Abstract
Free full text

Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast
Associated Data
Abstract
The Arp2/3 complex is implicated in actin polymerization-driven movement of Listeria monocytogenes. Here, we find that Arp2p and Arc15p, two subunits of this complex, show tight, actin-independent association with isolated yeast mitochondria. Arp2p colocalizes with mitochondria. Consistent with this result, we detect Arp2p-dependent formation of actin clouds around mitochondria in intact yeast. Cells bearing mutations in ARP2 or ARC15 genes show decreased velocities of mitochondrial movement, loss of all directed movement and defects in mitochondrial morphology. Finally, we observe a decrease in the velocity and extent of mitochondrial movement in yeast in which actin dynamics are reduced but actin cytoskeletal structure is intact. These results support the idea that the movement of mitochondria in yeast is actin polymerization driven and that this movement requires Arp2/3 complex.
Mitochondrial motility is required for asymmetric distribution of the organelle during cell differentiation, enrichment of the organelle at sites of high ATP utilization, and mitochondrial inheritance. In the budding yeast, mitochondria undergo a series of cell cycle-linked motility events that result in transfer of mitochondria from mother to bud. These events include polarization of mitochondria toward the site of bud emergence in the G1 phase; linear, polarized movement of some mitochondria from mother cells to developing buds in S phase; and retention of mitochondria in the bud and mother cell (1–3). Because mitochondria are essential organelles that are produced only from preexisting organelles, these motility events are critical for mitochondrial inheritance and daughter cell viability during cell division.
Previous studies support a role for the actin cytoskeleton in control of mitochondrial movement and inheritance in budding yeast. The actin cytoskeleton of Saccharomyces cerevisiae consist of two structures detected by light and electron microscopy, actin patches, and actin cables (4, 5). Actin patches are punctate structures believed to be F-actin-associated cortical membranes. Actin cables are bundles of F-actin. In polarized, budding yeast, actin patches are localized to sites of polarized cell surface growth in the bud, and actin cables are aligned along the mother-bud axis. The orientation of actin filaments within actin cables has not been determined. However, actin cables seem to serve as tracks for at least two myosin-driven polarized movements (6–9). These movements are believed to require filaments that are oriented in the same direction in the cables. Also, fimbrin (Sac6p), an actin-bundling protein that cross-links actin filaments in a parallel manner, colocalizes with and stabilizes actin cables (10–12). These notions support the idea of uniformly directed filaments in actin cables.
Three lines of evidence support the model that actin cables provide directional cues for polarized movement of mitochondria from mother cells to buds in dividing yeast. First, mitochondria colocalize with actin cables (13, 14). Second, mutations in TPM1 or MDM20 genes that destabilize actin cables without affecting actin patches cause defects in mitochondrial morphology and inhibition of mitochondrial inheritance (2, 15). Third, deletion of TPM1 results in loss of all directed mitochondrial movement but has no significant effect on either the extent or the velocity of mitochondrial movement (2). Other studies support direct interaction between mitochondria and actin filaments. Isolated yeast mitochondria display ATP-sensitive binding to the lateral surface of F-actin (14). This binding is mediated by at least two components: (i) mitochondrial actin-binding protein(s) (mABP), a salt extractable, peripheral mitochondrial outer membrane protein(s), and (ii) Mmm1p and Mdm10p, two integral mitochondrial membrane proteins, required for docking of mABP on mitochondria (16). These observations support the model that mitochondria use actin cables as tracks for movement from the mother cell to the bud.
Myosin motors are responsible for motility events, including cytoplasmic streaming in plants, transport of melanosomes in mouse melanocytes, vesicle and mRNA movement in yeast, and organelle transport in neurons and Drosophila embryos (17). However, because none of the myosin proteins of yeast localizes to mitochondria (18–21), and myosin mutations have no effect on the velocity of mitochondrial movement (1), the role of myosins in mitochondrial movement in yeast remains elusive.
Another mechanism for intracellular particle or organelle movement utilizes the Arp2/3 complex and the rearrangement of actin cytoskeleton. The Arp2/3 complex, a conserved complex consisting of two actin-related proteins, Arp2p and Arp3p, and five other proteins, is implicated in movement of pathogens including Listeria monocytogenes through the cytoplasm of infected cells (22–25). This complex binds to the lateral surfaces and pointed ends of actin filaments (26), localizes to sites of actin dynamics including the actin comet tail of motile L. monocytogenes (27), and is essential for reconstitution of Listeria and Shigella movement using purified proteins (28). The Arp2/3 complex is recruited to the surface of the bacterium by ActA, an L. monocytogenes-encoded plasma membrane protein, and acts synergistically with ActA to promote actin nucleation from the fast-growing barbed end (29). These findings support a model in which activation of the Arp2/3 complex at the surface of L. monocytogenes leads to site-specific actin polymerization and production of forces sufficient to drive bacterial movement.
In budding yeast, Arp2p and Arp3p are resolved as punctate structures enriched in the bud. A subset of Arp2p-containing structures colocalize with actin patches (30, 31). ARP2 mutations result in disorganization and dysfunction of the actin cytoskeleton. Moreover, mutation of the ARP3 gene compromises movement of actin patches (31). These observations support a role for the Arp2/3 complex in actin organization.
