Abstract
Free full text

Environment-responsive transcription factors bind subtelomeric elements and regulate gene silencing
Associated Data
Abstract
Subtelomeric chromatin is subject to evolutionarily conserved complex epigenetic regulation and is implicated in numerous aspects of cellular function including formation of heterochromatin, regulation of stress response pathways and control of lifespan. Subtelomeric DNA is characterized by the presence of specific repeated segments that serve to propagate silencing or to protect chromosomal regions from spreading epigenetic control. In this study, analysis of genome-wide chromatin immunoprecipitation and expression data, suggests that several yeast transcription factors regulate subtelomeric silencing in response to various environmental stimuli through conditional association with proto-silencing regions called X elements. In this context, Oaf1p, Rox1p, Gzf1p and Phd1p control the propagation of silencing toward centromeres in response to stimuli affecting stress responses and metabolism, whereas others, including Adr1p, Yap5p and Msn4p, appear to influence boundaries of silencing, regulating telomere-proximal genes in Y′ elements. The factors implicated here are known to control adjacent genes at intrachromosomal positions, suggesting their dual functionality. This study reveals a path for the coordination of subtelomeric silencing with cellular environment, and with activities of other cellular processes.
Introduction
It is well established that environmental conditions modulate gene expression through local binding of a variety of conditionally active transcription factors (TFs), each responsive to specific environmental cues. However, another prevalent mechanism of gene regulation in eukaryotic cells is the long-range control of groups of genes by chromatin modifications or other position-dependent mechanisms. One such phenomenon, subtelomeric silencing, is an important and evolutionarily conserved mode of regulation that has been linked to cellular lifespan (Dang et al, 2009; Mak et al, 2009). Silencing factors, Sir2p, Sir3p and Sir4p bind to telomere repeat regions at chromosomal ends through interactions with the telomere-binding protein Rap1p (Rusche et al, 2003). Cooperative binding leads to a spread of silencing activity toward the centromere, silencing the expression of nearby subtelomeric genes. Silencing is further augmented through the clustering and tethering of telomeres to the nuclear envelope, increasing the local concentration of silencing molecules near subtelomeric regions (Buhler and Gasser, 2009). However, additional anti- and proto-silencing mechanisms exist to exert complex control of genes positioned within these subtelomeric regions (Fourel et al, 2002). Elucidation of how subtelomeric silencing and other telomere-related processes are coordinated with both cellular environment and the activities of other cellular processes is fundamental to our understanding of human conditions including aging and cancer.
Nuclear chromosomes of Saccharomyces cerevisiae terminate with telomeric repeats characterized by stretches of the sequence TG1–3 (Shampay et al, 1984). Regions proximal to these repeats contain two different kinds of subtelomeric repeat regions (STRs) called X and Y′ elements (Pryde and Louis, 1997). X elements are short repetitive regions found on every telomere and are either adjacent to telomeric repeats, or are separated from telomeres by one or more Y′ elements. All X elements contain a 473-bp core sequence containing an ARS consensus sequence and usually a binding site for the general regulatory factor (GRF), Abf1p. Many X elements also contain repeat regions on their telomeric side that contain a binding site for the GRF, Tbf1p (Louis et al, 1994; Pryde et al, 1995). Y′ elements are highly polymorphic repetitive elements of ~5–7 kb that are found between terminal telomeric repeats and X elements on 17 of the 32 telomeres (Chan and Tye, 1983; Louis and Haber, 1990, 1992).
X elements are proto-silencers that both relay and enhance silencing from telomeres. In subtelomeric regions, maximal gene silencing and Sir2p, Sir3p and Rap1p binding occur at X elements and diminish toward centromeres (Fourel et al, 1999; Pryde and Louis, 1999; Zhu and Gustafsson, 2009). Proto-silencing by X elements is dependent on Sir2, -3 and -4, and yKu70, a protein involved in tethering telomeres to the nuclear envelope (Pryde and Louis, 1999). X elements are also involved in establishing chromatin domain boundaries that protect Y′ element genes from silencing. In this case, GRFs facilitate interactions between X elements and Rap1p–Sir complexes, resulting in highly repressed heterochromatin, which spreads toward centromeres, bypassing Y′ elements (Fourel et al, 1999, 2002; Pryde and Louis, 1999; Zhu and Gustafsson, 2009). It has been proposed that this discontinuous spread of silencing is facilitated by telomeres folding onto themselves resulting in contact between telomeres and subtelomeres, and looping out of Y′ elements (Strahl-Bolsinger et al, 1997; Pryde and Louis, 1999).
Subtelomeric regions (within 25 kb of telomeres) contain many uncharacterized genes, but are also enriched for genes encoding helicases implicated in telomere maintenance (present in Y′ elements) (Yamada et al, 1998), as well as genes involved in carbon source utilization (Pryde and Louis, 1997) and stress responses (Wyrick et al, 1999; Ai et al, 2002; Robyr et al, 2002). Subtelomeric gene expression and silencing can be dynamically regulated by environmental cues (Ai et al, 2002); however, the mechanisms controlling these activities remain to be understood. One study identified enrichment of TFs in subtelomeric regions in response to stress, and suggested that conditional binding of TFs upstream of genes in this region conditionally controlled their expression in a manner linked to histone deacetylase Hda1p (Mak et al, 2009).
In this study, we reveal a novel and fundamental mechanism for the regulation of subtelomeric silencing in response to environmental stimuli including stress and carbon source. Using chromosome position analysis of microarray-based chromatin immunoprecipitation (ChIP-chip) data for environment-responsive TFs and genome-wide gene expression data under the same conditions, we show that several environment-responsive TFs interact with subtelomeric X elements and conditionally regulate their proto- and anti-silencing activities in response to environmental stimuli. Investigation of this mechanism during the response to fatty acid exposure showed that conditional proto-silencing activity is dependent on Sir2p and independent of Hda1p. TFs implicated in this process have previously been shown to modulate the expression of adjacent genes in response to the same stimuli, and have not previously been implicated in the regulation of silencing. The potential of this mechanism to coordinate telomere biology with environmental stimuli and other cellular processes is discussed.
Results
Oaf1p conditionally binds subtelomeric X elements
Through network analysis of protein–DNA interactions from ChIP-chip data, we have previously shown that four TFs, Oaf1p, Pip2p, Oaf3p and Adr1p, dynamically cooperate in the presence of fatty acids (Smith et al, 2007; Ratushny et al, 2008). These factors control two distinct classes of genes: those conditionally targeted by Oaf1p, Oaf3p and Adr1p, which enrich for general stress response genes downregulated by the stimulus, and those conditionally targeted by all four factors, which enrich for genes involved in fatty acid metabolism and are upregulated by the stimulus. Oaf1p is both a repressor and an activator, providing specificity and coordination of the two responses; it heterodimerizes with Pip2p and directly upregulates genes involved in fatty acid metabolism, and it binds independently of Pip2p to negatively regulate the class of genes involved in general stress response.
We sought to characterize the repressive activity of Oaf1p. Chromosome position analysis revealed that targets of Oaf1p in the context of negative regulation (those conditionally targeted by Oaf1p, Adr1p and Oaf3p, but not Pip2p) are enriched at regions within 10 kb of telomeres (Figure 1A). We did not detect a regional preference for the subclass of genes positively regulated by Oaf1p and none of these intergenic regions were within 10 kb of telomeres (data not shown). A higher resolution analysis of subtelomeric regions revealed striking enrichment of the targets between 5 and 7 kb from telomeres (Figure 1B; blue bars). In all, 25–30% of probes in these regions were bound by the three factors, considerably more than the 1% was expected if targets had no positional preference (green vector).
