Abstract
Free full text

Structural Basis for Selective Small Molecule Kinase Inhibition of Activated c-Met
Abstract
The receptor tyrosine kinase c-Met is implicated in oncogenesis and is the target for several small molecule and biologic agents in clinical trials for the treatment of cancer. Binding of the hepatocyte growth factor to the cell surface receptor of c-Met induces activation via autophosphorylation of the kinase domain. Here we describe the structural basis of c-Met activation upon autophosphorylation and the selective small molecule inhibiton of autophosphorylated c-Met. MK-2461 is a potent c-Met inhibitor that is selective for the phosphorylated state of the enzyme. Compound 1 is an MK-2461 analog with a 20-fold enthalpy-driven preference for the autophosphorylated over unphosphorylated c-Met kinase domain. The crystal structure of the unbound kinase domain phosphorylated at Tyr-1234 and Tyr-1235 shows that activation loop phosphorylation leads to the ejection and disorder of the activation loop and rearrangement of helix αC and the G loop to generate a viable active site. Helix αC adopts a orientation different from that seen in activation loop mutants. The crystal structure of the complex formed by the autophosphorylated c-Met kinase domain and compound 1 reveals a significant induced fit conformational change of the G loop and ordering of the activation loop, explaining the selectivity of compound 1 for the autophosphorylated state. The results highlight the role of structural plasticity within the kinase domain in imparting the specificity of ligand binding and provide the framework for structure-guided design of activated c-Met inhibitors.
Introduction
The receptor tyrosine kinase c-Met is the cell surface receptor for hepatocyte growth factor (1, 2). Although c-Met plays normal roles in stem and progenitor cells, abnormal activity of c-Met resulting from overexpression, inappropriate ligand binding, or mutation is implicated in tumor growth, invasion, and metastasis (1, 2). Numerous preclinical studies with animal models support the notion that inhibition of c-Met provides a therapeutic approach to cancer treatment, and several biologic and small molecule agents are in clinical evaluation in humans (3).
The crystal structure of the unphosphorylated c-Met kinase domain has the canonical bilobal kinase fold typical of protein kinases (4). The unphosphorylated kinase domain adopts an autoinhibited conformation that is mediated largely by the activation loop, which is ordered and folds against active site features of the kinase (4). In particular, the ordered activation loop conformation obstructs access to the active site and places helix αC and conserved catalytic residues that depend on the orientation of helix αC in an inactive conformation (4).
Activation of c-Met begins with the binding of hepatocyte growth factor to the extracellular receptor domain of c-Met, which results in c-Met oligomerization and autophosphorylation of the kinase domain at Tyr-1234 and Tyr-1235 (1, 2). The structural basis of activation to date is limited to crystallographic studies of phosphorylation-mimetic mutants. This approach is exemplified by a triple mutant containing an activation loop mutation (Y1235D) present in certain carcinomas (5) and Y1194F and Y1234F mutations that were introduced to obtain homogenous unphosphorylated enzyme (6). In contrast to the fully ordered activation loop of the unphosphorylated enzyme (4), residues 1231–1244 of the activation loop of the putative phosphorylation-mimetic mutant Y1235D are disordered (6). One feature of activated kinases is the reorientation of helix αC that contributes to the formation of a viable active site (7). However, the Y1199/Y1234F/Y1235D mutant crystallized with helix αC in the inactive conformation (6). In addition, the Y1235D single-point mutant has a reduced kcat compared with the activated, autophosphorylated wild-type enzyme (8). It appears that the Y1235D activating mutant does not replicate the biologically relevant activation mechanism of c-Met autophosphorylation.
The clinical candidate MK-2461 is a novel, receptor tyrosine kinase inhibitor (3, 9). MK-2461 preferentially binds the phosphorylated c-Met kinase domain in vitro and preferentially inhibits phosphorylated c-Met in cancer cell lines (9). MK-2461 also inhibits the kinases Flt1 and Ron with similar potency to c-Met, but other kinases are 8–30-fold less sensitive to MK-2461 (9). The success of targeting kinases that are abnormally activated in disease (10) generates interest in understanding the structural basis of activated kinase inhibition exhibited by compounds such as MK-2461.