Here, we show that two subunits of the Arp2/3 complex, Arp2p and Arc15p, are associated with mitochondria and that mutations in these subunits interfere with mitochondrial motility. Supporting this finding, dampening actin dynamics decreases mitochondrial motility. Moreover, the presence of actin polymers around mitochondria depends on functional Arp2p. These studies support a second function for the Arp2/3 complex in budding yeast: control of actin-based mitochondrial movement.
Materials and Methods
Yeast Strains.
Yeast strains used in this study are listed in Table Table1.1. Yeast cell growth and manipulations were carried out according to Sherman (32).
Table 1
Yeast strains used in this study
| Strain | Genotype | Source of strain |
|---|---|---|
| D273-10B | MATα | * |
| YPH499 | MATa, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp-1Δ63, ura3-522 | † |
| YMW81 | MATa, arp2-H330L, ade2-101, his3-Δ200, leu2-Δ1, lys2-801 trp1-Δ63, ura3-52 | † |
| DAUL1 | MAT, ade2, ura3, lys2 | ‡ |
| DDY1493 | MATa, act1-159 HIS3, his3-Δ200,
tub2-101, ura3-52, leu2-3,112 HIS3, his3-Δ200,
tub2-101, ura3-52, leu2-3,112 | § |
| DDY1495 | MATa, ACT1 HIS3, his3-Δ200, tub2-101,
ura3-52, leu2-3,112 HIS3, his3-Δ200, tub2-101,
ura3-52, leu2-3,112 | § |
| CRY3 | MATa/α, ade2-1/ade2-1, his3-11/his3-11, leu2-3/leu2-3, trp1-1/trp1-1, ura3-1/ura3-1 | ¶ |
| DNY211 | MATa/α,
ARC15/arc15-GFP KANMX6, ADE2/ade2-1, leu2-3/leu2-3,
TRP1/trp1-1, ura3-1/ura3-1 KANMX6, ADE2/ade2-1, leu2-3/leu2-3,
TRP1/trp1-1, ura3-1/ura3-1 | ‖ |
| DNY226-4c | MATα, arc15-GFP KANMX6, ade2-1, leu2-3,
TRP1, ura3-1 KANMX6, ade2-1, leu2-3,
TRP1, ura3-1 | ‖ |
| DNY226-4d | MATa, ade2-1, leu2-3, trp1-1, ura3-1 | ‖ |
| DNY231 | MATa/α, ARC15/arc15 TRP1,
ade2-1/ade2-1, his3-11/his3-11, leu2-3/leu2-3, trp1-1/trp1-1,
ura3-1/ura3-1 TRP1,
ade2-1/ade2-1, his3-11/his3-11, leu2-3/leu2-3, trp1-1/trp1-1,
ura3-1/ura3-1 | ‖ |
| DNY108 | MATα, leu2-3, trp1-1, ura3-1 | ‖ |
| DNY262 | MATα, ARC15-MYC TRP1, leu2-3, trp1-1,
ura3-1 TRP1, leu2-3, trp1-1,
ura3-1 | ‖ |
Tagging of the ARC15 Gene.
The COOH terminus of Arc15p was tagged with 13 tandem copies of the Myc epitope or with green fluorescent protein (GFP) (S65T), by using PCR-based insertion into the chromosomal copy of ARC15 (33). PCR fragments for integration by homologous recombination were first amplified from plasmids pFA6a-13Myc-TRP1 and pFA6a-GFP(S65T)-kanMX6, with forward primer, 5′-CACACCTATCGTTCATTATATATCGGATAGAAGAATGTACGGATCCCCGG-GTTAATTAA-3′ and reverse primer, 5′-GTATATTTCTTTATACTAAGTTTTACTGGGCAGA-GTATCAGAATTCGAGCTCGTTTAAAC-3′. The GFP- and Myc-tagged yeast strains (DNY 211, DNY226[4a–d], and DNY 262) were characterized for correct integration of the tagging cassette at the ARC15 locus by using PCR and Western blot analysis.
Detection of Actin Clouds.
Actin clouds were detected in yeast treated with 0.1% sodium azide for 15 min at 22°C. Cells were fixed, labeled with Alexa-phalloidin, and visualized by using a fluorescence microscope. Optical sections were obtained at 200-nm intervals through the entire cell by using a piezoelectric focus motor mounted on the objective lens of the microscope (Polytech PI, Auburn, MA). The exposure time of each section was 500 msec. Each z-series was subjected to digital deconvolution, and each deconvolved z-series was merged in a single projection by using Inovision (Raleigh, NC) software on a Silicon Graphics O2 computer (Mountain View, CA).
Other Methods.
Methods including peptide sequencing; actin affinity chromatography and sedimentation assay for mABP activity; visualization of mitochondria, Arp2/3 subunits, and the actin cytoskeleton; quantitation of mitochondrial movement in vivo; description of the preparation of Arp2p antibody; and immunoprecipitation of Arc15p-myc and Arp2p are included in the supplementary material (on the PNAS web site, www.pnas.org).