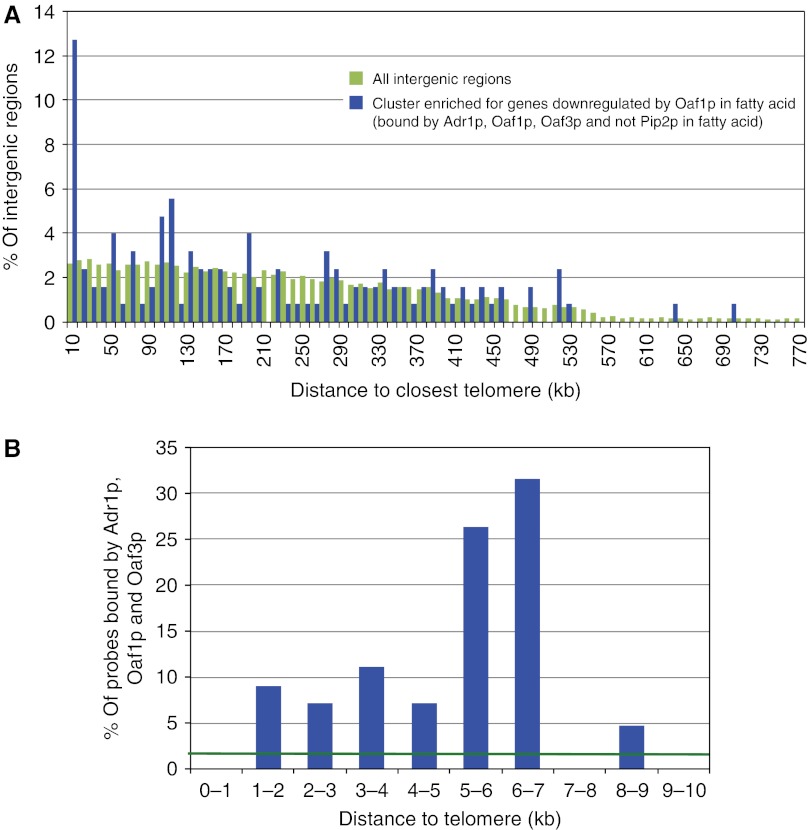
Network cluster negatively regulated by Oaf1p is enriched 5–7 kb from telomeres. (A) Histogram of distance to closest telomere for targets of Oaf1p (in a network cluster enriched for those downregulated by Oaf1p) (Smith et al, 2007). More than 12% of these targets were found within 10 kb of telomeres, whereas only about 2% of all intergenic regions on the microarray were in this position. In contrast, network clusters upregulated by Oaf1p were not found within 10 kb of telomeres (data not shown). (B) High-resolution histogram of DNA segments within 10 kb of telomeres shows that the same targets are enriched 5–7 kb from telomeres (blue bars). The expected binding frequency if targets had no positional preference is shown (green vector). The region within 1 kb of telomeres had no detectable interaction with the factors, but <10% of this region was present on the microarrays (data not shown).
The positional enrichment of Oaf1p binding prompted us to test whether subtelomeric targets of Oaf1p coincided with X elements, which are found either at the telomeric ends of subtelomeric regions or centromere-proximal to one or two 5–7 kb Y′ elements (Supplementary Figure S4) (Table I). Remarkably, 14 of the 15 X elements represented on the intergenic arrays were bound by Oaf1p, 79% of which had a binding pattern consistent with Oaf1p-negative regulation (bound by Adr1p, Oaf1p, Oaf3p in the presence of fatty acid) and none had a binding pattern consistent with Oaf1p activation (bound by Oaf1p and Pip2p). These data suggest that Oaf1p conditionally binds X elements, and functions as a negative regulator in this context.
Table 1
| Position of X element | Overlapping intergenic region | Extent of overlap (bp) | Interaction with TF in oleate | Interaction with TF in glucose |
|---|---|---|---|---|
| Shown are the 15 of the 32 X elements in the genome that overlap by 10 bp or more with intergenic region(s) on the microarrays analyzed in Figure 1. In all, 14 of these were bound by Oaf1p, 11 (79%) of which had a binding pattern consistent with Oaf1p-negative regulation (conditionally bound by Adr1p (A), Oaf1p (O) and Oaf3p (Y) in the presence of fatty acids, but not Pip2p (P); AOY topology in column 4). None of the probes had a binding pattern consistent with Oaf1p binding as an activator (bound by both Oaf1p and Pip2p). Only one X element on the array is not bound by Oaf1p. | ||||
| II-L | iYBL109W-0 | 483 | AOY | None |
| IV-R | iYDR542W | 64 | None | None |
| IV-R | iYDR543C | 162 | OY | None |
| V-R | iYERWomega2-0 | 491 | OY | None |
| V-R | iYERWomega2-1 | 239 | AOY | O |
| VI-L | iYFL064C | 246 | AOY | None |
| VII-R | iYGR295C-0 | 150 | None | None |
| VII-R | iYGR295C-1 | 561 | OY | None |
| VIII-L | iYHL049C-0 | 519 | OY | None |
| VIII-L | iYHL049C-1 | 34 | None | None |
| IX-L | iYIL177C-0 | 521 | AOY | AO |
| IX-L | iYIL177C-1 | 314 | None | None |
| X-L | iYJL225C-0 | 521 | AOY | AO |
| X-L | iYJL225C-1 | 314 | Y | None |
| XII-L | iYLL066C-1 | 367 | AOY | None |
| XII-R | iYLR461W-0 | 334 | None | None |
| XII-R | iYLR461W-1 | 415 | AOY | AO |
| XIII-L | iYML133C-0 | 378 | AOY | O |
| XIII-L | iYML133C-1 | 380 | AY | None |
| XIV-L | iYNL338W | 446 | AOY | AO |
| XV-R | TEL15R-2 | 15 | AOY | O |
| XVI-L | iYPL283C-0 | 445 | AOY | None |
| XVI-L | iYPL283C-1 | 253 | None | None |
| XVI-R | iYPR201W-1 | 375 | None | None |
Other environment-responsive TFs conditionally bind X elements
To determine whether this X element-binding property of Oaf1p is shared with other environment-responsive factors, a comprehensive ChIP-chip data set of 203 yeast TFs (Harbison et al, 2004) was analyzed. Factors that were found to interact with at least one X element-containing probe on the microarrays are shown in Figure 2. The top two graphs (Figure 2A and B) show factors that were analyzed in the absence and presence of a stress stimulus, while the bottom graph (Figure 2C) shows factors tested only in the absence of stress. Bars marked with asterisks correspond to factors that significantly enriched at X elements. This analysis identified 19 factors that appeared to (conditionally) concentrate at X elements, including known X element-binding proteins Rap1p (Zhu and Gustafsson, 2009) and Reb1p (Chasman et al, 1990).
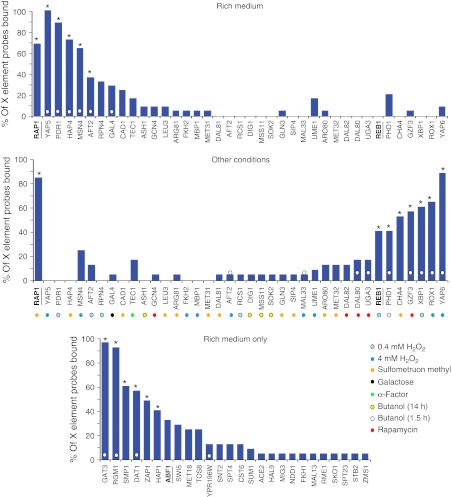
Many environment-responsive transcription factors (TFs) bind X elements. ChIP-chip data of 203 DNA-binding proteins were analyzed for factors that target X elements. Factors that bound at least one of the 25 probes on the arrays that overlap with X elements are shown. The top graph shows analysis of factors from cells grown in rich medium, and the middle graph shows the same factors from cells grown under other conditions. The bottom graph shows factors that were analyzed only in rich medium. Factors/conditions marked with a white dot on the bar are those shown to have enriched binding in subtelomeric regions (Mak et al, 2009). Underlined factors have been previously shown to bind X elements. The 19 factors with astrisks significantly enrich at X elements (i.e., they have hypergeometric distribution P<0.001 after multiple test correction, and bind to more than eight X element probes).
Some of the TFs that enriched at X elements have previously been shown to enrich in promoter regions of genes within subtelomeres (marked with a white circle on or above the bar) (Mak et al, 2009). Therefore, we determined the subtelomeric-binding profiles of the factors to determine whether they specifically enrich at X elements compared with the rest of the subtelomeric region (Figure 3). Binding profiles correlated with the position of X elements with Pearson's correlation coefficients >0.8 and P-values <1 × 10−6, with the exception of Phd1p, which had a coefficient of 0.51 and P=0.0045. This correlation of TF binding with X elements was not detected by a previous analysis (Mak et al, 2009) possibly because of the lower resolution of the previous study (which tested genomic fragments of ~390 kb compared with 10 kb used here). In addition, the previous study analyzed the positions of all X elements for correlation, whereas here, X elements that were not present on the microarrays were excluded from the analysis.

Binding profiles of transcription factors (TFs) correlate with X-element positions. Subtelomeric-binding profiles are shown for factors found to enriched at X elements in Figure 2. The densities of TF binding and X elements present on the microarrays are represented as heat maps. Binding conditions other than rich medium are marked with colored dots as in Figure 2. With the exception of Phd1p, all factors had maximal binding 5–7 kb from telomeres, matching the position of X element enrichment. Pearson's correlation coefficients (ρ) comparing each binding profile to the positions of X elements are shown at the right.
