Abstract
Free full text

COT/MAP3K8 drives resistance to RAF inhibition through MAP kinase pathway reactivation
Associated Data
Abstract
Oncogenic mutations in the serine/threonine kinase B-RAF are found in 50–70% of malignant melanomas1. Pre-clinical studies have demonstrated that the B-RAFV600E mutation predicts a dependency on the mitogen activated protein kinase (MAPK) signaling cascade in melanoma1–5—an observation that has been validated by the success of RAF and MEK inhibitors in clinical trials6–8. However, clinical responses to targeted anticancer therapeutics are frequently confounded by de novo or acquired resistance9–11. Identification of resistance mechanisms in a manner that elucidates alternative ‘druggable’ targets may inform effective long-term treatment strategies12. Here, we expressed ~600 kinase and kinase-related open reading frames (ORFs) in parallel to functionally interrogate resistance to a selective RAF kinase inhibitor. We identified MAP3K8 (COT/TPL2) as a MAPK pathway agonist that drives resistance to RAF inhibition in B-RAFV600E cell lines. COT activates ERK primarily through MEK-dependent mechanisms that do not require RAF signaling. Moreover, COT expression is associated with de novo resistance in B-RAFV600E cultured cell lines and acquired resistance in melanoma cells and tissue obtained from relapsing patients following treatment with MEK or RAF inhibition. We further identify combinatorial MAPK pathway inhibition or targeting of COT kinase activity as possible therapeutic strategies for reducing MAPK pathway activation in this setting. Together, these results provide new insights into resistance mechanisms involving the MAPK pathway and articulate an integrative approach through which high-throughput functional screens may inform the development of novel therapeutic strategies.
To identify kinases capable of circumventing RAF inhibition, we assembled and stably expressed 597 sequence-validated kinase ORF clones representing ~75% of annotated kinases (Center for Cancer Systems Biology (CCSB)/Broad Institute Kinase ORF Collection) in A375, a B-RAFV600E malignant melanoma cell line that is sensitive to the RAF kinase inhibitor PLX472013 (Fig. 1a, 1b, Supplementary Table 1, Supplementary Fig. 2). ORF-expressing cells treated with 1 µM PLX4720 were screened for viability relative to untreated cells and normalized to an assay-specific positive control, MEK1S218/222D (MEK1DD)14 (Supplementary Table 2 and summarized in Supplementary Fig. 1). Nine ORFs conferred resistance at levels exceeding two standard deviations from the mean (Fig. 1b and Supplementary Table 2) and were selected for follow-up analysis (Supplementary Fig. 3). Three of nine candidate ORFs were receptor tyrosine kinases, underscoring the potential of this class of kinases to engage resistance pathways. Resistance effects were validated and prioritized across a multi-point PLX4720 drug concentration scale in the B-RAFV600E cell lines A375 and SKMEL28. The Ser/Thr MAP kinase kinase kinases (MAP3Ks) MAP3K8 (COT/Tpl2) and RAF1 (C-RAF) emerged as top candidates from both cell lines; these ORFs shifted the PLX4720 GI50 by 10-600 fold without affecting viability (Supplementary Table 3 and Supplementary Fig. 4 and 5). Both COT and C-RAF reduced sensitivity to PLX4720 in multiple B-RAFV600E cell lines (Fig. 1c) confirming the ability of these kinases to mediate resistance to RAF inhibition.
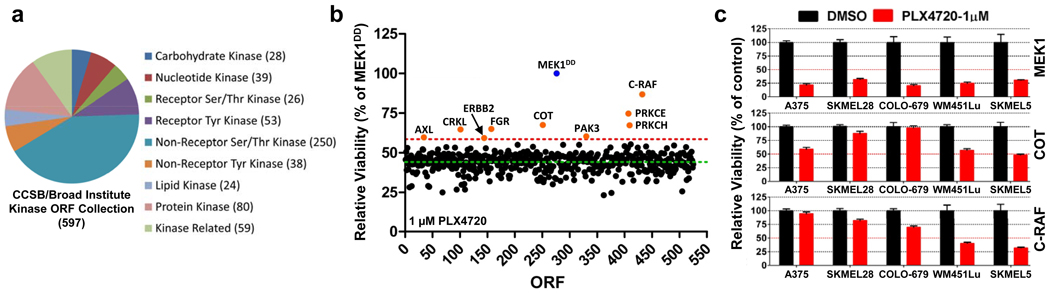
Overview of the CCSB/Broad Institute Kinase ORF collection. Kinase classification and number of kinases per classification are noted.
A375 expressing the CCSB/Broad Institute Kinase ORF collection were assayed for relative viability in 1 µM PLX4720 and normalized to constitutively active MEK1 (MEK1DD). Nine ORFs (orange circles) scored 2 standard deviations (red dashed line, 58.64%) from the mean of all ORFs (green dashed line, 44.26%).
Indicated ORFs were expressed in 5 B-RAFV600E cell lines and treated with DMSO or 1 µM PLX4720. Viability (relative to DMSO) was quantified after 4 days. Error bars represent standard deviation between replicates (n=6).
Next, we tested whether overexpression of these genes was sufficient to activate the MAPK pathway. At baseline, COT expression increased ERK phosphorylation in a manner comparable to MEK1DD, consistent with MAP kinase pathway activation (Fig. 2a and Supplementary Fig. 6). Overexpression of wild-type COT or C-RAF resulted in constitutive phosphorylation of ERK and MEK in the presence of PLX4720, whereas kinase-dead derivatives had no effect (Fig. 2a, Supplementary Fig. 7). Based on these results, we hypothesized that COT and C-RAF drive resistance to RAF inhibition predominantly through re-activation of MAPK signaling. Notably, of the nine candidate ORFs from our initial screen, a subset (3) did not show persistent ERK/MEK phosphorylation following RAF inhibition, suggesting MAPK pathway-independent alteration of drug sensitivity (Supplementary Fig. 8).
