast to the limb-based skeletal muscle, DMOD1 on chromosome 7 did not emerge as a QTL for fibrosis in the diaphragm and abdominal muscles. Instead, the diaphragm and abdominal muscles share an overlapping interval on chromosome 3, DMOD3, that influences fibrosis (Fig. 4). In diaphragm muscle, the DMOD3 interval extends from ~109.15 to 115.7 Mb with a peak LOD of 6.55. In the abdominal muscles, the interval extends from ~109.1472 to 117.7711 Mb with a peak LOD of 4.39. A second locus influencing fibrosis in the abdominal muscles was identified on chromosome 10, DMOD4, with a peak LOD of 3.71. These results suggest that fibrosis in the trunk muscles is regulated by a genetic program distinct from the limb-based muscles. Also, the genomic loci that regulate abdominal muscle fibrosis differ from those that regulate dye uptake, unlike what was observed for the quadriceps muscle.
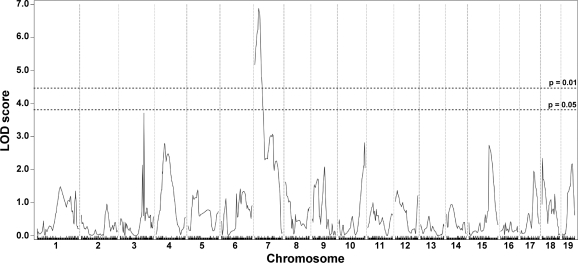
Fibrosis in quadriceps muscles is modified by DMOD1 on chromosome 7. A genomewide association scan for quadriceps collagen content was carried out with 270 SgcgD2/129 F2 progeny. Hydroxyproline (HOP) content was determined as a measure of collagen content. Autosomal chromosomes and the 330 informative SNP markers used are plotted on the x-axis; LOD scores are represented on the y-axis. A region on chromosome 7, DMOD1, showed highly significant association with HOP content in quadriceps, with a peak LOD score of 6.89.
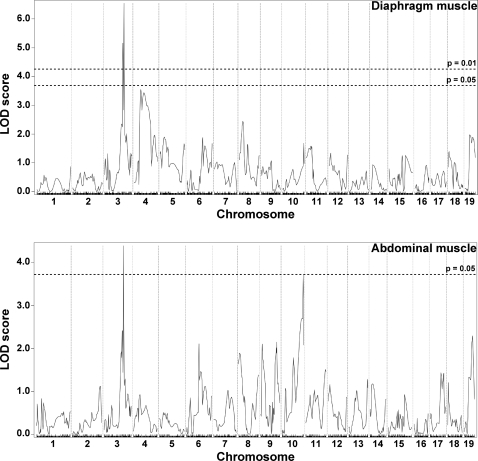
Fibrosis in the diaphragm and abdominal muscles is modified by a locus on chromosome 3, DMOD3. Genomewide association scans for fibrosis in SgcgD2/129 F2 progeny were performed in the diaphragm (n = 272) and abdominal (n = 270) muscles. Autosomal chromosomes and the 330 informative SNP markers used are plotted on the x-axis; LOD scores are represented on the y-axis. Overlapping regions on chromosome 3 showed highly significant association with fibrosis in diaphragm and in the abdominal muscles. The peak LOD score in the diaphragm interval was 6.55; the peak LOD score for the abdominal interval was 4.39. An additional locus, DMOD4, contributing to abdominal HOP content was identified on chromosome 10 with a peak LOD of 3.71.
Fibrosis in cardiac muscle.