Analysis of X element binding with high resolution tiled arrays
To further characterize the interaction of TFs with X elements, an in depth analysis of Oaf1p binding was conducted by repeating the ChIP of Oaf1p in the absence or presence of medium containing fatty acids, and analyzing the targets on tiled microarrays of the entire yeast genome (see Materials and methods). Subtelomeric-binding profiles under each condition were determined and are shown as histograms of the mean number of peaks per chromosome arm (Figure 4A, top two panels). These data revealed that Oaf1p targets in the presence of fatty acids were not only enriched 5–7 kb from telomeres, but were also enriched within 1 kb of telomeres, whereas in the absence of fatty acids the profile was similar to background (compare blue bars to red vector of mean binding frequency per kb in the entire genome). The Oaf1p-binding profile was strongly correlated with the positions of X elements (Figure 4A, third panel) in the presence of fatty acids, with a Pearson's correlation coefficient (ρ) of 0.98 (P<0.01), but not in the absence of fatty acids (ρ= −0.15). This strong correlation is supported by the fact that Oaf1p targeted almost all X elements in the genome with a false discovery rate (FDR) <0.001 in the presence of fatty acids (Figure 4B).
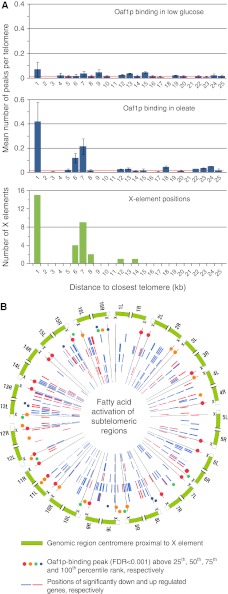
Oaf1p conditionally binds subtelomeric X elements. (A) Subtelomeric-binding profiles of Oaf1p in the absence and presence of fatty acids (from ChIP-chip using whole-genome tiling arrays with peak detection false discovery rate (FDR; <0.001). In the presence of fatty acids, binding of Oaf1p is highly enriched within 1 kb of telomeres and more subtly enriched 5–7 kb from telomeres, and matches the profile of X-element positions. Red vectors mark mean binding frequency per kb across the entire genome for the experiments. Error bars represent s.e. of three biological replicates of each experiment. (B) Visualization of fatty acid-activated subtelomeres using Circos visualization software (Krzywinski et al, 2009). In all, 32 subtelomeric regions in yeast are shown (peripheral ring) including X elements (marked with X and spokes extending toward center) and 25 kb of centromere-proximal sequence (green bars). Binding positions of Oaf1p as determined by ChIP-chip (FDR<0.001) are shown as colored circles inside of the peripheral ring. Red and blue bars show positions of significantly up and downregulated genes, respectively, in response to fatty acid exposure at 0.5, 1, 3, 6 and 9 h (from outer to inner ring) (Smith et al, 2002). Significantly downregulated genes are enriched in subtelomeric regions. See also Supplementary Figures S1 and S2.
The high-resolution ChIP-chip data set enabled analysis of specificity of Oaf1p binding at individual X elements (Supplementary Figure S1), which revealed that Oaf1p specifically binds to unique probes in most X elements in response to fatty acids. The data set also enabled identification of a putative DNA-recognition sequence for Oaf1p in the context of negative regulation (see Materials and methods). This sequence (G[not A]AGGGTAANNNNN[not C][not C]) is similar to the predicted binding motif of GRF, Reb1p (RTTACCCK) (Badis et al, 2008) and for the reasons outlined below, was termed subtelomeric Reb1-binding (SRB) motif. This motif occurs 238 times in the entire genome, and 65% of the motifs are within 100 bp of an Oaf1p-binding position (e.g., see Figure 6A). CPA analysis of the motif showed that it strongly enriches in subtelomeric regions at a frequency of >1 per arm, and colocalizes with X elements (Supplementary Figure S2). To characterize the interaction of Oaf1p with the SRB motif, an electromobility shift assay (EMSA) was performed (Supplementary Figure S2). Whereas the DNA-biding domain (DBD) of Reb1p bound the SRB motif, the DBD of Oaf1p did not, suggesting that Oaf1p does not interact directly with X elements and that this interaction might be mediated by Reb1p.
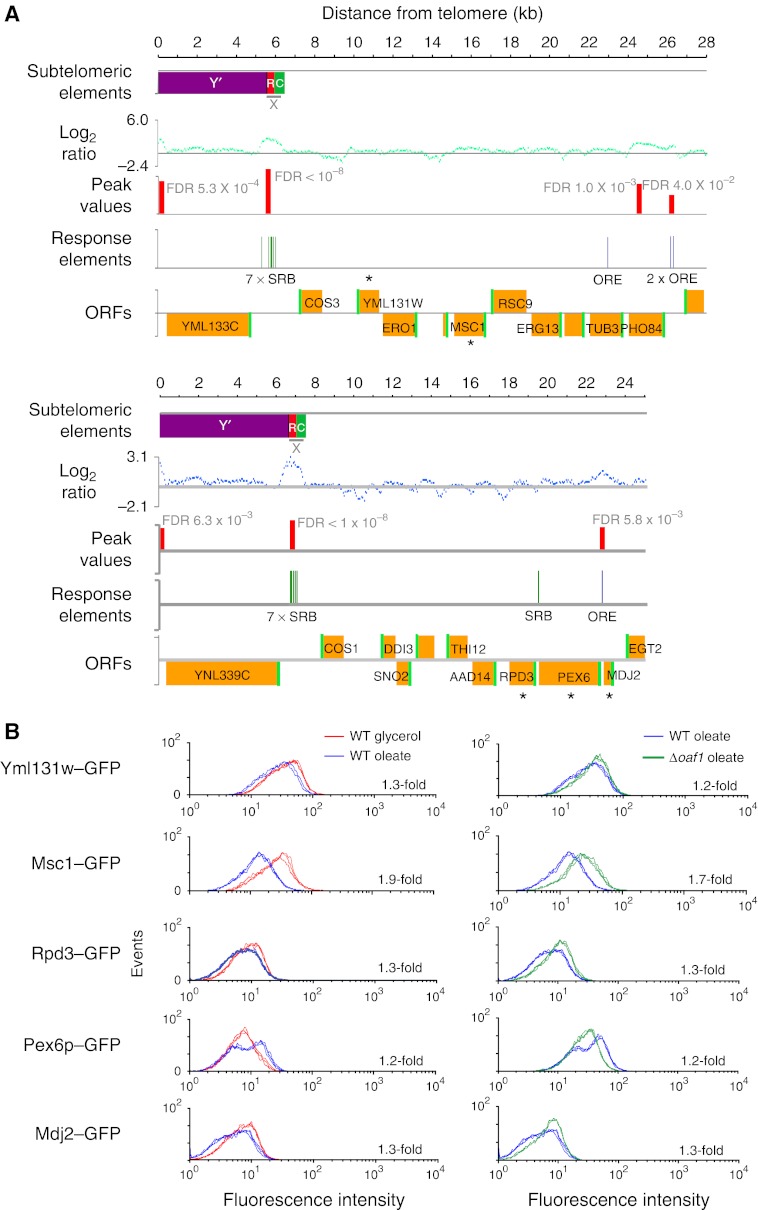
Oaf1p negatively regulates distally located subtelomeric genes. (A) Two subtelomeric regions (13 and 14 L) are shown. First row, positions of X and Y′ elements, with X element repeat regions (red) and cores (green) shown separately. Second row, log2 expression ratios (IP versus WCE) for each 50-mer probe for one replicate of Oaf1p ChIP-chip in the presence of the fatty acid oleate. Third row, Oaf1p-binding positions (false discovery rate; FDR <0.05) predicted from this data (red bars). Fourth row, positions of OREs, motifs that bind Oaf1p–Pip2p heterodimers that activate transcription (Rottensteiner et al, 2003), and subtelomeric Reb1-binding (SRB) motifs, predicted to bind Oaf1p without Pip2p to repress transcription (Supplementary Figure S2). Oaf1p binding at X elements coincide with SRB motifs, whereas other peaks coincide with OREs. Fifth row, ORFs with start sites in green. Genes analyzed in B are marked with asterisks. (B) FACS data showing abundance of subtelomerically encoded proteins fused to GFP in the presence and absence of fatty acids in wild-type (WT) and Δoaf1 strains. Three biological replicates are shown for each condition. All changes in levels between condition pairs are statistically significant with Student's t-test P-values <0.01 and mean fold changes are shown for each. Genes centromere-proximal to X elements appear to be negatively regulated by Oaf1p in the presence of oleate. An exception to this is PEX6, which is positively regulated by Oaf1p in oleate and also has an upstream ORE bound by Oaf1p (A). ORE, oleate response element.