Here we present the structural basis of preferential inhibition of activated c-Met. First, the origins of c-Met kinase domain activation are revealed by the crystal structure of the kinase domain in the bona fide and biologically relevant autophosphorylated state. The structure differs from those previously observed with activating mutations. Second, the structural basis of selective inhibition of the dually phosphorylated kinase domain is revealed by the crystal structure of autophosphorylated c-Met in complex with an MK-2461 analog that is 20-fold selective for the dually phosphorylated state. Taken together, the results provide a framework for the rational design of inhibitors that target the activated state of c-Met.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A PCR product encoding residues 1048–1348 of human c-Met with a C-terminal His tag was amplified from partial c-Met cDNA (Open Biosystems LifeSeq3489592) and cloned via Gateway cloning method into a custom destination vector designed for baculovirus recombination derived from the transfer vector pVL1393 (Pharmingen, San Diego, CA) (11). The generation of recombinant baculovirus using BacMagic (Novagen) was performed according to the manufacturer's procedures.
Virus harboring the c-Met kinase domain was used to infect Sf21 insect cells at a multiplicity of infection of 0.2. The cells were grown at 27 °C and harvested 72 h post-infection. The cells were lysed by microfluidization in 50 mm Tris, pH 8.0, 250 mm NaCl, 5 mm 2-mercaptoethanol, 25 mm imidazole, and 5% glycerol. The soluble lysate fraction was loaded on to a HisTrap FF crude column in 50 mm Tris, pH 8.0, 150 mm NaCl, 5 mm 2-mercaptoethanol, and 25 mm imidazole, and the enzyme was eluted with a gradient of 50–500 mm imidazole. The enzyme was loaded on to Q-Sepharose in 50 mm Tris, pH 8.5, 50 mm NaCl, and the flowthrough was loaded onto heparin-Sepharose in 50 mm HEPES, pH 7.0, 50 mm NaCl, followed by elution with a gradient to 1 m NaCl. Final purification was with gel filtration with a Superdex 75 column equilibrated in 50 mm Tris, pH 8.0, 150 mm NaCl. The purified enzyme was exchanged via a desalting column into crystallization buffer (50 mm HEPES, pH 7.0, 250 mm NaCl, 3 mm DTT, 5% glycerol) prior to concentration for crystallography.
Autophosphorylated enzyme was generated by incubation in 50 mm Tris, pH 8.0, 150 mm NaCl, 5 mm MgCl2, and 1 mm ATP at 4 °C for 30 min. A mass increase of 160 Da was verified by LC/MS, the reaction was quenched with 10 mm EDTA, and the enzyme was exchanged into crystallization buffer as above.
Calorimetry
ITC2 was performed at 25 °C with an ITC200 (GE Healthcare) by injecting 2 μl of 300 μm compound 1 in 50 mm HEPES, pH 7.0, 250 mm NaCl, 3 mm DTT, 5% glycerol, and 1% Me2SO into 40 μm c-Met kinase domain in the same buffer. The injection time was 4 s, and the injection delay was 180 s. The data were fit with nonlinear least squares to a single-site binding model with Origin for ITC v6.0 (MicroCal) by varying stoichiometry, ΔH, and the association constant (Ka).
Activity Assays
Inhibition of peptide phosphorylation was carried out as described elsewhere (9) using 25 pm enzyme in assay buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 250 μm ATP, 5 mm MgCl2, 5 mm DTT, and 0.05 mg/ml BSA) for 60 min at 22 °C. Inhibition of c-Met kinase domain autophosphorylation was performed by direct observation of the singly phosphorylated state with LC/MS. Autophosphorylation reactions contained 25 nm enzyme in assay buffer (see above) and varying concentrations of compound 1 in Me2SO with a final Me2SO concentration of 1%. The reactions were initiated by the addition of 250 μm ATP, incubated at 37 °C for 3 h, and quenched with EDTA (final concentration, 10 mm). The samples were analyzed by LC/MS on a Thermo LTQ MS, using a Varian PLRP-S column and a gradient from 18 to 63% acetonitrile containing 0.01% trifluroacetic acid over 10 min to separate the c-Met kinase domain from BSA. The spectra were analyzed with Promass (Novatia) to deconvolute and assign peak intensities to the singly phosphorylated (+80 Da) and unphosphorylated forms of the protein.
Crystallization
The crystals were obtained using 15.6 mg/ml autophosphorylated c-Met kinase domain mixed at a 1:1 ratio with the reservoir solution (150 mm malic acid, 20% PEG 3350, pH 7.0). The crystals were grown at 20 °C as sitting drops using microseeding and cryo-protected with 20% ethylene glycol. Crystals of the autophosphorylated c-Met kinase domain in complex with compound 1 were obtained by co-crystallization under the same conditions of 15.6 mg/ml enzyme in the presence of a 2-fold molar excess of compound 1.