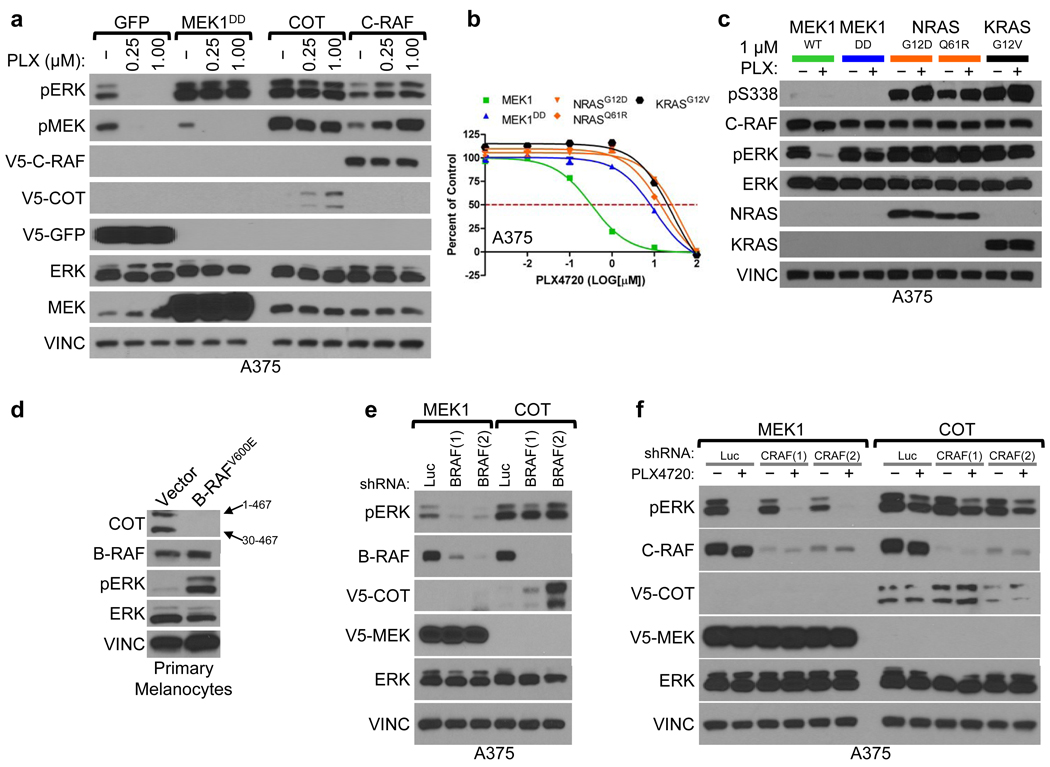
Indicated ORFs were expressed in A375. Levels of phosphorylated MEK and ERK were assayed following 18 h. treatment with DMSO (−) or PLX4720 (concentration noted).
Proliferation of A375 expressing indicated ORFs. Error bars represent standard deviation between replicates (n=6).
C-RAF (S338) and ERK phosphorylation in lysates from A375 expressing indicated ORFs.
COT expression in lysates from immortalized primary melanocytes expressing BRAFV600E or empty vector. COT mRNA has an internal start codon (30M) resulting in two protein products of different lengths; amino acids 1–467 or 30–467, noted with arrows.
COT and ERK phosphorylation in lysates from A375 expressing indicated ORFs following shRNA-mediated B-RAF depletion (shBRAF) relative to control shRNA (shLuc).
ERK phosphorylation in lysates from A375 expressing indicated ORFs following shRNA-mediated C-RAF depletion (shCRAF) or control shRNA (shLuc), following 18 h. treatment with DMSO (−) or 1 µM PLX4720 (+).
Several groups have shown that C-RAF activation and heterodimerization with B-RAF constitute critical components of the cellular response to B-RAF inhibition15–18. In A375 cells, endogenous C-RAF: B-RAF heterodimers were measurable and inducible following treatment with PLX4720 (Supplementary Fig. 9). However, endogenous C-RAF phosphorylation at S338—an event required for C-RAF activation—remained low (Supplementary Fig. 9). In contrast, ectopically expressed C-RAF was phosphorylated on S338 (Supplementary Fig. 9) and its PLX4720 resistance phenotype was associated with sustained MEK/ERK activation (Fig. 2a, Supplementary Fig. 9). Moreover, ectopic expression of a high-activity C-RAF truncation mutant (C-RAF(W22) was more effective than wild-type C-RAF in mediating PLX4720 resistance and ERK activation (Supplementary Fig. 10), further indicating that elevated C-RAF activity may direct resistance to this agent. Consistent with this model, oncogenic alleles of NRAS and KRAS conferred PLX4720 resistance in A375 cells (Fig. 2b) and yielded sustained C-RAF(S338) and ERK phosphorylation in the context of drug treatment (Fig. 2c). Thus, although genetic alterations that engender C-RAF activation (e.g., oncogenic RAS mutations) tend to show mutual exclusivity with B-RAFV600E mutation, such co-occurring events19, 20 might be favored in the context of acquired resistance to B-RAF inhibition.
To investigate the role of COT in melanoma, we first determined its expression in human melanocytes. We found that primary immortalized melanocytes (B-RAF wild-type) expressed COT (Fig. 2d), although ectopic B-RAFV600E expression reduced COT mRNA levels (Supplementary Fig. 11) and rendered COT protein undetectable (Fig. 2d). Conversely, whereas ectopically expressed COT was only weakly detectable in A375 cells (Fig. 2a, 2e), shRNA-mediated depletion of endogenous B-RAFV600E caused an increase in COT protein levels that correlated with the extent of B-RAF knockdown (Fig. 2e). Moreover, treatment of COT-expressing A375 cells with PLX4720 led to a dose-dependent increase in COT protein (Fig. 2a) without affecting ectopic COT mRNA levels (Supplementary Fig. 11). Thus, oncogenic B-RAF may antagonize COT expression largely through altered protein stability (Fig. 2a, d, e, and Supplementary Fig. 11), and B-RAF inhibition may potentiate the outgrowth of COT-expressing cells during the course of treatment. Notably, neither C-RAF nor B-RAF alone or in combination was required for ERK phosphorylation in the context of COT expression, even in the presence of PLX4720 (Figure 2e, 2f and Supplementary Fig. 12), suggesting that COT expression is sufficient to induce MAP kinase pathway activation in a RAF-independent manner.
