| PMC full text: | Prepublished online 2010 Sep 17. doi: 10.1182/blood-2010-05-286328
|
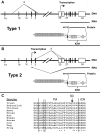
Figure 4

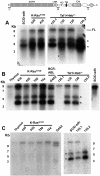
Evidence of RAG involvement in Notch1 rearrangements in murine T-ALL. (A) Rearrangements in Notch1 deduced from sequencing of PCR products generated from 11 cell lines with type 1 deletions (top) and 3 cell lines with type 2 deletions (bottom). GL is the sequence of the germline DNA flanking the breakpoints. Nucleotide positions are expressed relative to the ATG start codon in exon 1 of Notch1. Flanking sequences resembling RAG recognition sequences are boxed. Residues matching the consensus RAG signal sequence (a CACATGT heptamer followed by a 12- or 23-bp spacer and the nonameric sequence ACAAAAAAC) are denoted with an asterisk. N nucleotides and P nucleotides (underlined) are also shown. The point of joining in SCID-adh contains a single cytosine residue (underlined) of unknown origin. (B) Distribution of RAG2 binding and H3K4 trimethylation across the murine Notch1 locus. ChIP-Seq was performed with antibodies specific for RAG2 and H3K4-me3 on DNA immunoprecipitated from normal thymocytes (αRAG2 WT), thymocytes expressing a RAG1 D708A mutant (αRAG2 Mut) that binds chromatin but is catalytically inactive, and homozygous RAG2 knockout thymoctyes (αRAG2−/−).22 Histograms showing sequence reads that aligned to the murine genome are superimposed on a diagram of the Notch1 locus. The y-axis of each histogram corresponds to the number of aligned reads per 106 total reads.