Structure Determination
The data were collected at the Industrial Macromolecular Crystallography Association beamline (Advanced Photon Source, Argonne National Laboratories) using an x-ray wavelength of 1.00 Å. Oscillation frames of 0.5 degrees were collected over a total rotation range of 180 degrees using an ADSC Quantum 210 detector. The data were integrated and reduced with HKL2000 (12). The structures were determined by molecular replacement using the structure of the c-Met kinase domain in complex with K252a (Protein Data Bank code 1R1W) as a starting model for rigid body refinement with REFMAC as implemented in CCP4 (13). The model was rebuilt manually with Coot (14) and completed using iterative rounds of refinement with autoBUSTER version 1.7.3 (Global Phasing Limited) and rebuilding. SigmaA-weighted (15) omit maps were calculated with autoBUSTER version 1.7.3. Structure alignments were performed with SSM (16) and DALI (17). The figures were prepared with PyMOL (Schrödinger LLC).
RESULTS
Selective Inhibition of Autophosphorylated c-Met
The small molecule inhibitor MK-2461 exhibits a 6-fold preference for the phosphorylated c-Met kinase domain in direct binding measured with surface plasmon resonance (9). Compound 1 is an analog of MK-2461 that shares the same tricyclic core. The direct binding of compound 1 to the c-Met kinase domain in both the unphosphorylated and autophosphorylated states was characterized with ITC (Fig. 1A). Both states of the enzyme bind compound 1 with a 1:1 stoichiometry. The binding affinity of compound for the autophosphorylated kinase domain (Kd = 27 ± 7 nm) is ~20-fold higher than for the unphosphorylated kinase domain (Kd = 0.55 ± 0.14 μm), indicating a marked preference for the phosphorylated state of the kinase domain. The binding of compound 1 to autophosphorylated c-Met is enthalpically driven (ΔH = −9.4 ± 0.1 kcal/mol and TΔS = 0.98 kcal/mol), whereas binding is relatively more entropy-driven to the unphosphorylated state (ΔH = −5.8 ± 0.1 kcal/mol and TΔS = 2.8 kcal/mol).

Preferential binding of compound 1 to the phosphorylated c-Met kinase domain. A, direct binding monitored with ITC to the unphosphorylated and autophosphorylated forms of the c-Met kinase domain. B, inhibition by compound 1 and K252a of peptide phosphorylation by the autophosphorylated c-Met kinase domain. C, inhibition by compound 1 and K252a of c-Met kinase domain autophosphorylation.
Compound 1 also exhibits a preference for the autophosphorylated state in functional assays. In a fluorescence assay that measures inhibition of the autophosphorylated kinase domain activity with a peptide substrate, compound 1 exhibits an IC50 of 11 ± 5 nm (Fig. 1B). In a LC/MS assay that directly measures autophosphorylation of the kinase domain to the singly phosphorylated state, compound 1 exhibits an IC50 value of 1.7 ± 0.3 μm (Fig. 1C).
The staurosporine analog K252a is a c-Met inhibitor (18). In the same assays as used for compound 1, K252a exhibits IC50 values of 113 ± 12 nm for peptide phosphorylation by dually phosphorylated c-Met (Fig. 1B) and 81 ± 9 nm for autophosphorylation (Fig. 1C). The results for K252a suggest that the different values observed for compound 1 do not reflect assay methodology but do reflect the combined properties of compound 1 and the enzyme phosphorylation state.
Structure of the Autophosphorylated c-Met Kinase Domain
The 1.6 Å crystal structure of the human c-Met kinase domain phosphorylated at Tyr-1234 and Tyr-1235 was determined to understand the structural basis of c-Met activation by phosphorylation and to provide the framework for understanding selective inhibition of the activated kinase domain (Table 1). The presence of dual phosphorylation was confirmed with mass spectrometry and by direct observation of the phosphate groups of Tyr(P)-1234 and Tyr(P)-1235 in the ligand-bound crystal structure (see below). The general structure of the C lobe is largely unchanged upon activation loop phosphorylation (Cα r.m.s.d. of 0.98 Å) with larger changes prevalent in the N lobe and activation loop (Cα r.m.s.d. of 3.8 Å).