We predicted that cell lines expressing elevated COT in a B-RAFV600E background should exhibit de novo resistance to PLX4720 treatment. To identify such instances, we screened a panel of cell lines for evidence of MAP3K8/COT copy number gains coincident with the B-RAFV600E mutation. Of 534 cell lines that had undergone copy number analysis and mutation profiling, 38 cell lines (7.1%) contained the B-RAFV600E mutation. Within this subgroup, two cell lines—OUMS-23 (colon cancer) and RPMI-7951 (melanoma)—also showed evidence of chromosomal copy gains spanning the MAP3K8/COT locus (Fig. 3a, Supplementary Fig. 13) and robust COT protein expression (Fig. 3b, Supplementary Fig. 14). We also screened a panel of melanoma short-term cultures for COT protein expression. Only one of these lines expressed COT: M307, a short-term culture derived from a B-RAFV600E tumor that developed resistance to allosteric MEK inhibition following initial disease stabilization14 (Fig. 3c). All three cell lines were refractory to PLX4720 treatment, exhibiting GI50 values in the range of 8–10 µM (Fig. 3d) and showing sustained ERK phosphorylation in the context of B-RAF inhibition (Figs. 3e, 3f). OUMS-23 and RPMI-7951 are MAPK pathway inhibitor-naïve cell lines, implying that COT may confer de novo resistance to RAF inhibition (a phenomenon observed in ~10% of B-RAFV600E melanomas6).
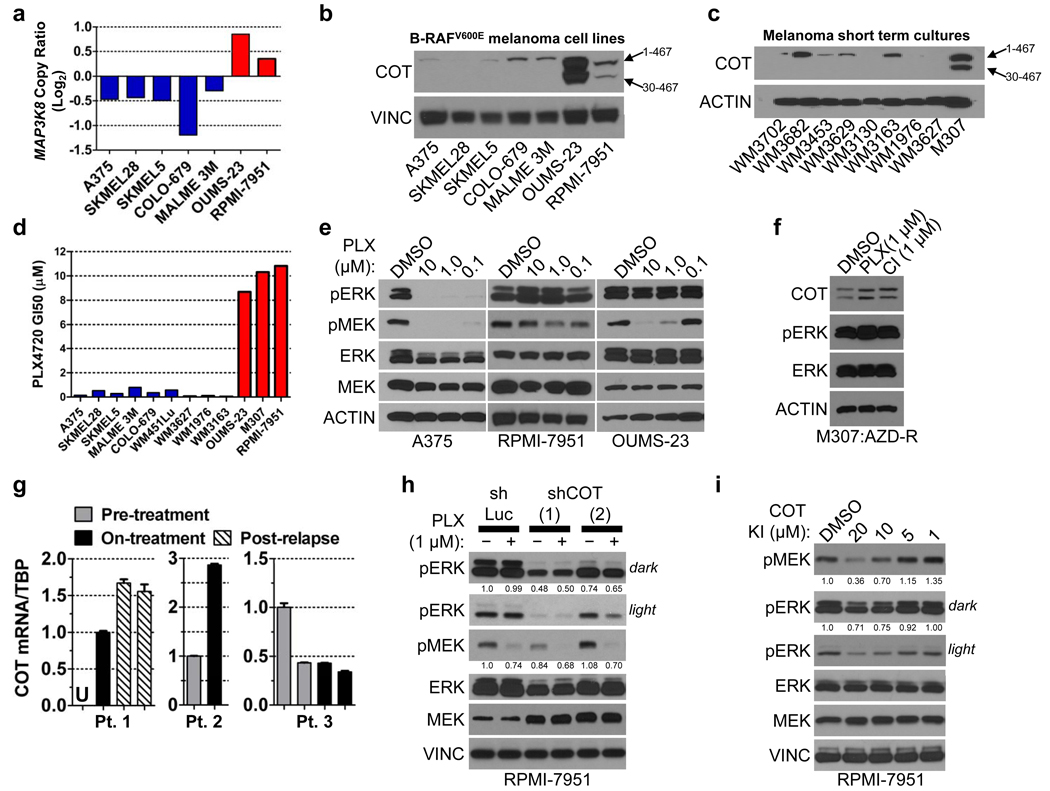
MAP3K8/COT copy number; red bars: COT amplification, blue bars: non-amplified COT.
COT expression in B-RAFV600E cell lines and
short-term cultures.
PLX4720 GI50 in B-RAFV600E cell lines. Colors as in (a).
MEK and ERK phosphorylation following treatment with DMSO or PLX4720 (concentration indicated).
ERK phosphorylation in M307 lysates (AZD-R; AZD6244-resistant) treated with DMSO or 1 µM PLX4720 (PLX) or CI-1040 (CI).
COT mRNA expression (QRT/PCR) in patient/lesion-matched PLX4032-treated metastatic melanoma tissue samples. Pts. 1 and 3 had multiple biopsies from the same lesion. Error bars represent SEM (n=3). U; undetermined/undetectable.
ERK and MEK phosphorylation in RPMI-7951 following shRNA-mediated COT depletion (shCOT) versus control (shLuc) and treatment with DMSO (−) or 1 µM PLX4720 (+). ERK and MEK phosphorylation are quantified.
ERK and MEK phosphorylation in RPMI-7951 following 1 h. treatment with a small molecule COT kinase inhibitor. ERK and MEK phosphorylation are quantified.