Results
Arc15p, a Subunit of the Arp2/3 Complex, Is Recovered as a Salt Extractable Peripheral Outer Membrane Protein.
Previously, we developed a sedimentation assay to study actin–mitochondrial interactions. In it, a mixture consisting of F-actin and mitochondria is subjected to low-speed centrifugation, which sediments mitochondria with bound actin but leaves unbound F-actin in the supernatant. Using this assay, we find that (i) untreated mitochondria have full actin binding activity (mABP); (ii) salt washing the organelle with 1 M KCl blocks this activity; and (iii) treatment of salt-washed mitochondria (SW) with the salt-extracted mitochondrial proteins (SE) restores full mABP activity. Thus, mABP activity is present in isolated mitochondria, is released from mitochondria with high salt wash, and binds reversibly to the organelle (16).
To identify mABP, proteins with ATP-sensitive actin-binding activity were isolated from salt-extracted mitochondrial peripheral membrane proteins by F-actin affinity chromatography (Fig. (Fig.11a). Most of the proteins applied to the F-actin column are recovered in the flow-through. However, a subset of 6–8 proteins remain bound to the column after washes with 0.2 M KCl, and can be eluted from the column with 0.1 mM ATP. We monitored the fractionation behavior of mABP using the sedimentation assay. Here, we tested the ability of each column fraction to restore ATP-sensitive actin binding to salt-washed mitochondria. We detect mABP activity in the F-actin column load (SE), column flow-through (FT), and the fraction eluted from the column with 0.1 mM ATP (A1). mABP activity is absent from the column wash (W) and from the 1-mM ATP eluate (A2) (Fig. (Fig.11b).
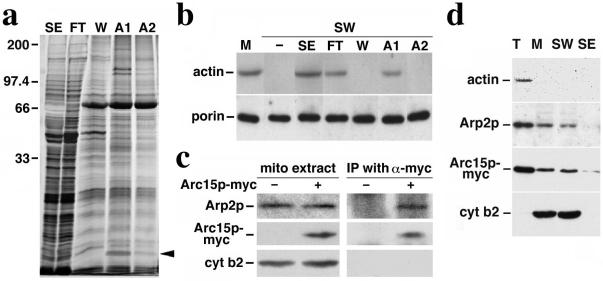
Association of Arc15p and Arp2p subunits of Arp2/3 complex with mitochondria. (a) Silver-stained SDS gel of proteins recovered by actin affinity chromatography. SE, salt-extracted mitochondrial membrane proteins loaded onto the F-actin column; FT, column flow-through; W, proteins eluted with 200 mM KCl wash; A1, proteins eluted with 0.1 mM ATP; and A2, proteins eluted with 1 mM ATP. The sizes shown were determined by using molecular weight standards; the arrow points to the 15-kDa Arc15p-containing band in A1. (b) Restoration of mitochondrial actin-binding activity with column fractions. SE and column fractions (FT, W, A1, and A2) were added to salt-washed mitochondria (SW). Mitochondria were then separated from the mixture by centrifugation at 10,000 × g at 4°C. mABP activity in control mitochondria (M) and salt-washed mitochondria incubated with various column fractions was measured by using a sedimentation assay (16). Proteins recovered in the mitochondrial pellet were identified by Western blot analysis using antibodies raised against the mitochondrial marker, porin, and monoclonal antibody raised against actin (c4d6). (c) Immunoprecipitation of Arc15p-myc and Arp2p. Purified mitochondria were solubilized in the presence of 0.5% digitonin. The solubilized mitochondria were incubated with affinity-purified anti-myc antibody prebound to protein G-Sepharose beads. Immunoprecipitated proteins on the beads were identified by SDS/PAGE and Western blot analysis. Lanes 1–2, 25-μg digitonin-solubilized mitochondrial extracts from cells expressing untagged (−) or myc-tagged (+) Arc15p (DNY108 and DNY262). Lanes 3–4, immuno-precipitated proteins from 350-μg mitochondrial extracts from cells expressing untagged (−) or myc-tagged (+) Arc15p. (d) Actin-independent association of Arp2p and Arc15p with mitochondria. Isolated mitochondria from ARC15-MYC (DNY262) cells were washed with buffer containing 1 M KCl. Total cell lysate (T), mitochondria (M), salt washed mitochondria (SW), and salt extracted membrane proteins (SE) were evaluated for the levels of actin, Arp2p, Arc15p-myc, and cytochrome b2, a mitochondrial marker protein, by Western blot analysis. Proteins (30 μg) were loaded for T, M, and SW. SE was precipitated by adding trichloroacetic acid (TCA) to 10%. Precipitated proteins were resuspended to a volume equal to that of mitochondria (M). SE was loaded in the same volume as M.
Tandem mass spectroscopy-based peptide sequencing was used to identify the proteins whose fractionation behavior in F-actin chromatography correlated with that of mABP activity. Our studies focused on the 15-kDa band recovered in the 0.1 mM ATP eluate. Tryptic fragments derived from this band contained the peptide LTAADLVPPYETTVTLQELQPR, a sequence that corresponds to amino acids 20–41 of Arc15p (ORF name: YIL062c), the 15-kDa component of the Arp2/3 complex of budding yeast. This is the homologue to the 14-kDa subunit observed in mammalian and other cells.