TABLE 1
Structure determination statistics
The values in parentheses are for the highest resolution bin.
| c-Met | c-Met complex with compound 1 | |
|---|---|---|
| Resolution (Å) | 50.0-1.60 (1.70-1.60) | 50.0-1.75 (1.81-1.75) |
| Unit cell parameters (Å) | a = 39.720, b = 63.809, c = 112.63 | a = 42.895, b = 64.091, c = 111.107 |
| Space group | P212121 | P212121 |
| c-Met molecules per asymmetric unit. | 1 | 1 |
| Rsym (%) | 5.4 (46.0) | 5.4 (54.1) |
| Completeness (%) | 99.0 (98.4) | 98.7 (100.0) |
| Redundancy | 6.9 | 7.0 |
| R | 18.4 | 20.5 |
| Rfree | 21.4 | 22.7 |
| MolProbity score | 1.7 | 2.0 |
| Residues in most favored region of Ramachandran plot (%) | 98.2 | 96.6 |
| Number of atoms | 2568 | 2603 |
    Protein Protein | 2312 | 2372 |
    Waters Waters | 245 | 160 |
    Compound 1 Compound 1 | 39 | |
| Average B-factors (Å2) | ||
    Main chain Main chain | 19.16 | 31.10 |
    Side chain Side chain | 26.39 | 39.01 |
    Waters Waters | 34.55 | 40.34 |
    Compound 1 Compound 1 | 29.72 | |
| Bond length r.m.s.d. (Å) | 0.01 | 0.01 |
| Bond angle r.m.s.d. (°) | 1.4 | 1.2 |
The activation loop of unphosphorylated c-Met is folded onto the enzyme in an autoinhibitory conformation (Fig. 2A). In contrast, the structure of unliganded, autophosphorylated c-Met shows that the activation loop is ejected from the autoinhibitory conformation seen in the unphosphorylated structure (Fig. 2B). Electron density for the activation loop between the DFG motif and Leu-1245 is not visible in the structure, suggesting that residues 1225–1244 of the activation loop are disordered following phosphorylation.

Structures of the c-Met kinase domain in various states. A, the structure of the unphosphorylated kinase domain (4). The activation loop is ordered and induces an autoinhibited conformation. B, the structure of the dually phosphorylated kinase domain. The activation loop is ejected from the active site and becomes disordered. The G loop adopts an extended structure that occupies the region previously occupied by the activation loop in the unphosphorylated state. C, the structure of the complex formed by compound 1 and the dually phosphorylated kinase domain. The G loop adopts an induced fit conformation to cap the binding site, and the activation loop becomes ordered, showing the presence of the phosphate groups of Tyr(P)-1234 and Tyr(P)-1235. The activation loop, G loop, helix αC, and compound 1 are shown in orange, cyan, straw yellow, and yellow, respectively.
The ejection of the activation loop from the active site is accompanied by a marked reorientation of helix αC (Fig. 2B). The movement of helix αC is similar to that observed in the structure of other activated protein kinases (7) and leads to partial alignment of the side chains of Glu-1127 and Lys-1110 with the expected active conformation. However, the distance between the side chains (5.8 Å) is longer than would be expected for the conserved salt bridge typically observed in the fully activated conformation (7). The conserved catalytic base Asp-1204 and the Mg2+ chelating residue Asn-1209 of the catalytic loop move ~1.6 Å toward the active site upon phosphorylation.
The ejection of the activation loop also has marked structural consequences for the G loop, which is the Gly-rich loop that binds ATP. In the unphosphorylated structure (4), residues Met-1229 to Glu-1233 of the activation loop pack against the G loop, placing it in a position that occludes the ATP-binding site (Fig. 3A). Upon phosphorylation and ejection of the activation loop, the G loop adopts a remarkably different conformation that involves a movement of 9–11 Å for the Cβ atoms of Arg-1086, His-1088, and Phe-1089 (Fig. 3A). The extended β-hairpin conformation of the G loop is incompatible with the presence of the activation loop in the unphosphorylated state. This structural transition removes the active site obstruction seen in the unphosphorylated structure by the G loop side chains of Arg-1086, His-1088, and His-1089 and thus relieves an autoinhibitory feature of the unphosphorylated kinase.
