Abstract
Free full text

Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells
Abstract
Transitions between pluripotent stem cells and differentiated cells are executed by key transcription regulators. Comparative measurements of RNA polymerase distribution over the genome's primary transcription units in different cell states can identify the genes and steps in the transcription cycle that are regulated during such transitions. To identify the complete transcriptional profiles of RNA polymerases with high sensitivity and resolution, as well as the critical regulated steps upon which regulatory factors act, we used genome-wide nuclear run-on (GRO-seq) to map the density and orientation of transcriptionally engaged RNA polymerases in mouse embryonic stem cells (ESCs) and mouse embryonic fibroblasts (MEFs). In both cell types, progression of a promoter-proximal, paused RNA polymerase II (Pol II) into productive elongation is a rate-limiting step in transcription of ~40% of mRNA-encoding genes. Importantly, quantitative comparisons between cell types reveal that transcription is controlled frequently at paused Pol II's entry into elongation. Furthermore, “bivalent” ESC genes (exhibiting both active and repressive histone modifications) bound by Polycomb group complexes PRC1 (Polycomb-repressive complex 1) and PRC2 show dramatically reduced levels of paused Pol II at promoters relative to an average gene. In contrast, bivalent promoters bound by only PRC2 allow Pol II pausing, but it is confined to extremely 5′ proximal regions. Altogether, these findings identify rate-limiting targets for transcription regulation during cell differentiation.
Mouse embryonic stem cells (ESCs) provide an excellent model for understanding the gene regulatory framework underlying self-renewal, pluripotency, and developmental progression. The core transcriptional factors OCT4, SOX2, and NANOG form a positive regulatory network that is specific to pluripotent ESCs and early embryos (Boyer et al. 2005; Loh et al. 2006). These complexes localize to many promoters and regulate the transcription of target genes important for maintaining stem cell identity (Jaenisch and Young 2008; Kim et al. 2008; Marson et al. 2008). Perturbations to the transcriptional network established by these key transcription factors can result in a cascade of events that ultimately leads to the differentiation of stem cells (Ivanova et al. 2006; Rizzino 2008; Chambers and Tomlinson 2009). Moreover, expression of these core transcription factors is critical in programming the transition of differentiated mouse embryonic fibroblasts (MEFs) to induced pluripotent stem cells, which have the properties and gene expression patterns of ESCs (Mikkelsen et al. 2008; Guenther et al. 2010). Quantitatively determining the complete transcriptional activity and the rate-limiting steps of transcription at all genes in both ESCs and differentiated cell lineages is critical to understanding the regulatory mechanisms used in maintaining and generating ESC pluripotency and in directing ESC differentiation to specific lineages.
A plethora of studies has revealed that transcription can be regulated at several stages in eukaryotes (Fuda et al. 2009). The RNA polymerase II (Pol II) transcription cycle is comprised of multiple steps, including (1) recruitment of general transcription factors, including hypophosphorylated Pol II, to the promoter forming the preinitiation complex (PIC); (2) initiation of transcription; (3) clearance of Pol II from the promoter-bound factors; (4) pausing of Pol II after it transcribes ~25–50 nucleotides (nt), which is facilitated by the DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) protein complexes; and (5) escape from the pause to productive elongation, which is driven by positive elongation factor b (P-TEFb). Although genes can be regulated at the step of Pol II recruitment (Nevado et al. 1999), global localization studies of Pol II indicate that a large number of genes in metazoans have a rate-limiting transcriptional step following the recruitment of Pol II to a promoter. More specifically, genome-wide chromatin immunoprecipitation (ChIP) studies in both human (Kim et al. 2005; Guenther et al. 2007; Rahl et al. 2010) and Drosophila melanogaster (Muse et al. 2007; Zeitlinger et al. 2007) cells uncovered significant fractions of active and inactive genes that maintain high levels of Pol II density at the 5′ end of the gene. Accumulation of Pol II at the 5′ end of the gene by ChIP may be the result of Pol II assembled in a PIC, a transcriptionally arrested complex, or a paused complex, each of which is at distinct transcriptional steps subsequent to Pol II recruitment. Unfortunately, these different Pol II complexes cannot be distinguished by ChIP (Rougvie and Lis 1988; Adelman et al. 2005) or methods that quantify small, processed RNA that are transcribed from promoter-proximal sequences (Seila et al. 2008).