Peptide sequencing did not reveal other subunits of the Arp2/3 complex. However, we find that the Arp2p subunit of the Arp2/3 complex is recovered with isolated mitochondria (Fig. (Fig.11c, lane 2) and that the association of Arp2p and Arc15p with mitochondria does not depend on actin (Fig. (Fig.11d). Moreover, Arp2p coimmunoprecipitates with Arc15p in digitonin-solubilized mitochondrial extracts (Fig. (Fig.11c, lane 4). This finding suggests that Arc15p recovered from mitochondria is part of the Arp2/3 complex.
Localization of Arp2p.
Arp2p was previously reported to form punctate structures mainly at sites of polarized cell surface growth (30). Using the same antibody and fixation protocol, we confirmed this localization for Arp2p (Fig. (Fig.22B). However, this fixation method uses methanol and acetone, which extract lipids and disrupt membrane structure. Therefore, to test whether Arp2p is localized to mitochondria, we reevaluated Arp2p localization by comparing the previous methanol-acetone protocol with one that preserves membrane integrity. Mitochondria in wild-type yeast are long tubular structures aligned along the mother-bud axis (Fig. (Fig.22 A and C). Immunostaining using the membrane-preserving fixation conditions reveals that Arp2p-containing punctate structures (Fig. (Fig.22D) colocalize with the tubular mitochondrial structures (Fig. (Fig.22C). Thus, Arp2p localizes both at sites of polarized cell surface growth and at mitochondria.
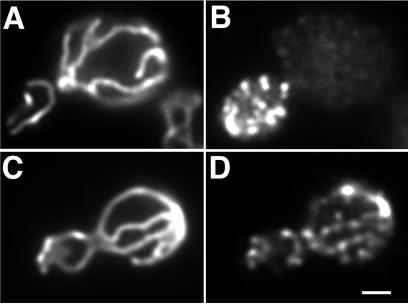
Localization of Arp2p in yeast. DAUL1 cells expressing a fusion protein consisting of the mitochondrial signal sequence of citrate synthase 1 fused to GFP were grown to mid-log phase, fixed with paraformaldehyde alone (C and D), or with additional methanol/acetone (A and B), converted to spheroplasts, and stained for Arp2p by using an antibody raised against a conserved peptide sequence found in Arp2p, but not in actin or any other actin-related protein (30). (A and C) Mitochondria visualized by using CS1-GFP; (B and D) Arp2p visualized by indirect immunofluorescence. Bar = 1 μm.
Arp2p and Arc15p Are Required for Mitochondrial Motility.
Insertion of GFP at the C terminus of the chromosomal copy of the ARC15 gene has no obvious effect on cell viability or actin polarization during polarized yeast cell growth and division (Fig. (Fig.33K; Table Table2).2). However, cells expressing Arc15p-GFP display defects in mitochondrial position, inheritance, and motility. Mitochondria in the Arc15p-GFP-expressing cells form abnormal aggregates and spherical structures (Fig. (Fig.33J). GFP insertion in ARC15 also results in a 4-fold increase in mitochondrial inheritance defects (Table (Table2).2). Finally, all linear, directed movement seen in wild-type yeast is lost in Arc15p-GFP-expressing cells (Fig. (Fig.33L). These observations suggest (i) that ARC15 is required for normal mitochondrial motility and inheritance, and (ii) that insertion of GFP into the C terminus of Arc15p compromises that function.
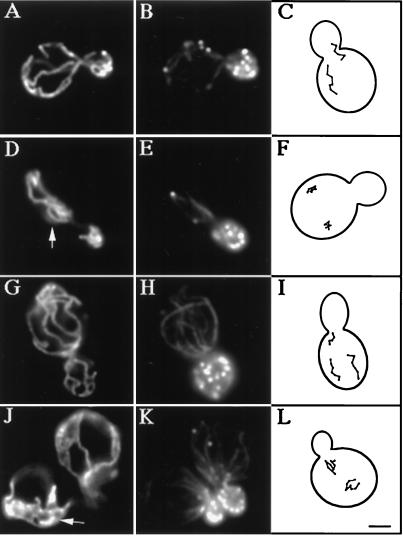
Mitochondrial morphology and motility are abnormal in arc15-GFP and arp2-1 mutants. The ARP2 parent (A–C), and arp2-1 mutant (D–F) were grown to mid-log phase at 22°C and shifted to 37°C for 1 h. The ARC15 wild-type (G–I) and arc15-GFP mutant cells (J–L) were grown to mid-log phase at 30°C. Mitochondrial morphology (A, D, G, and J) and actin organization (B, E, H, and K) were determined in fixed cells by indirect immunofluorescence by using an antibody raised against mitochondrial outer membrane proteins, and rhodamine phalloidin. Arrowheads point to examples of abnormal mitochondrial morphology. (C, F, I, and L) Tracings of the pattern of mitochondrial movement in living cells stained with the membrane potential sensing dye, DiOC6 relative to the boundary of dividing yeast. Movements of mitochondria were followed by marking the tip of motile organelles during the time in which they remained in the plane of focus in time lapse images. The points denote the position of organelles at 20-sec intervals. Bar = 1 μm.