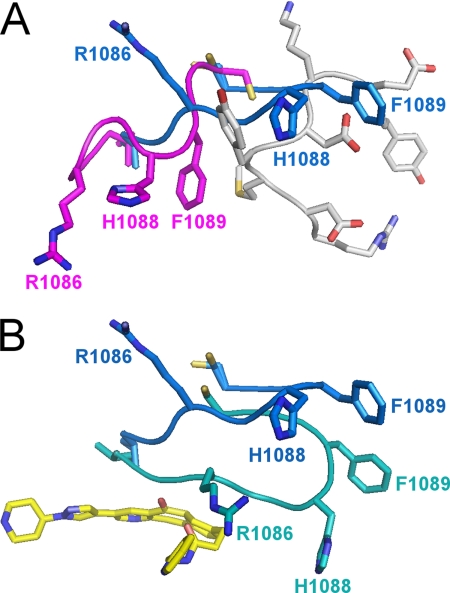
Conformation of the G loop of the c-Met kinase domain in various states. A, in the unphosphorylated state, the G loop (magenta) packs against the activation loop (gray) in a compact conformation that occludes the ATP-binding site. Upon phosphorylation, the G loop adopts an extended conformation (blue) that requires ejection of the activation loop (gray) and reveals the ATP-binding site. The side chains of Arg-1086, His-1088, and Phe-1089 undergo substantial conformational changes (9–11 Å for the Cβ atoms). B, in the ligand-bound state, the G loop undergoes an induced fit conformational change to cap the binding site. The side chains of Arg-1086, His-1088, and Phe-1089 undergo substantial conformational changes (6–20 Å), and His-1088 forms a new hydrogen bond with Asp-1204. The G loop in the unbound and bound states is shown in blue and teal, respectively.
Structure of the Autophosphorylated c-Met Kinase Domain Complex with Compound 1
The 1.75 Å crystal structure of dually phosphorylated c-Met bound to compound 1 was determined to understand the structural basis of selectivity for dually phosphorylated c-Met (Table 1). The conformation of the C lobe is largely unaltered upon ligand binding (Cα r.m.s.d. of 0.47 Å). In contrast, the N lobe experiences greater conformational changes (Cα r.m.s.d. of 1.3 Å), particularly for the G loop (see below). Phe-1223 is in a similar DFG-in orientation to the unbound phosphorylated form, as seen for Type I kinase inhibitors (19).
The observed electron density allows unambiguous positioning of compound 1 in the structure (Fig. 4A). The tricyclic ring of compound 1 occupies the ATP-binding site (Fig. 2C) in a predominantly hydrophobic environment formed by Ile-1084, Val-1092, Ala-1108, Lys-1110, Leu-1140, Tyr-1159, Met-1160, Met-1211, and Ala-1221 (Fig. 4B). The polar groups of the inhibitor make both direct and water-mediated hydrogen bonds with c-Met (Fig. 4C). A direct hydrogen bond is formed between the amide NH of the inhibitor and the backbone carbonyl of Arg-1086 (2.9 Å). The two carbonyl oxygens of compound 1 are coordinated, via a network of water-mediated hydrogen bonds, to the side chain and backbone amide nitrogen of Asp-1164 (Fig. 4C). Asp-1164 also forms a bridging hydrogen bond via a water molecule with the inhibitor pyridine moiety. Density for the pyridine group is not as clearly defined as for the rest of compound 1. However, the electron density, geometric constraints, and chemical environment are consistent with the modeled orientation.

Binding pose of compound 1. A, SigmaA-weighted omit maps showing the volunteer 2Fo − Fc 1 σ (blue) and Fo − Fc 3 σ (green) electron density for compound 1. B, hydrophobic interactions with the tricyclic core of compound 1 showing side chains of c-Met residues within 4 Å of the inhibitor (teal). C, polar interactions include a direct hydrogen bond with the Arg-1086 main chain carbonyl oxygen and several water-mediated hydrogen bonds, including one with Asp-1164. Water molecules are shown as red spheres. Compound 1 is shown in yellow, except the nitrogen and oxygen atoms are blue and red, respectively.
The G loop undergoes a conformational change upon binding compound 1 with the side chains of Arg-1086, His-1088, and Phe-1089 experiencing movements of 6–20 Å to form a cap to the binding pocket of compound 1 (Fig. 3B). The induced fit conformational change of the G loop also introduces new intramolecular interactions within c-Met (Fig. 5). Phe-1089 of the G loop forms a new packing interaction with helix αC, and Lys-1110 and Glu-1127 form a short salt bridge (2.8 Å). Also, the side chain of His-1088 of the G loop forms a new hydrogen bond with the side chain of Asp-1204 (2.5 Å).