X element-binding TFs conditionally regulate proto-silencing
Conditional binding of Oaf1p to X elements in the presence of fatty acids appeared to correlate with an overall repression of subtelomeric gene expression (compare ratio of down versus upregulated genes (blue versus red bars) in Figure 4B). This led us to hypothesize that X element-binding TFs regulate X element-mediated proto-silencing in response to environmental stimuli. To test this, expression profiles of subtelomeric genes centromere-proximal to X elements (within 20 kb) were analyzed during three responses that correspond to dynamic repositioning of factors at X elements in Figures 2 and and44 (Figure 5). The region was enriched for genes that significantly decreased in expression in response to fatty acid stress, but increased in expression in response to H2O2 or butanol stress (compared with the percentage of significantly up and downregulated genes in the entire genome, represented as green and red shading, respectively). Expression profiles that were determined to be significantly different from the global responses using a Student's t-test are marked with asterisks. Y′ element genes that are telomere-proximal to X elements were analyzed in the same way, and their responses were not significantly different from the global responses to the stimuli, except for a subtle enrichment of upregulated genes at 2 h of H2O2 exposure (3 versus 2.5% in the genome) (data not shown). These data suggest environmental regulation of X element-mediated proto-silencing.

Regulation of proto-silencing by X element binding TFs in response to stress. Microarray expression data were analyzed to determine the effects of environmental stresses on the expression of subtelomeric genes that are centromere-proximal to X elements, and the roles of X element-binding factors in this regulation. Three stress conditions resulting in differential binding of transcription factors (TFs) to X elements in Figures 1, ,22 and and33 were analyzed including exposure to 0.15% fatty acids (A), 0.32 mM H2O2 (B) and 1% butanol (C). For A and B, and C, there were a total of 227 and 230 genes, respectively, in the subtelomeric region of interest. The percentage of these genes that significantly increased (green bars) and decreased (red bars) in expression are shown, as well as the percentage of genes with significant changes in the entire genome (pink and green shading). Bars marked with asterisks have significantly different mean expression profiles than the entire genome (with Student's t-test P-values <0.001). The left panels show binding patterns of TFs that dynamically enrich at X elements in response to the stresses. The right panels show analyses of wild-type and TF deletion strains under the same conditions. OAF1 appears to conditionally enhance proto-silencing whereas ROX1 and PHD1 have the opposite effect. See also Supplementary Figure S3.
Next, the roles of TFs in proto-silencing were investigated by analyzing microarray expression profiles of subtelomeric genes in TF deletion strains (Figure 5). Growth in the presence of fatty acids resulted in decreased expression of subtelomeric genes centromere-proximal to X elements. Under the same condition, deletion of OAF1 resulted in increased expression of these genes compared with the wild-type strain, suggesting that Oaf1p is an enhancer of X element-mediated proto-silencing during this response. However, deletion of ROX1 or PHD1 resulted in enrichment of genes with significantly decreased expression in the presence of H2O2 or butanol, respectively, suggesting that Rox1p and Phd1p conditionally antagonize proto-silencing. To evaluate whether these activities were due to direct activation or repression by the TFs rather than regulation of proto-silencing at X elements, the analyses were repeated with direct targets of factors implicated in the responses excluded, and the results were very similar (Supplementary Figure S3). Together, these data suggest that conditional interaction of TFs with X elements regulate proto-silencing activity in response to the environment with long-range effects on genes that are not immediately adjacent to X elements.
To investigate these effects on distal genes, we analyzed the impact of Oaf1p regulation on specific subtelomeric genes (Figure 6). Panel A shows the binding patterns of Oaf1p in subtelomeric regions on chromosome arms 13 and 14 L. On both arms, X elements were detected (top row), bound by Oaf1p in the presence of fatty acids (second and third rows), and overlapped with SRB motifs, but not oleate response elements (OREs), which bind Oaf1p–Pip2p heterodimers that activate transcription (Rottensteiner et al, 2003) (fourth row). GFP fusion proteins encoded by genes not adjacent to X elements (open reading frames; ORFs marked with asterisks in fifth row) were monitored by fluorescence-activated cell sorting (FACS) in the absence or presence of fatty acids in wild-type and Δoaf1 cells (panel B). With the notable exception of Pex6–GFP (see below) the majority of proteins tested had reduced levels in the presence of fatty acid compared with glycerol (left), and this effect was dependent on the presence of Oaf1p (right). Negative regulation by Oaf1p was specific to the fatty acid response and was not apparent in glycerol-grown cells (data not shown). Altogether, 10 genes centromere-proximal to X elements were analyzed by FACS, 3 of which had significant decreases in expression in an OAF1 deletion strain by microarray analysis. In all, 7 of the 10 corresponding fusion proteins had significantly decreased abundance in fatty acid medium, and 6 had significant fatty acid-specific negative regulation by OAF1. These data support our interpretation that the binding of Oaf1p to X elements has a conditional role in controlling subtelomeric silencing.
A notable exception to the trend is PEX6 (shown in Panels A and B), which had increased protein levels in response to oleate and positive regulation by Oaf1p. A possible mechanism of upregulation is that, although Oaf1p is a proto-silencer at the X element on this chromosome arm, it also directly activates PEX6 by binding the upstream ORE as a heterodimer with Pip2p (Smith et al, 2007) (Figure 6A). These results are consistent with previous studies showing that PEX6 is upregulated by fatty acid exposure (Smith et al, 2002), and that transcriptional activators have access to promoters within otherwise silent chromatin (Xu et al, 2006; Mak et al, 2009). This phenomenon offers an explanation for why not all subtelomeric genes centromere-proximal to X elements were observed to be affected by Oaf1p-mediated silencing.
Oaf1p-mediated silencing is dependent on SIR2
To support the conclusion that Oaf1p interacts with X elements and mediates its effect on subtelomeric silencing from this position, we analyzed the expression of a reporter gene of subtelomeric silencing in the strains FEP318-23 and FEP318-19 (Loney et al, 2009). In these strains, a gene encoding Ura3–GFP is positioned in a subtelomeric region of chromosome end III-R or XI-L adjacent and centromere-proximal to the endogenous X element (Figure 7). The effects of fatty acid exposure and OAF1 deletion on this reporter were analyzed by FACS (Figure 7A). The data suggest that Oaf1p conditionally enhances silencing of the reporter gene in the presence of fatty acids. The reporter gene was also used to elucidate which histone deacetylase is involved in Oaf1p-mediated regulation of silencing. We tested for genetic interactions between OAF1 and two genes encoding histone deacetylases, SIR2, which is known to be required for X element-mediated proto-silencing (Pryde and Louis, 1999), and HDA1, which is involved in another subtelomeric silencing mechanism (Robyr et al, 2002) (Figure 7B). FACS analysis of reporter gene output in various genetic backgrounds suggests that Oaf1p-mediated silencing is dependent on Sir2p and not Hda1p. These data support the contention that environment-responsive TFs conditionally regulate Sir-mediated silencing from X elements and that this function is distinct from their roles linked to Hda1p at other subtelomeric loci (Mak et al, 2009).
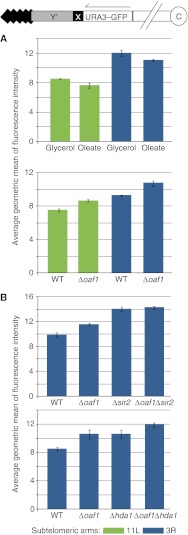
OAF1 conditionally regulates subtelomeric reporter gene expression and is dependent on SIR2. Strains with a subtelomeric gene encoding Ura3–GFP (Loney et al, 2009) were used to test the effects of Oaf1p on subtelomeric silencing by FACS analysis. Bars represent average geometric mean of fluorescence intensity of three to four biological replicates for each strain and error bars show s.d. values of the means. (A) Exposure of cells to the fatty acid oleate for 16 h results in a subtle but significant reduction of the levels of Ura3–GFP encoded in the subtelomeric region of arm 11L or 3R (top graph). Student's t-test P-values of the effects are 0.044 and 0.046 for arms 11L and 3R, respectively. In the presence of oleate, deletion of OAF1 results in a subtle, but significant increase of Ura3–GFP levels encoded on arm 11L or 3R, with t-test P-values of 0.002 and 0.009, respectively (bottom graph). (B) Comparison of a wild-type (WT) strain to isogenic deletion strains after growth in fatty acid medium for 16 h. Δsir2 appears to be epistatic to Δoaf1 (the effect of Δoaf1/Δsir2 double deletion is not significantly different from the effect of Δsir2 alone by Student's t-test). Masking of the Δoaf1 phenotype by Δhda1 was not detected (the effect of the two mutations together were approximately additive).