Table 2
Mitochondrial inheritance and movement are impaired in arp2-1 and arc15-GFP mutants
| Strain | Actin patch localization, % polarized | Mitochondrial morphology, % abnormal | Mitochondrial inheritance, % transfer | Mitochondrial movement
| |
|---|---|---|---|---|---|
| Velocity (nm/sec) | % polarized | ||||
| YPH499 (ARP2 wt) | 53.1 | 22.9 | ND | 26.7 ±
± 6.5 6.5 | 51.4 |
| YMW81 (arp2-1) | 41.6 | 57.8 | ND | 7.2 ±
± 2.9 2.9 | 3.1 |
| DNY226-4d (ARC15 wt) | 89.2 | 22.8 | 89.2 | 17.1 ± ± 5.0 5.0 | 43.9 |
| DNY226-4c (arc15-GFP) | 89.6 | 75.2 | 57.4 | 9.6 ±
± 2.9 2.9 | 8.2 |
The conditions for temperature shift of the temperature-sensitive arp2-1 mutant and the corresponding wild-type cell strain were used as for Fig. Fig.4.4. The actin cytoskeleton was scored as polarized if the cell showed no more than seven actin patches in the mother. Cells bearing long tubular mitochondria that aligned along the mother-bud axis were scored as normal for mitochondrial morphology. Cells bearing aggregated mitochondria were scored as abnormal. Inheritance represents the percentage of small buds that contain mitochondria. Long term shift of the arp2-1 strain produces severe actin disorganization and loss of cell viability. Therefore, inheritance was not determined (ND) for YPH499 and YMW81. Polarized movement refers to the percentage of motile mitochondria that showed a net displacement towards the bud. Velocity measurements were carried out as described in Supplementary Materials. The sample size for all measurements (n) was greater than 100.
More severe defects were observed in a strain bearing a temperature-sensitive mutation in ARP2 (30). The arp2-1 mutant used bears an H330L substitution in the ARP2 gene, and displays disorganization of the actin cytoskeleton and defects in actin-dependent processes, including polarized cell surface growth, endocytosis, and maintenance of mitochondrial morphology after shift to restrictive temperature (37°C) for 2 h. We find that a shorter shift of the arp2-1 mutant to 37°C, for 1 h, results in defects in mitochondrial morphology and loss of all directed mitochondrial movement (Fig. (Fig.33 D–F, and Table Table2).2). Mitochondrial movements in arp2-1 mutant (Fig. (Fig.33F) resemble the actin-independent, random oscillations observed on latrunculin-A-induced depolymerization of yeast F-actin (16). Incubation of yeast cells at 37°C results in transient depolarization of the actin cytoskeleton. As a result, we observe some depolarization in the temperature sensitive arp2-1 mutant and its wild-type parent strain. However, because actin polarization is compromised to a similar extent in the ARP2 wild-type and arp2-1 mutant strains, mitochondrial morphology defects are not a consequence of actin depolarization. Finally, although mitochondrial morphology and motility are compromised, mitochondria still colocalize with actin cables in arp2 and arc15 mutants.
Actin Dynamics Affect Mitochondrial Motility.
To test the role of actin dynamics in this process, we studied mitochondrial motility under conditions that dampen actin dynamics without affecting actin cable structures in yeast. These studies used the act1-159 mutant bearing a single amino acid substitution in the ACT1 gene (V159N). This mutant displays a decrease in the rate of actin filament depolymerization and, as a result, an increase in the steady state level of actin cables (12). This result can be seen by the more intense F-actin staining and the increased number of cables in the mutant (Fig. (Fig.44B) compared with wild-type cells (Fig. (Fig.44A). Actin dynamics in the act1-159 mutant were dampened further by addition of Lat-A, a G-actin-binding agent that blocks G-actin polymerization. Our Lat-A titration (see supplementary material) indicates that treatment of act1-159 cells with 15 μM Lat-A reduces the level of actin cables in the mutant to wild-type levels (Fig. (Fig.44C). Thus, using a mutation that inhibits F-actin depolymerization and an agent that blocks G-actin polymerization, we have identified conditions that suppress actin dynamics without affecting the level of actin cables in yeast.
Actin organization in wild-type cells and stable actin mutants. Actin structure in wild-type ACT1 cells (A), act1-159 mutants (B), and act1-159 mutants treated with 15 μM Lat-A (C). Actin was stained with Alexa-phalloidin, and mitochondria were visualized by using CS1-GFP. Incubation with 15 μM Lat-A reduces the level of actin cables in act1-159 cells to that observed in wild-type ACT1 cells. Bar = 2 μm.
We find that dampening of actin dynamics also decreases the velocity of mitochondrial movement. The velocity of mitochondrial movement in Lat-A-treated act1-159 cells (21.5 nm/sec; standard error = 1.35 nm/sec) is significantly lower (P < 0.001) than that in the wild-type control (32.9 nm/sec; standard error = 1.10 nm/sec). In addition, there is a 50% decrease in the proportion of moving mitochondria (P < 0.05). These findings suggest a role for actin dynamics in mitochondrial movement in budding yeast.