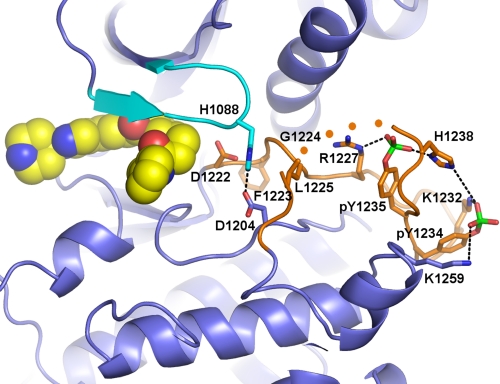
Stabilization of the activation loop of autophosphorylated c-Met. The induced fit conformation of the G loop (cyan) results in formation of a new hydrogen bond between His-1088 of the G loop and Asp-1204 and new packing interactions with Leu-1225 of the activation loop (orange). The two Tyr(P)-1234 and Tyr(P)-1235 residues form salt bridges with Lys-1232 and Lys-1259 and with Arg-1227 and His-1238, respectively. Asp-1222 of the DFG motif at the beginning of the activation loop is also shown, and the four disordered residues at the end of the activation loop are schematically represented with orange spheres.
One of the more striking differences observed between the unbound, phosphorylated c-Met structure and the structure observed in the presence of compound 1 is the nearly complete ordering of the activation loop, including the phosphate groups of Tyr(P)-1234 and Tyr(P)-1235 (Fig. 5). Only a short region of the activation loop remains disordered (residues 1240–1243). Upon loop stabilization, the phosphotyrosine residues form electrostatic interactions: Tyr(P)-1234 interacts with Lys-1232 and Lys-1253, and Tyr(P)-1235 interacts with Arg-1227 and His-1238 (Fig. 5).
DISCUSSION
Structural Basis of Activation by Autophosphorylation
The kinase activity of c-Met is stimulated by activation loop autophosphorylation at Tyr-1234 and Tyr-1235 following c-Met dimerization upon binding hepatocyte growth factor (1, 2). In the unphosphorylated enzyme, the activation loop is folded against the active site to disrupt the catalytically competent conformation of the kinase domain and occlude the ATP-binding site (4). The autoinhibitory features of c-Met are relieved by the ejection and disorder of the activation loop upon autophosphorylation. In particular, the ATP-binding site becomes exposed when the G loop adopts the extended conformation and helix αC and the catalytic loop reorients to form a viable active site. The side chain of Phe-1223 of the DFG motif is in the active DFG-in conformation in both the unphosphorylated and phosphorylated states. The change in helix αC orientation and the DFG-in motif mirror those seen in other activated protein kinases (7).
The ejection of the activation loop from the active site is most likely a consequence of the incompatibility of Tyr(P)-1234 and Tyr(P)-1235 with the autoinhibited conformation. In particular, Tyr-1235 is buried in the unphosphorylated structure and forms a hydrogen bond with Glu-1127 (4). It is likely that the activation loop rearrangement is driven by an unfavorable burial of the Tyr(P)-1235 phosphate (6).
It is notable that the active conformation of helix αC is not present in the triple mutant structure that includes the activation loop phosphorylation mimic Y1235D (6). Other structural studies of mutant c-Met kinase domains bound with inhibitors have also employed multiple mutations in the activation loop selected from single-point oncogenic mutations (20, 21). In these cases, the activation loop is disordered, as seen here upon phosphorylation, and formally consistent with the use of naturally occurring activating mutations. However, helix αC in the mutant structures (6, 20, 21) does not adopt the active orientation seen here for autophosphorylated c-Met. In the case of the ligand-bound mutant structures (19, 20), apo structures of the c-Met mutants have not been reported, so it is unknown whether the inhibitor induces the inactive helix αC conformation or that the orientation of helix αC results from the presence of the multiple mutations. Nonetheless, it is clear that bona fide autophosphorylated c-Met adopts an active conformation that is not necessarily induced by activation loop mutations.
Structural Basis of Selective Inhibition
The binding pose of compound 1 to the autophosphorylated kinase domain is incompatible with the presence of the autoinhibitory conformation of the activation loop of the unphosphorylated enzyme. In particular, the extension of the G loop following activation loop ejection upon phosphorylation is required to permit access to the ATP-binding site occupied by compound 1 (Fig. 2A). Moreover, the extended conformation of the G loop is primed in the dually phosphorylated state to bind compound 1. The G loop undergoes a marked induced fit conformational change of up to 20 Å in the presence of compound 1 that forms the binding pocket for compound 1 and facilitates new intramolecular interactions with other regions of the kinase domain. Ejection of the activation loop upon phosphorylation therefore allows the c-Met kinase domain to preferentially populate conformational states that are compatible with the binding of compound 1.