Additional studies have sought to determine the status of Pol II accumulated at the 5′ end of genes. The mapping of a promoter-proximal Pol II transcription bubble by permanganate sensitivity assays in Drosophila cells showed that dozens of tested genes display what appears to be a rate-limiting step at an early stage of elongation (Muse et al. 2007; Zeitlinger et al. 2007; Lee et al. 2008). Moreover, sequencing of short-capped transcripts in Drosophila demonstrated that the polymerases at promoters had initiated but were prevented from transitioning to productive elongation (Nechaev et al. 2010). Another study in human cells demonstrated that ~30% of all coding genes featured peaks of transcriptionally competent polymerases near the transcription start site (TSS), implicating pausing as a common rate-limiting step in transcription (Core et al. 2008). These transcripts at the promoter-proximal region are generated in nuclear run-on reactions by RNA Pol II that are engaged in and competent for transcription, and therefore cannot be backtracked and arrested (Rougvie and Lis 1988; Adelman et al. 2005). Therefore, we refer to these accumulated, engaged polymerases assayed by genome-wide nuclear run-on methodology (GRO-seq) on the mRNA-coding genes as “paused” Pol II. These independent assays are critical for defining the rate-limiting steps in transcription that are modulated in transitions between cell states.
In addition to the presence of Pol II and pausing-related proteins (Guenther et al. 2007; Rahl et al. 2010), gene promoters also have distinct chromatin signatures. More specifically, active promoters have an open (nuclease-sensitive) chromatin structure with adjacent nucleosomes that are trimethylated at Lys 4 of histone H3 (H3K4me3) (Guenther et al. 2007; Mikkelsen et al. 2007; Barrera et al. 2008). In contrast, genes that are repressed have nucleosomes enriched with trimethylation at Lys 27 of histone H3 (H3K27me3), a modification mediated by Polycomb-repressive complexes (PRCs). It has yet to be determined how these combinations of modifications and PRCs influence promoter-proximal pausing of Pol II in vivo.
Recent studies have shown that these two histone H3 modifications are not mutually exclusive, as promoters of ~15% of genes in ESCs retain “bivalent” domains featuring both active H3K4me3 and repressive H3K27me3 marks (Mikkelsen et al. 2007). During lineage specification, many bivalent marks are resolved to a monovalent mark that is indicative of their transcriptional activity in differentiated cells. Interestingly, developmental regulators are one category of genes enriched with bivalent marks and are targets of PRCs (Boyer et al. 2006; Lee et al. 2006; Ku et al. 2008). Because ESCs have the potential to activate any developmental regulator as needed upon differentiation, bivalent chromatin domains in ESCs may provide the flexibility needed to prime these lineage-specific genes for activation or repression, depending on the developmental lineage.
PRC-mediated repression at promoters can be a result of a block in any step from chromatin accessibility and Pol II initiation to release from the pause site. Previous results indicate that the regulation may occur at either Pol II recruitment or the pause release step in ESCs, as some ChIP studies did not detect Pol II at the promoters of PRC target genes, suggestive of PRC's role in preventing Pol II recruitment (Boyer et al. 2006; Lee et al. 2006). In contrast, another ChIP study detected Pol II near the 5′ end of the selective genes in a post-initiation form (Ser5-phosphorylated Pol II), indicating that repression occurs early in Pol II elongation (Stock et al. 2007). Resolving the mechanism by which PRCs repress transcription requires that the regulated step in the transcription cycle be defined for PRC-regulated genes.
GRO-seq maps the distribution of short transcripts generated by transcriptionally engaged RNA polymerase that are allowed to transcribe (run-on) a short distance and incorporate an affinity tag into the nascent RNAs. Sequencing of these RNAs and aligning to the genome provide a density and orientation map on mRNA-encoding genes of transcriptionally competent Pol II (Core et al. 2008). These Pol II include those on the body of genes that are caught in the process of transcriptional elongation, as well as those that accumulate as promoter-proximal paused Pol II.