Next, we examined COT expression in the context of resistance to the clinical RAF inhibitor PLX4032 by obtaining biopsy material from 3 patients with metastatic, B-RAFV600E melanoma. Each case consisted of frozen, lesion-matched biopsy material obtained prior to and during treatment (“pre-treatment” and “on-treatment”; Fig. 3g, Supplementary Table 4); additionally, one sample contained two independent biopsy specimens from the same relapsing tumor site (“post-relapse”; Fig. 3g). Consistent with the experimental models presented above, quantitative real-time RT-PCR (qRT/PCR) analysis revealed increased COT mRNA expression concurrent with PLX4032 treatment in 2 of 3 cases. COT mRNA levels were further increased in a relapsing specimen relative to its pre-treatment and on-treatment counterparts (Fig. 3g, Patient #1). An additional, unmatched relapsed malignant melanoma biopsy showed elevated COT mRNA expression comparable to levels observed in RAF inhibitor-resistant, COT-amplified cell lines (Supplementary Fig. 15). This specimen also exhibited robust MAPK pathway activation and elevated expression of B-RAF, C-RAF and COT relative to matched normal skin or B-RAFV600E cell lines (Supplementary Fig. 15). Sequencing studies of this tumor revealed no additional mutations in BRAF, NRAS or KRAS (data not shown). These analyses provided clinical evidence that COT-dependent mechanisms may be operant in at least some PLX4032-resistant malignant melanomas.
To determine if COT might actively regulate MEK/ERK phosphorylation in B-RAFV600E cells that harbor naturally elevated COT expression, we introduced shRNA constructs targeting COT into RPMI-7951 cells. Depletion of COT suppressed RPMI-7951 viability (Supplementary Fig. 16) and decreased ERK phosphorylation (Fig. 3h), implying that targeting COT kinase activity might suppress MEK/ERK phosphorylation in cancer cells with COT overexpression or amplification. Treatment of RPMI-7951 cells with a small molecule COT kinase inhibitor21–23 resulted in dose-dependent suppression of MEK and ERK phosphorylation, providing additional evidence that COT contributes to MEK/ERK activation in these cells (Fig. 3i).
We then considered whether COT-expressing cancer cells remain sensitive to MAPK pathway inhibition at a target downstream of COT or RAF. Here, we queried the OUMS-23 and RPMI-7951 cell lines for sensitivity to the MEK1/2 inhibitor CI-1040. Interestingly, both cell lines were refractory to MEK inhibition (Fig. 4a) and displayed sustained ERK phosphorylation even at 1 µM CI-1040 (Fig. 4b). Ectopic COT expression in A375 and SKMEL28 cells also conferred decreased sensitivity to the MEK inhibitors CI-1040 and AZD6244, suggesting that COT expression alone was sufficient to induce this phenotype (Fig. 4c, 4d, Supplementary Fig. 17). Similar to results observed with pharmacological MEK inhibitors, MEK1/2 knockdown only modestly suppressed COT–mediated ERK phosphorylation in A375 cells (Supplementary Fig. 18). In accordance with prior observations24, these data raised the possibility that COT may activate ERK through MEK-independent and MEK-dependent mechanisms. To test this hypothesis directly, we performed an in vitro kinase assay using recombinant COT and ERK1. Indeed, recombinant COT induced pThr202/Tyr204 phosphorylation of ERK1 in vitro (Supplementary Fig. 18) suggesting that in certain contexts, COT expression may potentiate ERK activation in a MEK-independent manner.
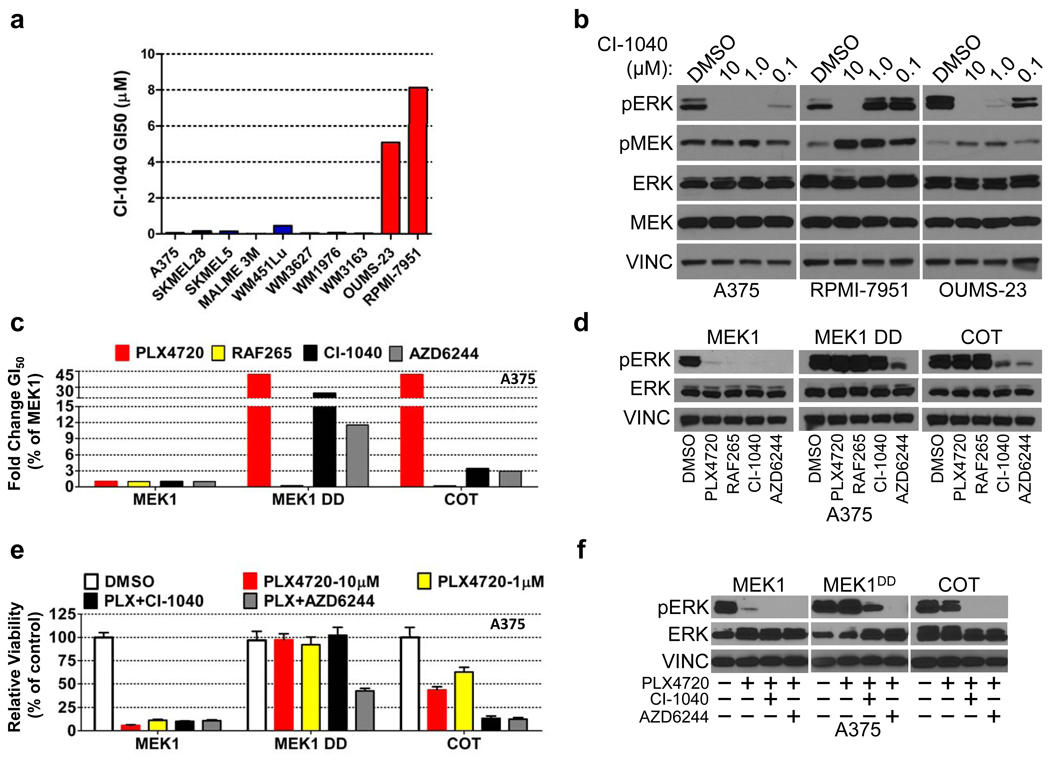
CI-1040 GI50 in a panel of B-RAFV600E cell lines; red bars: COT expression/amplification, blue bars: undetectable/non-amplified COT.