The ordering of the phosphorylated activation loop appears to be related to the conformational change of the G loop upon binding compound 1 and may reflect formation of a domino effect of interactions that originate with the G loop. The induced fit movement of the G loop upon binding compound 1 allows His-1088 at the top of the G loop to form a short (2.5 Å) hydrogen bond with Asp-1204 (Fig. 5). In turn, the His-1088–Asp-1204 pair forms packing interactions with Leu-1225, which is disordered in the unphosphorylated state, and may nucleate stabilization of the loop. Upon activation loop stabilization, Tyr(P)-1234 and Tyr(P)-1235 form new electrostatic interactions (Fig. 5). It is tempting to speculate that the G loop would adopt a conformation upon binding ATP similar to that induced by the occupation of the ATP-binding site by compound 1, leading to ordering of the phosphorylated activation loop that would then provide a scaffold for substrate recognition and binding.
The activation loops of protein kinases have been observed in both disordered conformations (as seen here for phosphorylated c-Met) and ordered conformations (as seen here for phosphorylated c-Met upon ligand binding). When viewed in isolation, the ordered activation loop of c-Met in the autophosphorylated, ligand-bound state appears to lack a regular structure (Fig. 6). When compared with the ordered activation loops seen in the crystal structures of the phosphorylated tyrosine kinase domains of IRK and Lck (22, 23), however, the phosphorylated activation loops of c-Met, IRK, and Lck adopt a clearly conserved structural motif (Fig. 6). The activation loops of Lck, c-Met, and IRK contain one, two, and three Tyr(P) residues, respectively, and share a conserved Tyr(P) (Tyr(P)-1235 in c-Met) (Fig. 6). The conserved Tyr(P) in each kinase forms an electrostatic interaction with a conserved Arg (Arg-1227 in c-Met) that forms the stapled base of the activation loop (Fig. 6).
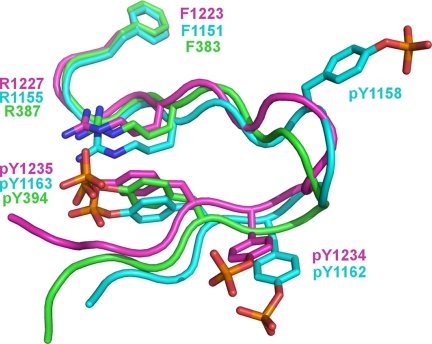
Structural conservation of ordered activation loops. The conserved interaction exemplified by Arg-1227–Tyr(P)-1235 of c-Met forms a conserved structural feature that stabilizes the activation loop upon phosphorylation. The phosphorylated activation loops are shown for c-Met (magenta), IRK (blue), and Lck (green).
IRK and c-Met contain a second conserved Tyr(P) residue (corresponding to Tyr(P)-1234 in c-Met; Fig. 6). The interactions involving Tyr(P)-1234 are not conserved as is the case for the Arg-1227–Tyr(P)-1235 interaction. IRK contains a third Tyr(P) at position 1158. Interestingly, Tyr-1158 of IRK aligns in sequence with Tyr-1230 of c-Met but with the side chains in a different orientation and with Tyr(P)-1158, making no contact with the rest of IRK (Fig. 6). It does not appear, therefore, that multiple phosphorylation of the activation loops of IRK and c-Met play a structural role in adopting the active conformation.
Thermodynamic Features of Selective Inhibition
Enthalpy drives binding of compound 1 to autophosphorylated c-Met, whereas binding to the unphosphorylated state is more entropy-driven. The enthalpy changes are consistent with activation loop ordering upon binding. This presumably contributes a favorable enthalpy term via the formation of new intramolecular interactions involving the activation loop including the ionic interactions with the phosphate groups as well as intermolecular bonding interactions with compound 1. In contrast, binding to the unphosphorylated form would involve ejection of the activation loop from the nucleotide-binding pocket leading to the loss or changes of intramolecular interactions of the activation loop in the unbound state. The absence of the phosphate group bonding network may also lead to a partial disorder of the activation loop, perhaps reflected in the difference in entropy changes upon binding to the two forms of c-Met.