To evaluate the cardiac muscle, both atria and the valvular structures were removed and the HOP assay was performed on the left and right ventricles as a single unit (n = 265). Although there is dye uptake in the heart in this and other models of muscular dystrophy, the heart cannot be bisected in an unbiased manner. Therefore, we only analyzed fibrosis in heart ventricles. Unlike the other data sets, the data for cardiac ventricle HOP could not be normalized with square root or log transformation since a relatively small number of individual hearts had a large degree of fibrosis leading to a nonnormal distribution of data. With nonparametric analysis, a modifier locus, DMOD5, influencing fibrosis was identified on chromosome 9 (Fig. 5). The approximate 95% confidence interval for this locus is 16.53485–36.88701 Mb, with a peak LOD score of 4.12. A summary of the dystrophic modifier loci for each trait and the regions linked to these loci is presented in Tables 1 and and22.
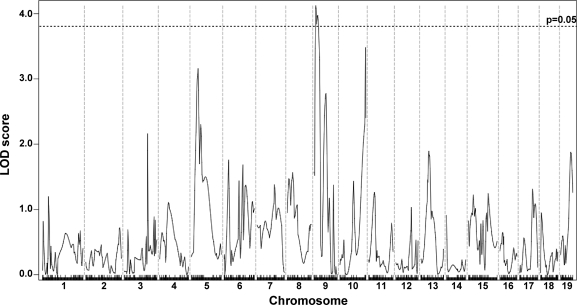
Fibrosis in cardiac muscle is modified by a locus on chromosome 9, DMOD5. A genomewide linkage scan for heart ventricle HOP content in 265 SgcgD2/129 F2 progeny was performed. Autosomal chromosomes and the 330 informative SNP markers used are plotted on the x-axis; LOD scores are represented on the y-axis. An ~20-Mb region on chromosome 9 showed significant linkage to HOP content in the heart with a peak LOD score of 4.12.
Table 1.
Modifier loci identified for membrane permeability and fibrosis in multiple muscle groups in muscular dystrophy
| Muscle Group | Membrane Permeability | Fibrosis |
|---|---|---|
| Limb muscles | ||
    Quadriceps Quadriceps | DMOD1 | DMOD1 |
    Gastrocnemius/soleus Gastrocnemius/soleus | DMOD1 | nd |
    Gluteus/hamstring Gluteus/hamstring | DMOD1 | nd |
    Triceps Triceps | DMOD1 | nd |
| Diaphragm | nd | DMOD3 |
| Abdominal | DMOD2 | DMOD3, DMOD4 |
| Heart ventricle | nd | DMOD5 |
nd, Not done.
Table 2.
Summary of modifier loci
| Modifier Locus | Chromosome | 95% Confidence Interval |
|---|---|---|
| DMOD1 | 7 | 27.10065–29.10065 Mb* |
| rs3724397–rs13479208 | ||
| DMOD2 | 18 | 6.6.2274–17.516539 Mb |
| rs6312389–rs3662641 | ||
| DMOD3 | 3 | 109.1472–117.7711 Mb |
| rs13477320–rs3718783 | ||
| DMOD4 | 10 | 69.15556–127.613 Mb |
| rs-6326263–rs3676330 | ||
| DMOD5 | 9 | 16.53485–36.88701 Mb |
| rs13480073–rs13480142 |
Relationship of phenotypes across muscle groups.
This QTL analysis identified unique loci for limb-based skeletal muscle versus trunk-based skeletal muscle. Because of this we examined the correlation between the phenotypes in these muscles using the primary data. Dye uptake in the quadriceps muscle was significantly positively correlated with dye uptake in the abdominal muscles as well as each of the limb-based skeletal muscles (Table 3). This indicates that high dye uptake in the dystrophic quadriceps muscle is a general indicator of higher dye uptake throughout the musculature. However, despite significantly nonzero correlations for each muscle, the phenotypes were not highly predictive in individual animals. This is especially apparent in the abdominal muscles, which show the least correlation with dye uptake in the quadriceps muscle. These data, as well as other comparisons made between limb-based skeletal muscles and abdominal muscles, suggest that, with some exceptions, the limb-based skeletal muscles have dye uptake more similar to each other than to the abdominal muscles. In light of these findings together with the differences seen in the QTL analysis, the abdominal muscles emerge as having additional, or potentially different, mechanisms contributing to the membrane permeability trait seen in muscular dystrophy.