The reporter gene used here can also be used in a 5FOA viability assay that can measure levels of silencing of a subtelomeric URA3 gene, which directly correlates with cell viability in the presence of 5FOA. This assay was used to test the effects of five TFs on the expression of Ura3–GFP under conditions of X element binding as determined by chromosome position analysis (Figure 2) using previously published ChIP-chip data (Harbison et al, 2004). The results of this approach suggest a role for Gzf3p in reducing X element-mediated proto-silencing in the presence of rapamycin (Supplementary Figure S4). This positive influence on transcription is in contrast to its known role as a negative regulator of nitrogen catabolic genes (Soussi-Boudekou et al, 1997). No effect on silencing was detected for the other four factors tested in the assay (Xbp1p, Yap6p, Rox1p and Cha4p) (data not shown) possibly because of the sensitivity of the assay, or the fact that the assay requires modification of the growth conditions used for chromosome position analysis in Figure 2.
Conditional insulation of Y′ element genes
The effects of conditional TF binding to X elements on Y′ element gene expression were also tested. First, analysis of two other microarray gene expression data sets (Smith et al, 2002, 2007) suggested that Y′ element genes are upregulated in response to two non-fermentative carbon sources (oleate and glycerol) and that this is mediated by the conditional binding of Adr1p to X elements (Figure 8B). Next, microarray data of TF deletion strains grown in rich medium (Hu et al, 2007) were analyzed for expression of Y′ element genes (Figure 2). The analysis suggested that six of the factors that bind X elements in rich medium are involved in anti-silencing of Y′ element genes under this condition, whereas little or no influence was detected for genes centromere-proximal to X elements under this condition (Figure 8A). It should be noted that Y′ elements share a very high degree of sequence homology with each other; therefore, it is not possible to address expression of individual Y′ element genes by this analysis. However, all ORFs in Y′ elements are thought to encode similar protein sequences with putative helicase motifs, and helicase activity of a Y′ element encoded protein has been characterized (Yamada et al, 1998). In addition, the negative effects of the six TF deletions on Y′ element gene expression identified in Figure 8A are dramatic as the mean expression values of Y′ element genes were at least sixfold lower than in wild-type strains. These data indicate that there is a dramatic influence of X element-bound TFs on Y′ element gene expression as a class of genes, suggesting that they are involved in functional regulation of Y′ element processes.
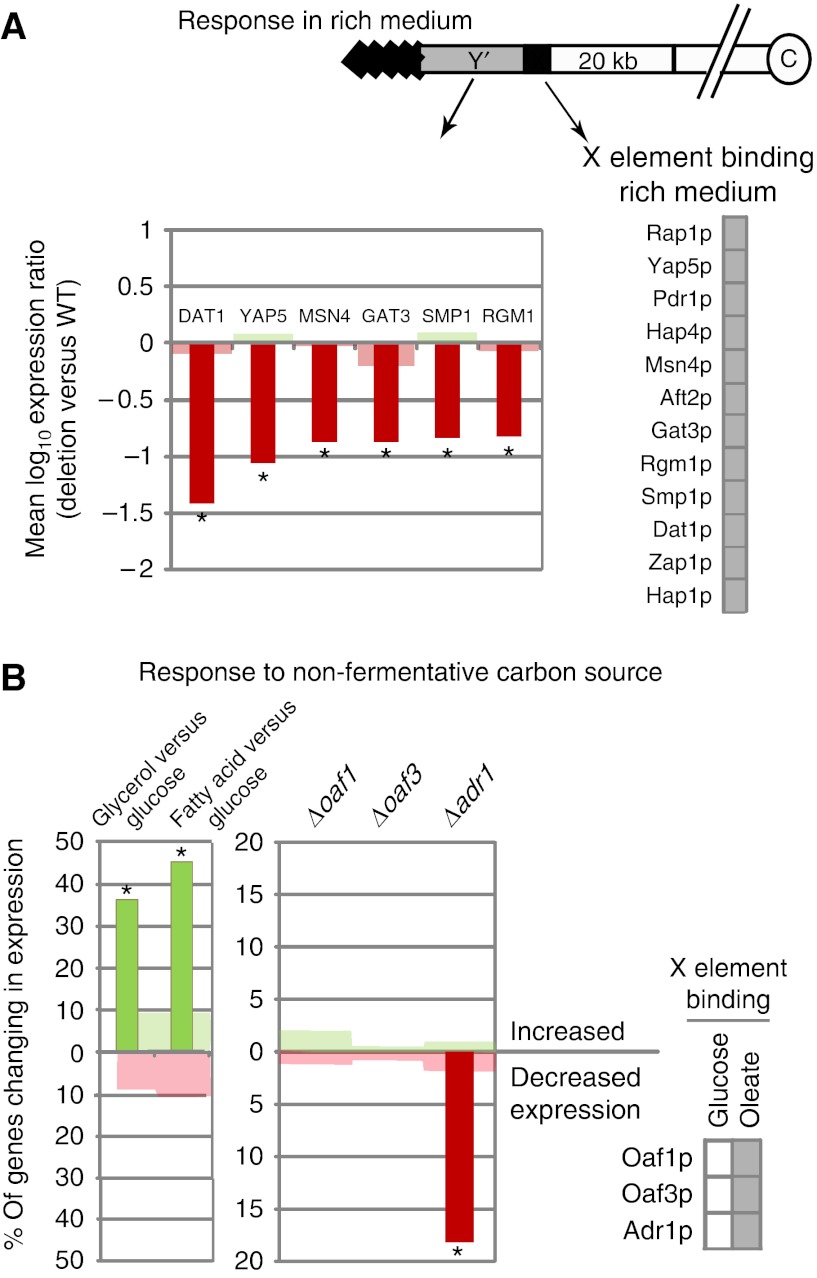
Several X element-binding transcription factors (TFs) positively regulate Y′ element genes. (A) Analysis of Y′ element gene expression in microarray data comparing cells grown in non-fermentative conditions (fatty acid or glycerol) to those grown in rich medium (glucose) shows that Y′ elements are significantly enriched for genes upregulated in non-fermentative conditions (left panel). Analysis of microarray data comparing TF deletions to wild-type (WT) strains after growth in fatty acids shows that Y′ elements are significantly enriched for genes positively regulated by Adr1p, suggesting that it enhances anti-silencing under this non-fermentative growth condition. (B) A microarray gene expression data set of TF deletion strains grown in rich medium was analyzed to determine the influence of X element-binding factors on Y′ element genes. In all, 11 of the 12 factors that bind X elements in rich medium (Figure 2), were in the data set and 7 appeared to significantly influence the expression of Y′ element genes. Bars represent significant changes in Y′ element gene expression, shading represents changes in the entire genome and asterisks mark profiles with means that are significantly different from background as described in the legend of Figure 5. Expression changes are reported as a percent of total number of genes analyzed in the genomic region of interest. There is a total of 33 Y′ element genes in the data sets used for both panels. For A and B, no significant effects were detected for genes centromere-proximal to X elements, except for subtle effects of Δsmp1and Δgat3 (data not shown).
Discussion
Subtelomeric gene expression has long been implicated in cellular responses to the environment and control of lifespan; however, the mechanisms underlying these links are only now being established. Subtelomeric chromatin silencing is mediated by the activity of the Sirtuin family of proteins (Michan and Sinclair, 2007). Sir2p establishes chromatin silencing in subtelomeric regions by histone H4 deacetylation and by recruiting other silencing proteins. A decline of Sir2p activity corresponds to compromised subtelomeric transcriptional silencing, which apparently leads to decreased longevity (Dang et al, 2009). Additionally, caloric restriction extends lifespan, and this activity is linked to both the TOR pathway and Sirtuin activities (Medvedik et al, 2007). Moreover, activation of the TOR pathway by rapamycin exposure, stimulates Sir3p phosphorylation and subtelomeric silencing (Ai et al, 2002).
In addition to Sir-mediated silencing, a second subtelomeric silencing mechanism involving the histone deacetylase Hda1p is active in yeast (Robyr et al, 2002). A recent study showed that environment-responsive TFs conditionally enrich in subtelomeric regions, and suggested that they bind upstream of target genes and regulate their expression in an Hda1p-linked manner (Mak et al, 2009). In this study, we show that control of subtelomeric gene expression is also mediated by a novel mechanism involving environment-responsive TF binding to subtelomeric X-elements, which leads to both control of proto-silencing at distally located sites and boundary activity (Figure 9).