MEK and ERK phosphorylation in lysates from indicated cell lines treated with DMSO or CI-1040 (concentration noted).
Fold change (relative to MEK1) GI50 of A375 ectopically expressing the indicated ORFs for PLX4720, RAF265, CI-1040 and AZD6244.
ERK phosphorylation in A375 expressing indicated ORFs following treatment with DMSO or 1 µM of PLX4720, RAF265, CI-1040 or AZD6244.
Viability of A375 expressing the indicated ORFs and treated with DMSO, PLX4720 (concentration indicated) and PLX4720 in combination with CI-1040 or AZD6244 (all 1 µM). Error bars represent the standard deviation (n=6).
ERK phosphorylation in A375 expressing indicated ORFs following treatment with DMSO, PLX4720 (1 µM) or PLX4720 in combination with CI-1040 or AZD6244 (all 1 µM).
In experimental models, the use of RAF and MEK inhibitors in combination can override resistance to single-agents14. We therefore reasoned that combined RAF/MEK inhibition might circumvent COT-driven resistance. In the setting of ectopic COT expression, exposure to AZD6244 or CI-1040 in combination with PLX470 (1 µM each) reduced cell growth and pERK expression more effectively than did single-agent PLX4720, even at concentrations of 10 µM (Fig. 4e, 4f and Supplementary Fig. 19). These data underscore the importance of this pathway in B-RAFV600E tumor cells and support earlier findings14 that dual B-RAF/MEK inhibition may help circumvent resistance to RAF inhibitors.
B-RAF mutations are found in ~8% of all cancers and at high frequencies in malignant melanoma, colon and thyroid cancers25. The clinical promise of selective RAF inhibitors has widespread ramifications for patient treatment, yet single agent targeted therapy is almost invariably followed by relapse due to acquired drug resistance. Our results suggest that ORF-based, systematic functional screening may offer a powerful means to identify clinically relevant resistance mechanisms that also specify novel treatment strategies. In particular, resistance to RAF inhibition can be achieved by multiple MAP3K-dependent mechanisms of MEK/ERK reactivation but might be intercepted through combined therapeutic modalities for MAPK pathway inhibition (e.g., RAF/MEK or RAF/COT combinations). Future systematic drug resistance studies may be expanded to a genome scale that encompasses many compounds, thereby enabling comprehensive identification of both therapy-specific resistance genes and drug targets of novel therapeutics.
Methods summary
The arrayed, lentiviral ORF screen was performed as previously described26. Effects of individual ORFs on drug resistance were determined by measuring differential viability (ratio of raw viability in 1 µM PLX4720: control) and subsequent normalization to an assay-specific positive control, MEK1DD. Secondary screens were performed with the top nine candidate ORFs in 96-well format in A375 and SKMEL28 cells. Prioritization was accomplished via generation of a GI50 for each ORF across a multi-point PLX4720 concentration range in both cell lines. The effects of identified resistance ORFs on MAPK pathway activation were demonstrated using both biochemical and cell biological approaches. Cell line copy number data was obtained as previously described27. Detailed descriptions of all procedures are included in Materials and Methods.
Materials and Methods
Center for Cancer Systems Biology (CCSB)/Broad Institute Kinase Open Reading Frame Collection
We assembled a library of 597 kinase ORFs in pDONR-223 Entry vectors (Invitrogen). Individual clones were end-sequenced using vector-specific primers in both directions. Clones with substantial deviations from reported sequences were discarded. Entry clones and sequences are available via Addgene (www.addgene.org/human_kinases). Kinase ORFs were assembled from multiple sources; 337 kinases were isolated as single clones from the ORFeome 5.1 collection (ude.dravrah.icfd.bdfroh); 183 kinases were cloned from normal human tissue RNA (Ambion) by reverse transcription and subsequent PCR amplification to add Gateway sequences (Invitrogen); 64 kinases were cloned from templates provided by the Harvard Institute of Proteomics (HIP); and 13 kinases were cloned into the Gateway system from templates obtained from collaborating laboratories. The Gateway-compatible lentiviral vector pLX-Blast-V5 was created from the pLKO.1 backbone. LR reactions were performed to introduce the 597 kinases into pLX-Blast-V5 according to manufacturers protocol (Invitrogen).
High throughout ORF screening
A375 melanoma cells were plated in 384-well microtiter plates (500 cells per well). The following day, cells were spin-infected with the lentivirally-packaged kinase ORF library in the presence of 8 ug/ml polybrene. 48 hours post-infection, media was replaced with standard growth media (2 replicates), media containing 1 µM PLX4720 (2 replicates, 2 time points) or media containing 10 ug/ml blasticidin (2 replicates). After four days and 6 days, cell growth was assayed using Cell Titer-Glo (Promega) according to manufacturer instructions. The entire experiment was performed twice.
Identification of candidate resistance ORFs
Raw luminescence values were imported into Microsoft Excel. Infection efficiency was determined by the percentage of duplicate-averaged raw luminescence in blasticidin selected cells relative to non-selected cells. ORFs with an infection efficiency of less than 0.70 were excluded from further analysis along with any ORF having a standard deviation of >15,000 raw luminescence units between duplicates. To identify ORFs whose expression affects proliferation, we compared the duplicate-averaged raw luminescence of individual ORFs against the average and standard deviation of all control-treated cells via the z-score, or standard score, below,
where x = average raw luminescence of a given ORF, μ = the mean raw luminescence of all ORFs and σ = the standard deviation of the raw luminescence of all wells. Any individual ORF with a z-score >+2 or <−2 was annotated as affecting proliferation and removed from final analysis. Differential proliferation was determined by the percentage of duplicate-averaged raw luminescence values in PLX4720 (1 µM) treated cells relative to untreated cells. Subsequently, differential proliferation was normalized to the positive control for PLX4720 resistance, MEK1S218/222D (MEK1DD), with MEK1DD differential proliferation = 1.0. MEK1DD normalized differential proliferation for each individual ORF was averaged across two duplicate experiments, with two time points for each experiment (day 4 and day 6). A z-score was then generated, as described above, for average MEK1DD normalized differential proliferation. ORFs with a z-score of >2 were considered hits and were followed up in the secondary screen.