Here, we used our GRO-seq technology to provide quantitative transcription maps of mouse ESCs and differentiated MEFs that have significantly higher sensitivity, lower backgrounds, much improved dynamic range, and higher resolution than corresponding Pol II ChIP-seq analyses. Importantly, the distribution of transcriptionally engaged RNA polymerase across the pre-mRNA transcription units supports the importance of transcription regulation at the step of promoter-proximal pausing. Our study shows that promoter-proximal paused RNA polymerase on mRNA-encoding genes is a rate-limiting step in transcription for at least 40% of genes in both ESCs and MEFs. Comparison of the changes in Pol II density in the paused peak relative to the Pol II along the body of the genes in different cell types supports the hypothesis that the transition of Pol II from the paused to the productive elongation stage of transcription is a major regulated step during early differentiation in mouse cells. In addition, this study shows that bivalent genes are modulated during both elongation and the stages prior to elongation, as bivalent genes with only PRC2 have reduced levels of transcriptionally engaged polymerase that are confined to a region close to the TSS, while those with both PRC2 and PRC1 complexes are strongly depleted of transcriptionally engaged RNA polymerase in both the promoter-proximal region (sense) orientation and the upstream divergent (antisense) orientation.
Results
Genome-wide densities of transcriptionally engaged RNA polymerases in ESCs and MEFs
We used our recently developed nuclear run-on method, GRO-seq, to generate a genome-wide view of the location, orientation, and density of engaged RNA polymerases at high resolution in mouse ESCs and MEFs. A GRO-seq experiment generates short extensions (~100 nt in length) of Br-UTP-labeled nascent transcripts by engaged RNA polymerases (Core et al. 2008). These engaged RNA polymerases include those on the body of genes that are caught in the process of transcriptional elongation as well as those that accumulate as promoter-proximal paused RNA polymerase. The presence of the detergent sarkosyl in the run-on reactions strips factors that normally impede transcription of paused RNA polymerase, allowing these RNA polymerase to elongate with an efficiency comparable with engaged polymerase on the body of genes (Rougvie and Lis 1988). These labeled RNAs are then base-hydrolyzed to ~100 nt and affinity-purified three times using an anti-BrdU antibody, which also binds tightly to Br-U-substituted RNA. Triple selection enriches Br-UTP-labeled transcripts 500,000-fold from bulk RNA, as estimated using spike-in transcript controls (Supplemental Material). Both ends of the Br-UTP-labeled transcripts were enzymatically processed, then ligated to distinct linkers for mass scale Illumina sequencing from the 5′ ends of their cDNA copies (Supplemental Figs. S1–S3) and aligned to the mouse genome (mm9 assembly). The levels of sequenced run-on transcripts reflect the density of transcriptionally competent polymerases genome-wide.
We obtained ~19 million and 15 million nascent transcript sequences of 35- to 36-base lengths from two to three biological replicates in mouse ESCs and MEFs, respectively, 45% of which mapped uniquely to the mouse genome (Supplemental Table S1). GRO-seq is extremely sensitive in detecting low copy numbers of transcripts and highly selective for nascent transcripts with little contamination from processed mRNA accumulated in the cell, based on estimates using spike-in controls (Supplemental Material). Based on the total mass of RNA in each fraction, the overall purity of Br-UTP-labeled transcripts is 99.98% for our libraries, or a background level of 0.02%. Replicates in each group yielded strong correlations to each other (Supplemental Table S2). Thus, the reads represent run-on transcripts that are highly purified from bulk RNA, and the protocol is highly reproducible.
The density of engaged RNA polymerase in the body of a gene and on its coding strand is a quantitative measure of that gene's nascent transcriptional activity. Additionally, peaks in the GRO-seq density across a primary transcription unit represent slow steps in the transcription of that gene. Several notable general features of transcription units were uncovered by GRO-seq analysis of ESCs and MEFs, as exemplified in Figure 1, A and B. (1) The GRO-seq assay precisely measures the differential transcriptional activity in ESCs and MEFs along the primary transcription unit from initiation to termination, including polymerases that are transcribing through exons and introns. For example, the Tgfb3 and Esrrb genes exhibit strikingly different levels of transcriptional activity in ESCs versus MEFs. While, in most cases, known start sites align with the start of GRO-seq densities, the Esrrb gene shown here uses an alternative promoter that is not annotated in RefSeq. Recent Pol II ChIP analysis confirms the use of this alternative promoter in ESCs (Barrera et al. 2008), and microarray analysis by another group confirms high expression levels of Esrrb in ESCs and Tgfb3 in MEFs but not in the other cell types (Mikkelsen et al. 2007). (2) Transcriptionally engaged RNA polymerase (the bulk of which is Pol II) (Seila et al. 2009; Rahl et al. 2010) accumulates at the 5′ ends of many mRNA-encoding genes. The level of promoter-proximal Pol II at the 1700019E19Rik gene is higher than the Pol II level in the gene body in MEFs, and in ESCs to a lesser degree, indicating that Pol II's transition from this promoter-proximal pause to productive elongation is a rate-limiting step during transcription of this gene. (3) A divergent polymerase peak in the antisense direction is observed upstream of many active genes in both cell types (Core et al. 2008; Seila et al. 2008; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009). 1700020O03Rik is a clear example. The Pol II peak detected in a ChIP assay using an antibody against hypophosphorylated CTD (8WG16) (Seila et al. 2008) for the Nom1 gene is resolved into two distinct peaks of upstream divergent and paused polymerase in the GRO-seq analysis.