Table 3.
Correlation of dye uptake in each muscle group with dye uptake in quadriceps muscle
| Muscle | r2 | Slope | P Value |
|---|---|---|---|
| Abdominal | 0.0407 | 0.1186 ± 0.03449 | 0.0007 |
| Gastrocnemius/soleus | 0.1728 | 0.3381 ± 0.04429 | <0.0001 |
| Gluteus/hamstring | 0.1389 | 0.2037 ± 0.03036 | <0.0001 |
| Triceps | 0.1172 | 0.2486 ± 0.04086 | <0.0001 |
In the analysis of fibrosis, HOP contents in the quadriceps, abdominal, and diaphragm muscles and heart were compared (Table 4). Again, HOP content in each muscle was significantly positively correlated with quadriceps muscle HOP, but the muscles varied in their degree of correlation. Specifically, heart ventricle HOP showed the least correlation with quadriceps muscle compared with abdominal muscle and diaphragm muscle.
Table 4.
Correlation of hydroxyproline content in each muscle group with quadriceps hydroxyproline content
| Muscle | r2 | Slope | P Value |
|---|---|---|---|
| Abdominal | 0.2245 | 0.3177 ± 0.03620 | <0.0001 |
| Diaphragm | 0.2500 | 0.4177 ± 0.04420 | <0.0001 |
| Heart | 0.1110 | 0.3735 ± 0.06517 | <0.0001 |
DISCUSSION
Multiple modifier loci for muscular dystrophy.
In humans with muscular dystrophy, the onset of disease is usually first noted as muscle weakness. Despite the fact that >30 different genes have been identified that cause muscular dystrophy, most patients now receive a genetic diagnosis to explain their weakness (19). The specific genetic diagnosis is highly meaningful in that it predicts the likely tempo of disease and also predicts whether there is likely to be accompanying cardiac muscle disease or extramuscle findings. Despite this success in genetic testing, often the identical gene mutation is associated with a range of phenotypic outcome. The specific genetic diagnosis often cannot predict the timing at which ambulation will be lost and which muscle groups will be most affected. Because there is a single common Sgcg mutation that causes LGMD type 2C (LGMD2C), this inheritance eliminates allelic heterogeneity, and the phenotypic range with this one mutation argues strongly for the presence of genetic modifiers (13, 18). Such variability is also seen with dystrophin gene mutations, where brothers often have a distinct disease course.
We conducted a study to map genetic modifiers since the identification of such modifier genes may point to new pathways that can regulate outcome. Our study was conducted with a mouse model of LGMD2C, Sgcg mice. Murine models allow control of environmental variables such as diet and housing conditions. Whether these same loci will also affect humans with LGMD2C or other forms of muscular dystrophy remains to be shown. There are inherent differences in muscle function between humans and mice given the bipedal nature of humans. We evaluated both fore- and hindlimb muscle groups as well as muscles of the trunk since we expect that many of these functions, and therefore modifiers, will be conserved in humans with muscular dystrophy. Additionally, since the model of muscular dystrophy we used in our study derives its phenotype from disruption of the dystrophin complex, it is possible that these modifiers act only on dystrophin- or even sarcoglycan-mutant forms of muscular dystrophy. This may be especially true for the trait of membrane leakage, since the dystrophin complex directly mediates plasma membrane stability in muscle.
Dye uptake and fibrosis in limb-based skeletal muscle is primarily modified by DMOD1.