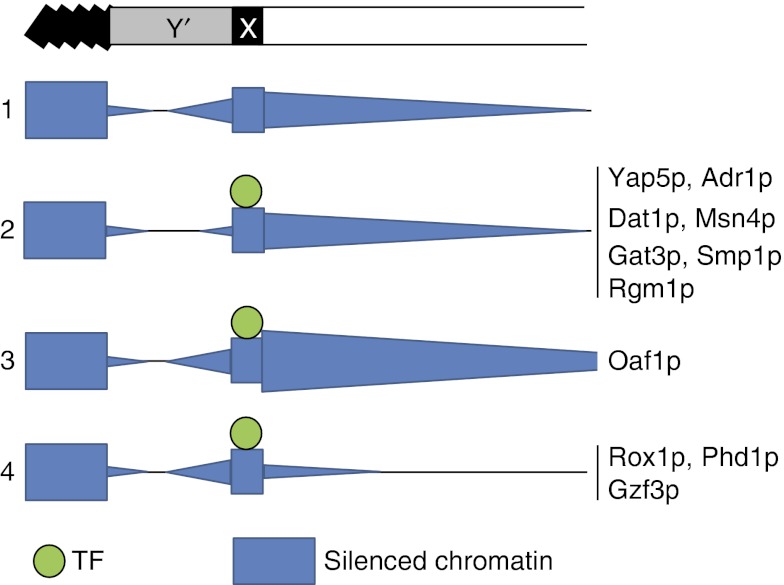
Model of conditional regulation of subtelomeric silencing. A chromosome end is shown with the telomere positioned at the left. Row 1 shows levels of silenced chromatin (blue) in the same genomic region. Rows 2–4 show qualitative effects of X element-binding transcription factors (TFs) on silencing. All effects are relative to silencing levels in the absence of each TF in the first row. TFs in group 2 affect expression of telomere-proximal genes whereas TFs in groups 3 and 4 affect those that are centromere-proximal to X elements.
Proto-silencing activities of stress-responsive TFs
The spread of silencing in yeast subtelomeres is not continuous. Subtelomeric X elements interact with telomeric silencing molecules and relay Sir2p-mediated silencing to centromere-proximal genes. This proto-silencing activity is known to be mediated by the GRF, Abf1p (Pryde and Louis, 1999). The data presented here suggest that proto-silencing mediated by X elements is regulated in response to the cellular environment by multiple TFs that conditionally interact with X elements. In particular, Oaf1p binds X elements and enhances this silencing in response to fatty acid exposure, and Rox1p, Gzf3p and Phd1p bind and conditionally antagonize silencing in response to exposure to H2O2, rapamycin and butanol, respectively (Figure 5 and Supplementary Figure S4). Analysis of conditional Oaf1p-mediated silencing showed that it is dependent on SIR2 but not HDA1, consistent with a novel role for TFs in X element-mediated silencing that is distinct from their roles related to Hda1p at other subtelomeric loci (Mak et al, 2009).
Although the coregulatory mechanism identified here conditionally regulates a group of positionally and functionally related genes in subtelomeric regions that make up ~5% of the yeast genome, the consequences of this regulation remain elusive. This is consistent with our previous study showing that genes that transcriptionally respond to fatty acid exposure are largely not (measurably) required for fitness when grown on this carbon source (Smith et al, 2006). Some of these responses likely reflect the many fundamental and complex relationships between cells and their environment that enable long-term survival of the species that are not easily measurable in the laboratory. These responses might result in a subtle growth advantage or other desirable property, which can influence the evolution of the organism. It will be interesting to learn to what extent this regulatory mechanism impacts aging, evolution and other ‘broad-scale’ cellular processes.
Anti-silencing activities of stress-responsive TFs
Y′ elements are known to be protected from silencing by telomere-directed anti-silencing activity mediated by GRFs bound to X elements including Tbf1p (Fourel et al, 1999, 2001), Rap1p and Abf1p (Fourel et al, 2002). Y′ element gene expression is responsive to meiosis (Burns et al, 1994), but little is known about the regulation of Y′ element gene expression in response to environmental cues. In this study, we show that a large number of TFs interact with X elements (Figure 2) and appear to positively regulate Y′ element genes in rich medium (Figure 8A). These genes are further upregulated after the cells are switched from glucose (a fermentative carbon source that does not require the Kreb's cycle for metabolism) to non-fermentative growth conditions (with higher metabolic oxidation from active respiration) (Schuller, 2003) (Figure 8B). Our data suggest that Adr1p conditionally binds X elements (Figure 1) and is involved in activating these genes during non-fermentative growth (Figure 8B). Importantly, these data are consistent with a previous study showing that transcriptional activation domains of several TFs, when tethered to tandem DNA sites in X elements, can act as insulators of telomere-proximal genes and that this mechanism of regulation is distinct from transcriptional activation (Fourel et al, 2001). Therefore, we propose that X element-bound TFs regulate Y′ element gene expression by increasing insulation from silencing propagating from X elements.
The role of metabolic control of Y′ element gene expression is not known. Y′ elements have been implicated in an alternative lengthening of telomeres (ALT) mechanism that is active in the absence of functional telomerase, as survivors of telomerase mutants have been found to have amplification of these repeats, which may physically buffer chromatin degradation (Lundblad and Blackburn, 1993). This amplification coincides with increased expression of Y′ element genes, which encode helicases (Yamada et al, 1998) and involves active Ty1 transposable elements which also lead to gene duplications elsewhere in the genome. Therefore, it has been suggested that survivors of telomerase mutants with Y′ element amplification, likely also have increased genetic variation and are more suited to the stressful environment (Maxwell et al, 2004). This mechanism may be evolutionarily conserved, as recent data suggest that some cancer cells use a similar survival strategy to amplify subtelomeric repeat elements to lengthen telomeres (Marciniak et al, 2005). Considering these data, it will be interesting to determine whether conditional regulation of Y′ element gene expression by X element-binding TFs is linked to adaptation.
Coordination of telomere biology with other cellular responses
It is noteworthy that Oaf1p (Smith et al, 2007) and other X element-binding TFs, including Yap5p, Yap6p (Tan et al, 2008) and Pdr1p (MacPherson et al, 2006) are implicated as dual function activator/repressor proteins, which are also properties of X element-binding GRFs (Rap1p, Abf1p and Tbf1p) and GRFs known to nucleate heterochromatin formation in other systems such as ikaros in mouse and Cft1p in human cells (Gasser, 2001). Bifunctionality enables X element-binding TFs to coordinate subtelomeric silencing with other cellular responses. One way to accomplish this is through context-specific cooperation amongst factors, which has been suggested for many of the X element-binding TFs (MacPherson et al, 2006; Smith et al, 2007; Tan et al, 2008). For example, Oaf1p heterodimerizes with Pip2p to activate genes involved in fatty acid metabolism; however, Pip2p does not appear to be involved in regulation of X element-mediated proto-silencing. Thus, the distinct roles of Oaf1p can be coordinated by controlling the abundance of Pip2p (Smith et al, 2007). Similarly, other X element-binding factors appear to have context-specific binding partners: Pdr1p and Msn4p each conditionally binds to X elements, but X element interactions were not detected with their other known protein partners, (Pdr3p/Stb5p and Msn2p, respectively) (Mamnun et al, 2002; MacPherson et al, 2006). It will be interesting to determine whether dual functionality of these and other factors have similar roles in coordinating silencing with other cellular processes.
It is also possible that X element-association of at least some of the TFs may significantly impact the activity of the factor, regardless of the impact on subtelomeric silencing. For example, the interaction may serve to keep the factors positioned at the nuclear periphery to sequester the factor from intrachromosomal targets, or to receive signals transduction across the nuclear membrane, roles which have recently been attributed to the nuclear lamina underlying the inner nuclear membrane in metazoans, which is not present in yeast (Andres and Gonzalez, 2009).
Repression of subtelomeric genes through the spreading of silencing molecules from telomeres toward the centromere is an evolutionarily conserved phenomenon. In this study, we show using chromosome position analysis of genome-wide ChIP-chip and expression data, that silencing is regulated in response to stress and metabolism by a group of TFs that also modulate intrachromosomal gene expression in response to the same stimuli. These findings provide a critical link in establishing the mechanisms by which telomere biology is coordinated with other cellular processes including responses to environmental stimuli, aging and adaptation.
Materials and methods
Strains and cell culture
GFP fusion strains are from the BY4741 (S65T) GFP collection, and deletion strains are from the BY4742 deletion library available from Invitrogen (Carlsbad, CA) except for Δoaf1, which has previously been described (Smith et al, 2007). Haploid tagged strains with OAF1 deletions were generated by mating, sporulation and tetrad dissection. Strains were grown in 1% yeast extract, 2% peptone and 2% glucose (YPD), SCIM (1.7 g yeast nitrogen base without amino acids and ammonium sulfate (YNB-aa-as)/l, 0.5% yeast extract, 0.5% peptone, 0.79 g complete supplement mixture/l, 5 g ammonium sulfate/l) containing either 0.1% glucose (SCIM-O) or 0.5% Tween 40 (w/v) and 0.15% (w/v) oleate (SCIM-D), or YPB (0.3% yeast extract, 0.5% potassium phosphate (pH 6.0), 0.5% peptone) with either 2% glucose (YPBD), 3% glycerol (YPBG) or 0.5% Tween 40 (w/v) and 0.15% (w/v) oleate (YPBO).