ORF and shRNA expression
ORFs were expressed from pLX-Blast-V5 (lentiviral) or pWZL-Blast, pBABE-Puro or pBABE-zeocin (retroviral) expression plasmids. For lentiviral transduction, 293T cells were transfected with 1 µg of pLX-Blast-V5-ORF or pLKO.1-shRNA, 900 ng Δ8.9 (gag, pol) and 100 ng VSV-G using 6 µl Fugene6 transfection reagent (Roche). Viral supernatant was harvested 72 h post-transfection. Mammalian cells were infected at a 1:10–1:20 dilution of virus in 6-well plates in the presence of 5 µg/ml polybrene and centrifuged at 2250 RPM for 1 h at 37° C. Twenty-four hours after infection blasticidin (pLX-Blast-V5, 10 µg/ml) or puro (pLKO.1, 0.75 µg/ml) was added and cells were selected for 48 hrs. For retrovirus production, 293T were transfected with 1 µg of retroviral plasmid-ORF, 1 µg pCL-AMPHO and 100 ng VSV-G, as described above. Cells were infected with retrovirus containing supernatant at a 1:2 dilution in 5 µg/ml polybrene overnight, followed by media change to growth medium. Infection was repeated once more (twice total), followed by selection, above.
Secondary Screen
A375 (1.5 × 103) and SKMEL28 cells (3 × 103) were seeded in 96-well plates for 18 h. ORF-expressing lentivirus was added at a 1:10 dilution in the presence of 8 µg/ml polybrene, and centrifuged at 2250 RPM and 37° C for 1 h. Following centrifugation, virus-containing media was changed to normal growth media and allowed to incubate for 18 h. Twenty-four hours after infection, DMSO (1:1000) or 10× PLX4720 (in DMSO) was added to a final concentration of 100, 10, 1, 0.1, 0.01, 0.001, 0.0001 or 0.00001 µM. Cell viability was assayed using WST-1 (Roche), per manufacturer recommendation, 4 days after the addition of PLX4720.
Cell lines and reagents
A375, SKMEL28, SKMEL30, COLO-679, WM451lu, SKMEL5, Malme 3M, SKMEL30, WM3627, WM1976, WM3163, WM3130, WM3629, WM3453, WM3682 and WM3702 were all grown in RPMI (Cellgro), 10% FBS and 1% penicillin/streptomycin. M307 was grown in RPMI (Cellgro), 10% FBS and 1% penicillin/streptomycin supplemented with 1 mM sodium pyruvate. 293T and OUMS-23 were grown in DMEM (Cellgro), 10% FBS and 1% penicillin/streptomycin. RPMI-7951 cells (ATCC) were grown in MEM (Cellgro), 10% FBS and 1% penicillin/streptomycin. Wild-type primary melanocytes were grown in HAM’s F10 (Cellgro), 10% FBS and 1% penicillin/streptomycin. B-RAFV600E-expressing primary melanocytes were grown in TIVA media [Ham’s F-10 (Cellgro), 7% FBS, 1% penicillin/streptomycin, 2mM glutamine (Cellgro), 100 uM IBMX, 50 ng/ml TPA, 1mM dbcAMP (Sigma) and 1 µM sodium vanadate]. CI-1040 (PubChem ID: 6918454) was purchased from Shanghai Lechen International Trading Co., AZD6244 (PubChem ID: 10127622) from Selleck Chemicals, and PLX4720 (PubChem ID: 24180719) from Symansis. RAF265 (PubChem ID: 11656518) was a generous gift from Novartis Pharma AG. Unless otherwise indicated, all drug treatments were for 16 h. Activated alleles of NRAS and KRAS have been previously described28, 29.
Pharmacologic Growth Inhibition Assays
Cultured cells were seeded into 96-well plates (3,000 cells per well) for all melanoma cell lines; 1,500 cells were seeded for A375. Twenty-four hours after seeding, serial dilutions of the relevant compound were prepared in DMSO added to cells, yielding final drug concentrations ranging from 100 µM to 1 × 105 µM, with the final volume of DMSO not exceeding 1%. Cells were incubated for 96 h following addition of drug. Cell viability was measured using the WST1 viability assay (Roche). Viability was calculated as a percentage of control (untreated cells) after background subtraction. A minimum of six replicates were performed for each cell line and drug combination. Data from growth-inhibition assays were modeled using a nonlinear regression curve fit with a sigmoid dose–response. These curves were displayed and GI50 generated using GraphPad Prism 5 for Windows (GraphPad). Sigmoid-response curves that crossed the 50% inhibition point at or above 10 µM have GI50 values annotated as >10 µM. For single-dose studies, the identical protocol was followed, using a single dose of indicated drug (1 µM unless otherwise noted).
Immunoblots and immunoprecipitations
Cells were washed twice with ice-cold PBS and lysed with 1% NP-40 buffer [150 mM NaCl, 50 mM Tris pH 7.5, 2 mM EDTA pH 8, 25 mM NaF and 1% NP-40] containing 2× protease inhibitors (Roche) and 1× Phosphatase Inhibitor Cocktails I and II (CalBioChem). Lysates were quantified (Bradford assay), normalized, reduced, denatured (95 °C) and resolved by SDS gel electrophoresis on 10% Tris/Glycine gels (Invitrogen). Protein was transferred to PVDF membranes and probed with primary antibodies recognizing pERK1/2 (T202/Y204), pMEK1/2 (S217/221), MEK1/2, MEK1, MEK2, C-RAF (rabbit host), pC-RAF (pS338) (Cell Signaling Technology; 1:1,000), V5-HRP (Invitrogen; (1:5,000), COT (1:500), B-RAF (1:2,000), Actin (1:1,000), Actin-HRP (1:1,000; Santa Cruz)), C-RAF (mouse host; 1:1,000; BD Transduction Labs), Vinculin (Sigma; 1:20,000), AXL (1:500; R&D Systems). After incubation with the appropriate secondary antibody (anti-rabbit, anti-mouse IgG, HRP-linked; 1:1,000 dilution, Cell Signaling Technology or anti-goat IgG, HRP-linked; 1:1,000 dilution; Santa Cruz), proteins were detected using chemiluminescence (Pierce). Immunoprecipitations were performed overnight at 4° C in 1% NP-40 lysis buffer, as described above, at a concentration of 1 µg/μl total protein using an antibody recognizing C-RAF (1:50; Cell Signaling Technology). Antibody: antigen complexes were bound to Protein A agarose (25 µL, 50% slurry; Pierce) for 2 hrs. at 4° C. Beads were centrifuged and washed three times in lysis buffer and eluted and denatured (95 °C) in 2× reduced sample buffer (Invitrogen). Immunoblots were performed as above. Phospho-protein quantification was performed using NIH Image J.