Arp2/3 Complex-Dependent Formation of Actin Clouds Around Mitochondria.
We find that treatment of yeast with 0.1% sodium azide results in fragmentation of mitochondria and loss of actin cables. Actin patches were detected in sodium azide-treated cells; however, they were depolarized and found in the mother cell and bud (Fig. (Fig.5).5). The results obtained with sodium azide treatment are similar to those observed on deletion of the tropomyosin I gene: both conditions result in fragmentation of mitochondria, loss of actin cables, and depolarization of actin patches (2, 9).
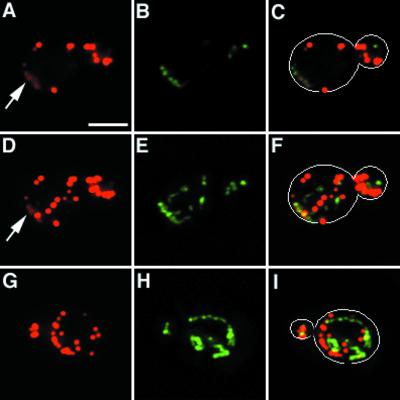
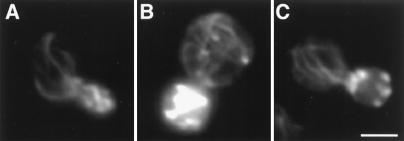
Actin cloud formation around mitochondria. The arp2-1 mutant and its wild-type parent expressing CS1-GFP were grown to mid-log phase at 22°C and treated with 0.1% sodium azide for 15 min. Cells were then fixed and stained with Alexa-phalloidin. Actin and mitochondria were visualized by optical sectioning, digital deconvolution, and three-dimensional reconstruction. A single optical section of a sodium azide-treated wild-type cell is shown in A–C. Three-dimensional projection of the same cell is shown in D–F. Three-dimensional projection of an arp2-1 mutant treated with sodium azide is shown in G–I. (A, D and G) Alexa-phalloidin-labeled F-actin; (B, E, and H) GFP-labeled mitochondria; (C, F, and I) overlay of A and B, D and E, and G and H, respectively. Outlines of cells are superimposed over fluorescent images in C, F, and I. Arrows point to actin clouds surrounding mitochondria. Bar = 2 μm.
Optical sectioning, digital deconvolution, and three-dimensional reconstruction of F-actin and mitochondria in sodium azide-treated cells revealed areas of diffuse F-actin staining (Fig. (Fig.5,5, arrow). Examination of deconvolved images and optical sections indicated that this diffuse fluorescence was not out-of-focus fluorescence emanating from actin patches in a different plane of focus. Moreover, actin clouds are distinct from actin patches. First, actin clouds show less intense staining with Alexa-phalloidin compared with actin patches. Second, actin patches are always spherical in structure whereas actin clouds are more diffuse and amorphous in shape. Third, in contrast to actin patches, which do not colocalize with mitochondria, we detect association of actin clouds with mitochondria. Not all mitochondria are associated with actin clouds, suggesting that the fluorescence signal of actin clouds is not due to bleed-through from GFP-labeled mitochondria. However, all actin clouds detected are associated with mitochondria.
Localization of actin clouds around mitochondria suggests that actin polymerization and/or assembly may be occurring at these sites. Consistent with this finding, we see far fewer actin clouds on treatment of the arp2-1 mutant with sodium azide (Fig. (Fig.5).5). In contrast to wild-type cells, which show mitochondria associated with clouds in 62.3% of sodium azide-treated cells tested (n = 101), arp2-1 mutants show an 84% decrease in actin cloud formation (10.1%. n = 109). This loss of actin clouds occurs at the permissive temperatures for growth of the arp2-1 mutant, conditions that compromise other Arp2/3 complex-dependent functions, including osmoregulation and endocytosis (30). Actin patches in arp2-1 mutants and wild-type cells treated with sodium azide are indistinguishable, suggesting that the loss of actin clouds in sodium azide-treated arp2-1 mutants is not due to global defects in actin organization.
Discussion
Here, we provide evidence for a role of the Arp2/3 complex and actin polymerization in movement of an organelle, the mitochondrion. The first link between the Arp2/3 complex and mitochondria was the observation that a subunit of this complex, Arc15p, was extracted from salt-washed mitochondria, and isolated from the salt extract by actin affinity chromatography. Thus, Arc15p is a mitochondrial peripheral membrane protein that is or associates with a protein with actin-binding activity.
Three lines of evidence suggest that intact Arp2/3 complex is present on isolated yeast mitochondria. First, although Arp2/3 complex is present in and fractionates with other yeast compartments (e.g., actin patches; refs. 30 and 31), Arp2p and Arc15p are recovered with mitochondria on subcellular fractionation. Second, association of Arp2p and Arc15p with mitochondria is tight and actin-independent: over 90% of Arp2p and Arc15p remain associated after salt washing or LatA treatment of mitochondria. Whereas the majority of Arc15p remains associated with mitochondria after salt washing, the small amount that is released is sufficient to be isolated by actin affinity chromatography. Third, Arp2p and Arc15p coimmunoprecipitate from detergent-extracted mitochondria.