Comparison with Other c-Met Inhibitors
Compound 1 is a Type I inhibitor in which the c-Met kinase domain adopts the active DFG-in conformation with helix αC in the active conformation. The Type I binding mode of compound 1 is clearly distinct from Type I modes seen for unambiguously unphosphorylated c-Met in which the activation loop is in the autoinhibitory conformation (24,–28). In these examples, crystal structures show that inhibitors form hydrogen bond interactions with the hinge region and π-stacking interactions with Tyr-1230 of the activation loop. Such inhibitors are potentially specific for the unphosphorylated state given the ejection of the activation loop upon phosphorylation and are potentially sensitive to the Y1230H mutation observed in certain cancers. A different Type I inhibitor is exemplified by AM7 (29). In the crystal structure of putatively unphosphorylated c-Met bound to AM7, the activation loop is disordered, and AM7 packs against helix αC, which is in the inactive conformation. The position of helix αC in the dually phosphorylated state is incompatible with the AM7 binding pose observed in the putative unphosphorylated state. Clearly, the ability of c-Met to adopt different conformations, including induced fit changes that may prove elusive to computational approaches, allows for a variety of inhibition modes that rely on the presence or absence of specific conformational features of both the activation and G loops.
Implications for Kinase Drug Design
The conserved nature of protein kinases introduces challenges to the discovery of selective inhibitors. Consequently, small molecule inhibitors for one kinase typically exhibit activity toward other kinases when profiled across the kinome (30). The conformational plasticity of kinase domains can result in distinct conformations of a given kinase. In the case of c-Met, selective binding of compound 1 to the phosphorylated kinase domain depends on a large (up to 20 Å) induced fit change of the G loop. Moreover, the previously unobserved ordering of the phosphorylated activation loop of c-Met upon inhibitor binding contributes to favorable binding thermodynamics and presents an unprecedented c-Met scaffold for the rational design of selective c-Met inhibitors. Our results support the notion that kinase inhibitor specificity can arise from details of structural plasticity that can introduce additional three-dimensional diversity to the overall kinase fold.
The atomic coordinates and structure factors (codes 3Q6U and 3Q6W) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
2The abbreviations used are:
- ITC
- isothermal titration calorimetry
- IRK
- insulin receptor kinase
- r.m.s.d.
- root mean square deviation.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m110.204404
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/article/S0021925820537447/pdf
Free to read at intl.jbc.org
http://intl.jbc.org/cgi/content/abstract/286/13/11218
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/content/full/286/13/11218
Free after 12 months at intl.jbc.org
http://intl.jbc.org/cgi/reprint/286/13/11218.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Biophysical and structural characterization of the impacts of MET phosphorylation on tepotinib binding.
J Biol Chem, 299(11):105328, 06 Oct 2023
Cited by: 0 articles | PMID: 37806493 | PMCID: PMC10654029
The mechanism of cancer drug addiction in ALK-positive T-Cell lymphoma.
Oncogene, 39(10):2103-2117, 05 Dec 2019
Cited by: 7 articles | PMID: 31804622 | PMCID: PMC7060126
Pharmacophore Mapping Approach for Drug Target Identification: A Chemical Synthesis and in Silico Study on Novel Thiadiazole Compounds.
J Clin Diagn Res, 11(5):KF01-KF08, 01 May 2017
Cited by: 4 articles | PMID: 28658808 | PMCID: PMC5483710
Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25(6):867-877.e3, 18 May 2017
Cited by: 14 articles | PMID: 28528776 | PMCID: PMC5507626
Approved and Experimental Small-Molecule Oncology Kinase Inhibitor Drugs: A Mid-2016 Overview.
Med Res Rev, 37(2):314-367, 24 Oct 2016
Cited by: 26 articles | PMID: 27775829
Review
Go to all (17) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (3)
-
(1 citation)
PDBe - 1R1WView structure
-
(1 citation)
PDBe - 3Q6UView structure
-
(1 citation)
PDBe - 3Q6WView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLN518.
Blood, 104(9):2867-2872, 15 Jul 2004
Cited by: 48 articles | PMID: 15256420
The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J Biol Chem, 292(38):15705-15716, 19 Jul 2017
Cited by: 11 articles | PMID: 28724631 | PMCID: PMC5612104
Structure of the Tie2 RTK domain: self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail.
Structure, 8(11):1105-1113, 01 Nov 2000
Cited by: 77 articles | PMID: 11080633
Identification of functional domains in the hepatocyte growth factor and its receptor by molecular engineering.
J Biotechnol, 37(2):109-122, 01 Sep 1994
Cited by: 27 articles | PMID: 7765452
Review