We found that the muscles of the limbs, including muscle groups in the forelimbs and hindlimbs, were significantly modified by a single locus on chromosome 7. We previously reported this locus as containing an insertion/deletion polymorphism in Ltbp4 encoding a transforming growth factor (TGF)-β binding protein (9). Our present data suggest that this accounts for 9–37% of the variance, depending on the limb muscle group evaluated. The DMOD1 locus on chromosome 7 associates with both features of muscular dystrophy, membrane leakiness measured by dye uptake and fibrosis measured as collagen deposition or HOP content. The finding that both these traits are linked to the same locus is surprising. Membrane leakiness is thought to be a reflection of the inherent instability of the muscle membrane in the absence of a normal dystrophin complex (24). That the Ltbp4 gene can influence this property was unanticipated and suggests that there could be an interaction between this extracellular protein and membrane components of the dystrophin complex and potentially identifies a novel function for TGF-β.
Trunk-based muscles are modified by alternative loci.
Limb-based muscles are subject to repetitive exercise in voluntary ambulation. Physiologically, such muscles are quiescent or inactive until recruited by motoneurons into action. The sudden movement of a previously inactive muscle may actually render more damage, and therefore DMOD1 may specifically be activated in response to this physiology. In contrast to the finding of membrane permeability in limb-based skeletal muscle, we found that the abdominal wall musculature was modified by a region on chromosome 18 called DMOD2. Although this region is very large and contains many genes, several genes of interest fall within the interval. This region contains Lama3, a gene encoding an extracellular matrix protein. Lama2 encodes a protein known to interact directly with the dystrophin complex, and mutations in Lama2 lead to muscular dystrophy (8, 29). Aqp4 encodes aquaporin 4, a protein noted to be reduced in multiple forms of muscular dystrophy (1, 6). Also in this region is Mib1, the mammalian ortholog of Drosophila Mib2, a gene linked to muscle growth (3). The physiology of posture maintenance results in a distinct activity pattern, one in which there is slower and more continual recruitment of muscle groups. The innervation of these muscles is also partly under involuntary control. Therefore, the use and innervation patterns could account for the presence of distinct genetic modifiers for limb versus axial musculature. However, it should be noted that there are developmental differences between limb- and trunk-based muscles that also could account for these findings (20). The development of limb-based muscles is from the ventral lateral region of the somite. Here muscle regulatory factors such as MyoD play a key role in promoting myogenesis. In contrast, the dorsal medial aspect of the somite gives rise to the epaxial muscles including the paraspinal, intercostal, and abdominal muscles. This origin is heavily influenced by additional muscle regulatory factors such as Myf5. Similarly, we noted that diaphragm and abdominal muscles fibrosis was modified by a locus on chromosome 3 called DMOD3. Within this locus is Ntng1; the netrin pathways were previously suggested to modify the dystrophin complex function in a Drosophila-based screen (15).
A novel locus for cardiac muscle fibrosis in muscular dystrophy.
Cardiac fibrosis in the ventricles was also modified by a unique locus, DMOD5, on chromosome 9. This region contains a number of potentially important genes including Dnm2 encoding dynamin-2, a gene linked to centronuclear myopathy (2). Adamts8, a disintegrin and metallopeptidase, is also in this interval; ADAM12 overexpression protects against muscular dystrophy (14). It is noteworthy the DMOD1/Ltbp4 interval did not emerge as a significant modifier for cardiac fibrosis given the high expression of Ltbp4 in the heart and its association with cardiomyopathy (12, 23). We also previously noted (9) that SgcgD2 mice had elevated TGF-β signaling within the heart, indicating that that DMOD1/Ltbp4 likely does influence cardiac fibrosis. Future analysis will need to genetically separate these modifier loci to determine their independent effects.