Chromosome position analysis (CPA)
Unless otherwise stated, all mapping of features to chromosome positions was done using Map Peak tool of NimbleScan software version 2.4. (Roche NimbleGen, Madison, WI). Chromosome ends were the first and last base pair of each chromosome sequence, which does not include telomeric repeats (from the Saccharomyces genome database (SGD) website (www.yeastgenome.org) on 9 September 2008). The positions of ORFs and start sites were from the S. cerevisiae annotation file from NimbleGen (created on 2 February 2007). The positions of X and Y′ elements and other genomic features were from SGD (31 December 2008). For generation of CPA histograms, features spanning more than one bin were counted only once and placed in the bin closest to the telomere. When CPA profiles were compared, Pearson's product moment correlation function of R 2.3.0 was used.
CPA of targets in a fatty acid-responsive network in Figure 1
ChIP-chip analysis of four TFs from cells grown in low glucose medium (SCIM-O), or 5 h after transferring the cells to medium containing fatty acid (SCIM-D) using intergenic microarrays has been described (Smith et al, 2007). Conditional protein–DNA interaction networks were clustered based on network topologies to identify groups of intergenic regions targeted by the same subset of TFs (Smith et al, 2007). For CPA, targets in each cluster were binned by their distance to closest telomere using the Dcount database function of Microsoft Excel. To obtain a background profile, the analysis was also done for all intergenic regions on the microarrays. The expected frequency of probes bound by Adr1p, Oaf1p and Oaf3p (but not Pip2p) was the total number of intergenic regions with this network topology/total number of intergenic regions on the array. To determine the representation of each subtelomeric region on the microarrays, individual 1 kb segments were scored as represented if there was any overlap with one or more intergenic region on the array. In all, 12.5% of chromosomes within 1 kb of telomeres were represented on the arrays; the number rose to ~75% between 5 and 10 kb from telomeres.
CPA of ChIP-chip data in Figures 2 and and33
ChIP-chip data of 204 DNA-binding factors (Harbison et al, 2004) was from the Pvalbyintergenic_9.2_forpaper file at http://jura.wi.mit.edu/young_public/regulatory_code/files_for_paper.zip. It was determined by CPA that 25 probes in the data set overlap with X elements. For each set of target probes (with interaction P-values <0.01), the number of X element-containing probes were determined. Significant enrichment of X element probes for each experiment was determined by calculating hypergeometric distribution P-values using R and applying a Bonferroni's multiple test correction of 352, corresponding to the number of experiments in the study. The heat maps of the binding profiles and X-element positions shown in Figure 3 were generated using MeV 4.5 (Saeed et al, 2006).
ChIP-chip analysis using tiled microarrays
ChIP-chip analysis of Oaf1p was performed in the presence (SCIM-D) and absence of fatty acids (SCIM-O) as previously described (Smith et al, 2007) with the changes described below. Three biological replicates of each experiment were performed. For each replicate, linkers were annealed to DNA ends in whole-cell extract and IP fractions, and fragments were amplified by ligation-mediated PCR with high-fidelity Taq polymerase, and shipped to NimbleGen Systems of Iceland for labeling, hybridization, scanning and preliminary analysis as described below: Equal amounts of DNA in whole-cell extracts and IP fraction were labeled with Cy3 and Cy5, respectively, combined and co-hybridized at 42°C to microarrays of 50-mer DNA probes that span both strands of the entire genome positioned every 64 bp (resulting in 14 bases of DNA between probes). Data was extracted using NimbleScan software and the log2 ratios of IP versus whole-cell extract for each probe was determined. Log2 ratios of adjacent probes were analyzed together to find peak regions of TF binding along with FDR using Find Peaks function of NimbleScan software. The chromatin localization data have been submitted to Gene Expression Omnibus database under accession number GSE21852.
CPA of tiling array ChIP-chip data in Figure 4
CPA was performed on peak regions of TF binding (FDRs <0.001). Histograms were generated, showing the mean number of peaks per chromosome arm in 1 kb segments of subtelomeric regions. This was calculated by dividing the number of peaks per segment by the number of chromosome arms (32) and by the number of strands analyzed (2). For each experiment, three biological replicates were analyzed separately and used to determine average number of peaks at each position and s.e.
Gene expression microarrays
The time course data set comparing cells grown in glycerol medium (YPBG) to those switched to oleate medium (YPBO) (Figure 5A) have been described (Smith et al, 2002). Comparison of TF deletion strains to a wild-type strain after 5 h of growth in medium containing fatty acids (SCIM-D) (Figures 5A and and7B)7B) have been described (Smith et al, 2007). The time course data set of 0.3 mM H2O2 exposure (Figure 5B) has been described (Gasch et al, 2000). Comparisons of TF deletion strains to a wild-type strain during growth in rich medium (YPD) (Figure 8A) have been described (Hu et al, 2007). Comparisons of cells grown in glucose (YPBD), glycerol (YPBG), or oleate (YPBO) (Figure 8B) have been described (Smith et al, 2002).
For the TF deletion analyses in Figure 5B and C, TF deletion strains were compared with an isogenic wild-type strain by microarray analysis under conditions used for ChIP-chip analysis of the factors (Harbison et al, 2004). All mutant and wild-type strains were grown in YPD to a density of 1 × 107 cells/ml, treated with stimulus, and compared. For Δrox1, Δxbp1 and Δphd1 analyses, cells were exposed to 4 mM H2O2 for 30 min, 0.4 mM H2O2 for 20 min, or 1% butanol for 90 min, respectively. To prepare samples for microarray analyses, total RNA was isolated by hot acid phenol extraction and then purified on Qiagen RNeasy columns according to the manufacturer's instructions. Next, cDNA was synthesized and labeled using a Superscript indirect cDNA labeling system (Invitrogen). Equal amounts of differently labeled cDNA samples were mixed and hybridized to S.cerevisiae oligonucleotide gene expression arrays (Agilent). All experiments were performed with duplicate biological and technical replicates of each condition, and included replicates in each labeling orientation. Replicates were merged and analyzed for significantly differentially expressed genes using maximum-likelihood analysis (Ideker et al, 2000). Significantly differentially expressed genes were those with a lambda value above 20.74, which was determined empirically to correspond to a false positive rate of 0.001. These gene expression data have been submitted to Gene Expression Omnibus database under accession number GSE21926.
FACS analysis
Cells were grown overnight in YPD, transferred to glycerol medium (YPBG), and grown overnight to a cell density of ~3 × 106 cells/ml. Cells were harvested by centrifugation, resuspended in oleate (YPBO) or glycerol medium (YPBG) and grown for an additional 8–18 h. For each sample, fluorescence intensities of 10 000 cells were measured using a FACS Caliber flow cytometer (BD Biosciences, San Jose, CA) with a forward scatter threshold of 18. Data were analyzed with WinMDI 2.8 (at http://FACS.scripps.edu/) with smoothing of 20 units. Three or four biological replicates of each experiment were performed and used for Student's t-tests of statistical significance.