Previously, we identified mABP, an ATP sensitive, reversible, salt sensitive, presumably peripheral membrane activity identified that mediates actin–mitochondrial interactions. Although Arc15p cofractionates with mABP activity during actin affinity chromatography, three findings suggest that Arp2/3 complex is not mABP. First, binding of purified Arp2/3 complex to actin is not ATP sensitive (24). Second, over 90% of Arp2p and Arc15p remain associated with mitochondria under salt wash conditions that completely abolish mABP activity. Third, we find that mitochondria colocalize with actin cables in arp2 and arc15 mutants. Thus, the Arp2/3 complex is required for mitochondrial motility and morphology, but is not required for association of mitochondria with actin in yeast. Rather, our findings support the idea that mABP and Arp2/3 complex are both required for mitochondrial movement in yeast (see below).
To address the physiological relevance of Arp2/3 complex association with mitochondria, we examined (i) localization of Arp2/3 complex subunits in intact yeast, (ii) the effect of mutation of Arp2/3 complex subunits on mitochondrial motility, and (iii) the effect of dampening actin dynamics and polymerization on mitochondrial motility. Using a conventional immunofluorescence protocol that preserves membrane integrity, we find that Arp2p is localized to punctate structures found in two locations. In confirmation of a previous report (30), Arp2p-containing structures are enriched in the bud. In addition, almost all Arp2p-containing patches detected in the mother cell coalign along mitochondria. Whereas Arp2/3 complex binding to actin cables may also contribute to its localization in mother cells, we find that Arp2p localization along mitochondria is lost under fixation conditions that extract membranes but preserve actin cytoskeletal structures. This finding supports direct, actin-independent interactions between this Arp2/3 complex subunit and mitochondria in vivo.
We also observe Arp2/3 complex-dependent formation of actin clouds around mitochondria. Previous studies showed that purified Arp2/3 complex promotes assembly of dense “clouds” of actin on non-motile bacteria. Addition of cytosolic extracts stimulated actin cloud formation, actin comet tail formation, and bacterial movement. These studies suggest that actin clouds are an intermediate in actin comet tail formation during Arp2/3 complex-mediated movement of bacteria (27, 29). If Arp2/3 complex-mediated actin polymerization provides the force for mitochondrial movement, conditions that partially compromise this process could produce actin clouds around the organelle. Our findings that actin clouds form around mitochondria in the absence of actin cables, and that Arp2/3 complex is required for this process, provide additional evidence for direct interactions between mitochondria and the Arp2/3 complex.
Mutation of Arp2p or Arc15p causes defects in mitochondrial morphology and loss of all linear, polarized mitochondrial movement. Indeed, mitochondrial movement in the arp2-1 mutant is indistinguishable from that of the actin-independent, apparently random oscillations observed in cells treated with Lat-A (16). Because these defects in mitochondrial motility are observed under conditions that do not have a detectable effect on actin cables or patches, it is likely that they are not an indirect effect of Arp2/3 complex mutations on actin cytoskeletal organization. Our studies support functional interactions between components of the Arp2/3 complex and yeast mitochondria, and suggest that one consequence of this interaction is control of mitochondrial motility.
Finally, we tested whether mitochondrial movement is coupled to F-actin assembly in vivo. These studies were carried out in act1-159 mutant cells, which carry an actin mutation that stabilizes actin in its filamentous state. These stable actin mutants were treated with Lat-A to suppress actin dynamics further and to reduce the total amount of actin cables to that observed in wild-type cells. We find that dampening actin dynamics, under conditions that reproduce the actin cable content of wild-type cells, results in a decrease in both velocity and extent of mitochondrial movement. Thus, actin dynamics are required for normal mitochondrial motility.
We observe common features underlying motility of yeast mitochondria and L. monocytogenes. Both motility events are actin based and require actin dynamics. In addition, both yeast mitochondria and L. monocytogenes are associated with the Arp2/3 complex subunits, show Arp2p-dependent actin cloud formation, and require functional Arp2/3 complex subunits for movement. These observations suggest some common features for movement of yeast mitochondria and a bacterial pathogen.
Whereas there are similarities in the movements of yeast mitochondria and L. monocytogenes, there are also some fundamental differences. First, previous studies indicate that the Arp2/3 complex-mediated actin polymerization occurs only on one tip of L. monocytogenes (27). In contrast, we find that Arp2p is present along the entire surface of mitochondrial tubules. Therefore, whereas L. monocytogenes induces actin assembly only at one end of the bacterial cell, yeast mitochondria can presumably initiate actin assembly at multiple sites along the whole length of the organelle. Second, motile L. monocytogenes shows an actin comet tail almost as wide and many times longer that the bacterium itself. Such a tail clearly does not exist behind motile mitochondria. Third, Arp2/3 complex-mediated movement of L. monocytogenes has no obvious direction or track dependence. In contrast, yeast mitochondria colocalize with actin cables and show linear, polarized, actin cable-dependent movement from the mother cell to the bud. Thus, in contrast to L. monocytogenes, yeast mitochondria show directed, apparently track-dependent movement.