Le Corvoisier and colleagues (17) mapped modifiers of heart failure, using a calsequestrin-expressing transgenic model of cardiomyopathy, and this study included the DBA strain as one of the background strains studied. With this approach, seven different loci were identified and one was identified as Tnni3K, a gene encoding cardiac troponin I interacting kinase (27). This gene was implicated in several traits linked to cardiac function such as fractional shortening and ventricular diameter. These data fit well with the observation that Tnni3K is a gene expressed in cardiomyocytes. Genetic modifier loci identified in the present study may encode genes expressed in fibroblasts. This may point to fibroblast function and suggest that fibroblast gene expression may differ for different organs or even unique muscle groups. For example, fibroblasts from the heart may have distinct properties of proliferation and gene expression. The previous identification of Ltbp4 as a modifier of fibrosis reflects its expression in both myofibers and fibroblasts and its overall deposition into the matrix, a property that may change over time with progressive scarring of the muscle (9). The identification of the genes responsible for these dystrophic modifiers will be helpful in understanding why and how these loci exert their effect.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grants R01-HL-61322 and T32-HL-07381 and the Muscular Dystrophy Association.
REFERENCES
Articles from Physiological Genomics are provided here courtesy of American Physiological Society
Full text links
Read article at publisher's site: https://doi.org/10.1152/physiolgenomics.00172.2010
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3026347
Free after 12 months at intl-physiolgenomics.physiology.org
http://intl-physiolgenomics.physiology.org/cgi/content/abstract/43/1/24
Free after 12 months at intl-physiolgenomics.physiology.org
http://intl-physiolgenomics.physiology.org/cgi/content/full/43/1/24
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1152/physiolgenomics.00172.2010
Article citations
Development of myofibers and muscle transcriptomic analysis in growing Yili geese.
Poult Sci, 103(2):103328, 14 Dec 2023
Cited by: 0 articles | PMID: 38157792 | PMCID: PMC10790089
Anti-latent TGFβ binding protein 4 antibody improves muscle function and reduces muscle fibrosis in muscular dystrophy.
Sci Transl Med, 13(610):eabf0376, 08 Sep 2021
Cited by: 17 articles | PMID: 34516828 | PMCID: PMC9559620
Dusp6 is a genetic modifier of growth through enhanced ERK activity.
Hum Mol Genet, 28(2):279-289, 01 Jan 2019
Cited by: 11 articles | PMID: 30289454 | PMCID: PMC6322066
Non-Glycanated Biglycan and LTBP4: Leveraging the extracellular matrix for Duchenne Muscular Dystrophy therapeutics.
Matrix Biol, 68-69:616-627, 23 Feb 2018
Cited by: 22 articles | PMID: 29481844 | PMCID: PMC6015525
Review Free full text in Europe PMC
Genetic modifiers of muscular dystrophy act on sarcolemmal resealing and recovery from injury.
PLoS Genet, 13(10):e1007070, 24 Oct 2017
Cited by: 24 articles | PMID: 29065150 | PMCID: PMC5669489
Go to all (23) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Modifiers of heart and muscle function: where genetics meets physiology.
Exp Physiol, 99(4):621-626, 08 Nov 2013
Cited by: 15 articles | PMID: 24213858 | PMCID: PMC3975685
Review Free full text in Europe PMC
The superhealing MRL background improves muscular dystrophy.
Skelet Muscle, 2(1):26, 05 Dec 2012
Cited by: 26 articles | PMID: 23216833 | PMCID: PMC3534636
Fibrosis and inflammation are greater in muscles of beta-sarcoglycan-null mouse than mdx mouse.
Cell Tissue Res, 356(2):427-443, 11 Apr 2014
Cited by: 12 articles | PMID: 24723230
Mutations in the delta-sarcoglycan gene are a rare cause of autosomal recessive limb-girdle muscular dystrophy (LGMD2).
Neurogenetics, 1(1):49-58, 01 May 1997
Cited by: 29 articles | PMID: 10735275
Funding
Funders who supported this work.
NHLBI NIH HHS (2)
Grant ID: R01-HL-61322
Grant ID: T32-HL-07381
NIDA NIH HHS (1)
Grant ID: R01 DA021336
NINDS NIH HHS (1)
Grant ID: P01 NS072027
 1,2
1,2