Acknowledgments
We thank Dr David Dilworth for insightful suggestions. We thank Dr Edward Louis for the kind gift of URA3–GFP reporter strains. This work was funded by grants NIH/NIGMS R01 GM075152, NIH P50 GM076547 and NIH U54 RR022220. We also thank the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg for support. Author contributions:JS and JA conceived initial concepts and designed the research. JS, LM and LV conducted experiments. JS performed data analysis. YW contributed unpublished data. JS, RK and LM processed and displayed data for presentation. JS and JA wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF (2002) Regulation of subtelomeric silencing during stress response. Mol Cell 10: 1295–1305 [Abstract] [Google Scholar]
- Andres V, Gonzalez JM (2009) Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol 187: 945–957 [Europe PMC free article] [Abstract] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [Europe PMC free article] [Abstract] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. ProcInt Conf Intell Sys Mol Biol 2: 28–36 [Abstract] [Google Scholar]
- Buhler M, Gasser SM (2009) Silent chromatin at the middle and ends: lessons from yeasts. EMBO J 28: 2149–2161 [Europe PMC free article] [Abstract] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M (1994) Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev 8: 1087–1105 [Abstract] [Google Scholar]
- Chan CS, Tye BK (1983) A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J Mol Biol 168: 505–523 [Abstract] [Google Scholar]
- Chasman DI, Lue NF, Buchman AR, LaPointe JW, Lorch Y, Kornberg RD (1990) A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev 4: 503–514 [Abstract] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [Europe PMC free article] [Abstract] [Google Scholar]
- Deng Y, He Z, Van Nostrand JD, Zhou J (2008) Design and analysis of mismatch probes for long oligonucleotide microarrays. BMC Genomics 9: 491. [Europe PMC free article] [Abstract] [Google Scholar]
- Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, Simmen KC, Muller K, Li R, Mermod N, Gilson E (2001) An activation-independent role of transcription factors in insulator function. EMBO Rep 2: 124–132 [Europe PMC free article] [Abstract] [Google Scholar]
- Fourel G, Miyake T, Defossez PA, Li R, Gilson E (2002) General regulatory factors (GRFs) as genome partitioners. J Biol Chem 277: 41736–41743 [Abstract] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [Europe PMC free article] [Abstract] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [Europe PMC free article] [Abstract] [Google Scholar]
- Gasser SM (2001) Positions of potential: nuclear organization and gene expression. Cell 104: 639–642 [Abstract] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [Abstract] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [Europe PMC free article] [Abstract] [Google Scholar]
- Hu Z, Killion PJ, Iyer VR (2007) Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet 39: 683–687 [Abstract] [Google Scholar]
- Ideker T, Thorsson V, Siegel AF, Hood LE (2000) Testing for differentially-expressed genes by maximum-likelihood analysis of microarray data. J Comput Biol 7: 805–817 [Abstract] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [Europe PMC free article] [Abstract] [Google Scholar]
- Loney ER, Inglis PW, Sharp S, Pryde FE, Kent NA, Mellor J, Louis EJ (2009) Repressive and non-repressive chromatin at native telomeres in Saccharomyces cerevisiae. Epigenetics Chromatin 2: 18. [Europe PMC free article] [Abstract] [Google Scholar]
- Louis EJ, Haber JE (1990) The subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics 124: 533–545 [Europe PMC free article] [Abstract] [Google Scholar]
- Louis EJ, Haber JE (1992) The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 131: 559–574 [Europe PMC free article] [Abstract] [Google Scholar]
- Louis EJ, Naumova ES, Lee A, Naumov G, Haber JE (1994) The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics 136: 789–802 [Europe PMC free article] [Abstract] [Google Scholar]
- Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360 [Abstract] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B (2006) A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70: 583–604 [Europe PMC free article] [Abstract] [Google Scholar]
- Mak HC, Pillus L, Ideker T (2009) Dynamic reprogramming of transcription factors to and from the subtelomere. Genome Res 19: 1014–1025 [Europe PMC free article] [Abstract] [Google Scholar]
- Mamnun YM, Pandjaitan R, Mahe Y, Delahodde A, Kuchler K (2002) The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol Microbiol 46: 1429–1440 [Abstract] [Google Scholar]
- Marciniak RA, Cavazos D, Montellano R, Chen Q, Guarente L, Johnson FB (2005) A novel telomere structure in a human alternative lengthening of telomeres cell line. Cancer Res 65: 2730–2737 [Abstract] [Google Scholar]
- Maxwell PH, Coombes C, Kenny AE, Lawler JF, Boeke JD, Curcio MJ (2004) Ty1 mobilizes subtelomeric Y′ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol Cell Biol 24: 9887–9898 [Europe PMC free article] [Abstract] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA (2007) MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biology 5: e261. [Europe PMC free article] [Abstract] [Google Scholar]
- Michan S, Sinclair D (2007) Sirtuins in mammals: insights into their biological function. Biochem J 404: 1–13 [Europe PMC free article] [Abstract] [Google Scholar]
- Pryde FE, Huckle TC, Louis EJ (1995) Sequence analysis of the right end of chromosome XV in Saccharomyces cerevisiae: an insight into the structural and functional significance of sub-telomeric repeat sequences. Yeast (Chichester, England) 11: 371–382 [Abstract] [Google Scholar]
- Pryde FE, Louis EJ (1997) Saccharomyces cerevisiae telomeres. A review. Biochemistry 62: 1232–1241 [Abstract] [Google Scholar]
- Pryde FE, Louis EJ (1999) Limitations of silencing at native yeast telomeres. EMBO J 18: 2538–2550 [Europe PMC free article] [Abstract] [Google Scholar]
- Ratushny AV, Ramsey SA, Roda O, Wan Y, Smith JJ, Aitchison JD (2008) Control of transcriptional variability by overlapping feed-forward regulatory motifs. Biophys J 95: 3715–3723 [Europe PMC free article] [Abstract] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [Abstract] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [Abstract] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol 16: 939–945 [Abstract] [Google Scholar]
- Rottensteiner H, Hartig A, Hamilton B, Ruis H, Erdmann R, Gurvitz A (2003) Saccharomyces cerevisiae Pip2p-Oaf1p regulates PEX25 transcription through an adenine-less ORE. Eur J Biochem 270: 2013–2022 [Abstract] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [Abstract] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193 [Abstract] [Google Scholar]
- Schuller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43: 139–160 [Abstract] [Google Scholar]
- Shampay J, Szostak JW, Blackburn EH (1984) DNA sequences of telomeres maintained in yeast. Nature 310: 154–157 [Abstract] [Google Scholar]
- Smith JJ, Marelli M, Christmas RH, Vizeacoumar FJ, Dilworth DJ, Ideker T, Galitski T, Dimitrov K, Rachubinski RA, Aitchison JD (2002) Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol 158: 259–271 [Europe PMC free article] [Abstract] [Google Scholar]
- Smith JJ, Ramsey SA, Marelli M, Marzolf B, Hwang D, Saleem RA, Rachubinski RA, Aitchison JD (2007) Transcriptional responses to fatty acid are coordinated by combinatorial control. Mol Syst Biol 3: 115; DOI: 10.1038/msb4100157 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Smith JJ, Sydorskyy Y, Marelli M, Hwang D, Bolouri H, Rachubinski RA, Aitchison JD (2006) Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol Syst Biol 2: 2006.0009; DOI: 10.1038/msb4100051 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Soussi-Boudekou S, Vissers S, Urrestarazu A, Jauniaux JC, Andre B (1997) Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol 23: 1157–1168 [Abstract] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93 [Abstract] [Google Scholar]
- Tan K, Feizi H, Luo C, Fan SH, Ravasi T, Ideker TG (2008) A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci USA 105: 2934–2939 [Europe PMC free article] [Abstract] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421 [Abstract] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR (2006) Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell 23: 219–229 [Abstract] [Google Scholar]
- Yamada M, Hayatsu N, Matsuura A, Ishikawa F (1998) Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J Biol Chem 273: 33360–33366 [Abstract] [Google Scholar]
- Zhu X, Gustafsson CM (2009) Distinct differences in chromatin structure at subtelomeric X and Y′ elements in budding yeast. PloS One 4: e6363. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Molecular Systems Biology are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/msb.2010.110
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3049408?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/msb.2010.110
Article citations
Telomeres are shorter in wild Saccharomyces cerevisiae isolates than in domesticated ones.
Genetics, 223(3):iyac186, 01 Mar 2023
Cited by: 4 articles | PMID: 36563016 | PMCID: PMC9991508
Spt3 and Spt8 Are Involved in the Formation of a Silencing Boundary by Interacting with TATA-Binding Protein.
Biomolecules, 13(4):619, 30 Mar 2023
Cited by: 1 article | PMID: 37189367 | PMCID: PMC10136053
A novel yeast hybrid modeling framework integrating Boolean and enzyme-constrained networks enables exploration of the interplay between signaling and metabolism.
PLoS Comput Biol, 17(4):e1008891, 09 Apr 2021
Cited by: 11 articles | PMID: 33836000 | PMCID: PMC8059808
RNA Sequencing Reveals Specific TranscriptomicSignatures Distinguishing Effects of the [SWI⁺] Prion and SWI1 Deletion in Yeast Saccharomyces cerevisiae.
Genes (Basel), 10(3):E212, 12 Mar 2019
Cited by: 7 articles | PMID: 30871095 | PMCID: PMC6471900
Gene regulation in Kluyveromyces marxianus in the context of chromosomes.
PLoS One, 13(1):e0190913, 18 Jan 2018
Cited by: 2 articles | PMID: 29346402 | PMCID: PMC5773181
Go to all (17) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE21852
- (1 citation) GEO - GSE21926
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sir2 and Reb1 antagonistically regulate nucleosome occupancy in subtelomeric X-elements and repress TERRAs by distinct mechanisms.
PLoS Genet, 18(9):e1010419, 22 Sep 2022
Cited by: 2 articles | PMID: 36137093 | PMCID: PMC9531808
Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast.
J Cell Biol, 192(3):417-431, 01 Feb 2011
Cited by: 46 articles | PMID: 21300849 | PMCID: PMC3101097
A SIR-independent role for cohesin in subtelomeric silencing and organization.
Proc Natl Acad Sci U S A, 116(12):5659-5664, 06 Mar 2019
Cited by: 7 articles | PMID: 30842278 | PMCID: PMC6431164
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae.
Genetics, 203(4):1563-1599, 01 Aug 2016
Cited by: 88 articles | PMID: 27516616 | PMCID: PMC4981263
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: U54 RR022220
NIGMS NIH HHS (3)
Grant ID: P50 GM076547
Grant ID: P50GM076547
Grant ID: R01 GM075152