Lysates from tumor and matched normal skin were generated by mechanical homogenization of tissue in RIPA [50 mM Tris (pH 7.4), 150 mM NaCl, 1mM EDTA, 0.1% SDS, 1.0% NaDOC, 1.0% Triton X-100, 25 mM NaF, 1mM NA3VO4] containing protease and phosphatase inhibitors, as above. Subsequent normalization and immunoblots were performed as above.
Biopsied melanoma tumor material
Biopsied tumor material consisted of discarded and de-identified tissue that was obtained with informed consent and characterized under protocol 02-017 (paired samples, Massachusetts General Hospital) and 07-087 (unpaired sample, Dana-Farber Cancer Institute). For paired specimens, ‘on-treatment’ samples were collected 10–14 days after initiation of PLX4032 treatment (Supplementary Table 4).
Inhibition of COT kinase activity
Adherent RPMI-7951 cells were washed twice with 1× PBS and incubated overnight in serum-free growth media. Subsequently, 4-(3-Chloro-4-fluorophenylamino)-6-(pyridin-3-yl-methylamino)-3-cyano-[1,7]-naphthyridine (EMD;TPL2 inhibitor I; Cat#: 616373, PubChem ID: 9549300), suspended in DMSO at the indicated concentration, was added to cells for 1 hour, after which protein extracts were made as described above.
Quantitative RT/PCR
mRNA was extracted from cell lines and fresh-frozen tumors using the RNeasy kit (Qiagen). Total mRNA was used for subsequent reverse transcription using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen) for cell lines and unpaired tumor samples, and the SuperScript VILO cDNA synthesis kit (Invitrogen) for paired frozen tumor samples. 5 µl of the RT reaction was used for quantitative PCR using SYBR Green PCR Master Mix and gene-specific primers, in triplicate, using an ABI 7300 Real Time PCR System. Primers used for detection are as follows; COT forward: 5’- CAA GTG AAG AGC CAG CAGTTT -3’; COT reverse: 5’- GCA AGC AAATCC TCC ACA GTT C -3’; TBP forward: 5’- CCC GAA ACG CCG AAT ATA ATC C -3’; TBP reverse: 5’- GAC TGT TCT TCA CTC TTG GCT C -3’; GAPDH forward: 5’- CAT CAT CTC TGC CCC CTC T –3’; GAPDH reverse: 5’-GGT GCT AAG CAG TTG GTG GT -3’.
In vitro kinase assay
In vitro kinase assays were performed as previously described14 using 1 µg each of COT (amino acids 30–397, R&D Systems) and inactive ERK1 (Millipore).
Cellular viability assays
Adherent RPMI-7951 cells were infected with virus expressing shRNAs against COT or Luciferase as described above. Following selection, cells were plated (1.5×105 cells/well) onto a 24-well plate in quadruplicate. Viable cells were counted via trypan blue exclusion using a VI-CELL Cell Viability Analyzer, per manufacturer’s specifications. Quadruplicate cell counts were averaged and normalized relative to that of the control shRNA.
The Cancer Cell line encyclopedia (CCLE)
The Cancer Cell line encyclopedia (CCLE) project is a collaboration between the Broad Institute, the Novartis Institutes for Biomedical Research (NIBR) and the Genomics Institute of the Novartis Research Foundation (GNF) to conduct a detailed genetic and pharmacologic characterization of a large panel of human cancer models, to develop integrated computational analyses that link distinct pharmacologic vulnerabilities to genomic patterns and to translate cell line integrative genomics into cancer patient stratification. All OncoMap mutation data and SNP-array derived copy number data is available online at www.broadinstitute.org/ccle.
Expression profiling of cancer cell lines
We carried out oligonucleotide microarray analysis using the GeneChip Human Genome U133 Plus 2.0 Affymetrix expression array (Affymetrix, Santa Clara, CA). Samples were converted to labeled, fragmented, cRNA per the Affymetrix protocol for use on the expression microarray.