Our results support the model that Arp2/3 complex-mediated actin nucleation and subsequent polymerization are the force for mitochondrial movement. Recent results show that actin polymerization-driven particle movements are not restricted to pathogens. Actin tail-like structures have been detected on endosomes in mast cells, and unidentified endogenous vesicles in HeLa cells and Xenopus egg extracts (34–36). In addition, Arp2/3 subunits colocalize with structures believed to be organelles in Acanthamoeba, Swiss 3T3 fibroblasts, and fission yeast (24, 25, 37). Therefore, it is possible that an actin polymerization-based mechanism is used for intracellular movement of organelles and vesicles in diverse cell types. However, the mediator that links this actin polymerization-based force generator to the cell polarity machinery to produce linear, directed movement is yet to be determined.
The simplest model for mitochondrial movement in yeast that emerges from our studies invokes two actin-dependent activities on the mitochondria surface: mABP and Arp2/3 complex. Because mABP mediates (i) reversible binding of mitochondria to the lateral surface of F-actin in vitro and (ii) association of mitochondria with actin cables in vivo, we propose that mABP mediates binding of mitochondria to actin cables. Because actin cables are believed to be polarized, this binding is critical because it links mitochondria and their associated Arp2/3 complex with the cell polarity machinery. In addition, we propose that Arp2/3 complex on mitochondria mediates actin nucleation and polymerization. Finally, we propose that newly polymerized F-actin is cross-linked to actin cables. This cross-linking results in polarization of forces produced by actin polymerization at the mitochondria-actin cable interface, and unidirectional movement of mitochondria along actin cables. Ongoing studies are designed to test this hypothesis.
Acknowledgments
We thank Dr. F. Chang and members of the Pon laboratory for critical evaluation of the manuscript; Drs. D. Drubin, T. Fox, and B. Winsor for yeast strains; Dr. R. Butow for the mitochondria-targeted GFP; Drs. J. Lessard, G. Schatz, and B. Winsor for antibodies; Dr. P. Crews for latrunculin-A; and Drs. M. Longtine and J. Pringle for tagging cassette DNA. This work was supported by research grants to L.P. from the National Institutes of Health (GM45735), and to J.Y. from the National Science Foundation (BIR921482) and the National Institutes of Health (RR11832). Digital deconvolution was performed at the Optical Microscopy Facility of the Herbert Irving Comprehensive Cancer Center, which is supported by grants from the National Institutes of Health (S10RR10506 and P30CA13696).
Abbreviations
| mABP | mitochondrial actin-binding protein |
| GFP | green fluorescent protein |
| SW | salt-washed mitochondria |
| SE | salt-extracted mitochondrial proteins |
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.051494698
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc30624?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Improved tools for live imaging of F-actin structures in yeast.
Mol Biol Cell, 35(9):mr7, 18 Jul 2024
Cited by: 1 article | PMID: 39024291 | PMCID: PMC11449393
Beyond fission and fusion-Diving into the mysteries of mitochondrial shape.
PLoS Biol, 22(7):e3002671, 01 Jul 2024
Cited by: 0 articles | PMID: 38949997 | PMCID: PMC11216622
Review Free full text in Europe PMC
Collagen VI Deficiency Impairs Tendon Fibroblasts Mechanoresponse in Ullrich Congenital Muscular Dystrophy.
Cells, 13(5):378, 22 Feb 2024
Cited by: 1 article | PMID: 38474342 | PMCID: PMC10930931
Inhibitors of Rho/MRTF/SRF Transcription Pathway Regulate Mitochondrial Function.
Cells, 13(5):392, 24 Feb 2024
Cited by: 1 article | PMID: 38474356 | PMCID: PMC10931493
Mitochondria: A Promising Convergent Target for the Treatment of Amyotrophic Lateral Sclerosis.
Cells, 13(3):248, 29 Jan 2024
Cited by: 5 articles | PMID: 38334639 | PMCID: PMC10854804
Review Free full text in Europe PMC
Go to all (115) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits.
Proc Natl Acad Sci U S A, 96(13):7288-7293, 01 Jun 1999
Cited by: 136 articles | PMID: 10377407 | PMCID: PMC22078
A mutant of Arp2p causes partial disassembly of the Arp2/3 complex and loss of cortical actin function in fission yeast.
Mol Biol Cell, 10(12):4201-4215, 01 Dec 1999
Cited by: 51 articles | PMID: 10588653 | PMCID: PMC25753
The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex.
Mol Biol Cell, 10(10):3521-3538, 01 Oct 1999
Cited by: 114 articles | PMID: 10512884 | PMCID: PMC25621
Abp1p and cortactin, new "hand-holds" for actin.
J Cell Biol, 154(4):679-682, 01 Aug 2001
Cited by: 39 articles | PMID: 11514584 | PMCID: PMC2196464
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P30 CA013696
Grant ID: P30CA13696
NCRR NIH HHS (2)
Grant ID: RR11832
Grant ID: S10RR10506
NIGMS NIH HHS (2)
Grant ID: GM45735
Grant ID: R01 GM045735