shRNA constructs used (pLKO.1)
| shRNA | TRC Identifier | NM No. | Sequence (5'→3') |
|---|---|---|---|
| shLuc | TRCN0000072243 | n/a | CTTCGAAATGTCCGTTCGGTT |
| shBRAF(1) | TRCN0000006289 | NM_004333.2-1106s1c1 | GCAGATGAAGATCATCGAAAT |
| shBRAF(2) | TRCN0000006291 | NM_004333.2-2267s1c1 | GCTGGTTTCCAAACAGAGGAT |
| shCRAF(1) | TRCN0000001066 | NM_002880.x-1236s1c1 | CGGAGATGTTGCAGTAAAGAT |
| shCRAF(2) | TRCN0000001068 | NM_002880.x-1529s1c1 | GAGACATGAAATCCAACAATA |
| shMEK1(1) | TRCN0000002332 | NM_002755.x-1015s1c1 | GATTACATAGTCAACGAGCCT |
| shMEK1(2) | TRCN0000002329 | NM_002755.x-455s1c1 | GCTTCTATGGTGCGTTCTACA |
| shMEK2(1) | TRCN0000007007 | NM_030662.2-1219s1c1 | TGGACTATATTGTGAACGAGC |
| shMEK2(2) | TRCN0000007005 | NM_030662.2-847s1c1 | CCAACATCCTCGTGAACTCTA |
| shCOT(1) | TRCN0000010013 | NM_005204.x-1826s1c1 | CAAGAGCCGCAGACCTACTAA |
| shCOT(2) | TRCN0000196518 | NM_005204.2-2809s1c1 | GATGAGAATGTGACCTTTAAG |
Acknowledgments
We thank members of the Broad Institute/Novartis Cancer Cell Line Encyclopedia (CCLE) for contributing cell line genomic data, expression data and pharmacological cell line sensitivity data; Joseph Thibault and Aaron Shipway for CCLE related tissue culture; Ron Depinho, Gavin Dunn, Stephen Ethier, Heidi Greulich, Andrew Henderson, David Kaplan, Ross Levine, Chris Miller, Helen Piwnica-Worms, Hiroyuki Suzuki, Marc Vigny, Douglas Vollrath and the Harvard Institute of Proteomics for contributing templates for the kinase collection; Jinyan Du and Douglas B. Wheeler for assistance with functional testing of kinases; David A. Barbie for helpful discussions, Susan E. Moody and Hiu Wing Cheung for technical assistance, and Jennifer K. Grenier and Serena J. Silver for compiling and annotating the list of kinases. This work was supported by the NIH Director's New Innovator Award (L.A.G.), grants from the Novartis Institutes for Biomedical Research (L.A.G.), Melanoma Research Alliance (L.A.G.), Starr Cancer Consortium (L.A.G.) the US National Cancer Institute (R33 CA128625, RC2 CA148268) (W.C.H.), NIH (CA134502) (J.J.Z) the Swiss National Foundation (310040-103671) (R.D.), the Gottfried and Julia Bangerter Rhyner Stiftung (R.D.) the Ellison Foundation (M.V. and CCSB) and institute sponsored research funds from the DFCI Strategic Initiative (M.V. and CCSB).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author contributions C.M.J., J.S.B. and L.A.G. designed the experiments, with help from C.M.E. C.M.J., S.Y.K. and L.W. performed primary screen, supervised by W.C.H. L.A.J. helped perform drug sensitivity profiling. J.S.B. designed and created the CCSB/Broad Institute Kinase ORF Collection in collaboration with S.R.T., H.H., R.R.M., K.S-A, J.J.Z., M.V., T.M.R., T.G., D.E.H. and W.C.H. Additional help with ORF experiments from X.P., D.E.R. and O.A. Clinical samples were collected or experiments performed by C.M.J., A.P.C., K.T.F., R.D., J.A.W. Large scale cell genomic and expression profiling along with pharmacological screening efforts were designed and data analyzed by, N.S., J.B., G.C., S.K., J.H., C.J.W., V.E.M., P.M.F., B.W., W.R.S., R.S., L.A.G. C.M.J. and L.A.G. wrote the manuscript. All authors discussed results and edited the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompanying the full-text HTML version of the paper at www.nature.com/nature.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature09627
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3058384
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101771917
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature09627
Article citations
When less is more - a fast TurboID knock-in approach for high-sensitivity endogenous interactome mapping.
J Cell Sci, 137(16):jcs261952, 28 Aug 2024
Cited by: 0 articles | PMID: 39056144 | PMCID: PMC11385326
Dual Chemotherapeutic Loading in Oxalate Transferrin-Conjugated Polymersomes Incorporated into Chitosan Hydrogels for Site-Specific Targeting of Melanoma Cells.
Pharmaceuticals (Basel), 17(9):1177, 06 Sep 2024
Cited by: 0 articles | PMID: 39338339 | PMCID: PMC11434979
Loss of symmetric cell division of apical neural progenitors drives DENND5A-related developmental and epileptic encephalopathy.
Nat Commun, 15(1):7239, 22 Aug 2024
Cited by: 0 articles | PMID: 39174524 | PMCID: PMC11341845
Deciphering NF-kappaB pathways in smoking-related lung carcinogenesis.
EXCLI J, 23:991-1017, 19 Aug 2024
Cited by: 1 article | PMID: 39253534 | PMCID: PMC11382301
Review Free full text in Europe PMC
The TRIM4 E3 ubiquitin ligase degrades TPL2 and is modulated by oncogenic KRAS.
Cell Rep, 43(9):114667, 22 Aug 2024
Cited by: 0 articles | PMID: 39178114 | PMCID: PMC11472288
Go to all (954) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database (Showing 6 of 6)
- (2 citations) RefSeq - NM_030662.2
- (2 citations) RefSeq - NM_004333.2
- (2 citations) RefSeq - NM_002880
- (2 citations) RefSeq - NM_002755
- (1 citation) RefSeq - NM_005204.2
- (1 citation) RefSeq - NM_005204
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation.
Nature, 468(7326):973-977, 24 Nov 2010
Cited by: 1400 articles | PMID: 21107323 | PMCID: PMC3143360
Drug discovery: How melanomas bypass new therapy.
Nature, 468(7326):902-903, 01 Dec 2010
Cited by: 31 articles | PMID: 21164474
Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma.
J Biol Chem, 287(33):28087-28098, 22 Jun 2012
Cited by: 119 articles | PMID: 22730329 | PMCID: PMC3431627
Mechanisms of resistance to RAF inhibition in melanomas harboring a BRAF mutation.
Am Soc Clin Oncol Educ Book, 01 Jan 2013
Cited by: 17 articles | PMID: 23714462
Review
Funding
Funders who supported this work.
NCI NIH HHS (7)
Grant ID: CA134502
Grant ID: K08 CA115927
Grant ID: RC2 CA148268
Grant ID: K08 CA115927-05
Grant ID: R01 CA134502
Grant ID: R33 CA128625
Grant ID: P50 CA093683
NIH HHS (2)
Grant ID: DP2 OD002750
Grant ID: DP2 OD002750-01