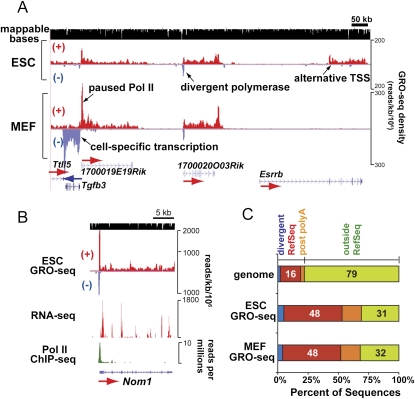
Densities of nuclear run-on transcripts in ESCs and MEFs analyzed by GRO-seq. (A) GRO-seq map in mouse ESCs and MEFs is presented in a strand-specific manner, with run-on transcripts along the top/forward (red +) and bottom/reverse (blue −) strands. GRO-seq reads are aligned to the genome at 1-nt resolution and the positions of their 5′ ends are displayed. GRO-seq densities are plotted here and hereafter as the number of reads per 1 kb per 1 million total uniquely mapped sequences in each library. Mappable regions are depicted as black bars in the top row, and RefSeq gene annotations are shown in the bottom row. (B) The mouse ESC GRO-seq map of the Nom1 gene is compared with maps of other genome-wide assays in ESCs. Y-axes show the total sequence reads per 1 kb per 1 million total uniquely mapped sequences (GRO-seq and RNA-seq) (Cloonan et al. 2008) or per million reads (Pol II ChIP-seq) (Seila et al. 2008). (C) The distributions of all mapped GRO-seq reads inside or outside (green) of RefSeq gene annotation are given for ESCs and MEFs. Annotated transcription units (red) are extended by 10 kb downstream on the same strand to include post-polyA transcription (orange), and by 5 kb upstream on the opposite strand to include the peak of divergent polymerase (blue). The percentages of the genome and GRO-seq libraries with respect to each relevant annotation are given in the bar graph (with divergent peaks accounting for 2%, 5%, and 4% of the genome, ESC, and MEF libraries, respectively, and post-polyA transcription accounting for 3%, 16%, and 16% as well).
In both ESCs and MEFs, ~50% of the mappable GRO-seq reads map to the coding strand and strictly within the boundaries of RefSeq gene annotations (Fig. 1C). Another ~20% of the GRO-seq reads appear to be from either divergent polymerase at annotated promoters (5%) or post-polyA transcription prior to termination (16%). These three types of regions contain 69% of reads but represent <21% of the genome; therefore, the bulk of RNA polymerase molecules that are engaged in transcription are intimately associated with annotated genes. In contrast to a view drawn from microarray analysis of mRNAs (Efroni et al. 2008), pervasive transcription (“transcriptional noise”) of the bulk of the genome is not as obvious using GRO-seq to quantify the distribution of transcriptionally competent RNA polymerases, and known gene deserts generally show extremely low levels of transcription with this very sensitive assay (Supplemental Fig. S4).
To investigate the occupancy and density of engaged RNA polymerases along all genes, we plotted the GRO-seq densities from both sense and antisense directions for all mappable RefSeq genes (Fig. 2A). The number of transcriptionally active genes was determined by using an experimentally determined background of 0.02% of reads at random throughout the genome (Supplemental Material). Statistical analysis shows that ~85% of genes in ESCs and 80% of genes in MEFs experience transcription above background (Figs. 2A, ,3A).3A). Additionally, we assessed whether each gene showed a significant level of paused Pol II relative to the transcription in the body of the gene by using the Fisher's exact test to compare the read density in the promoter-proximal region relative to the density in the body of each gene. A false discovery rate (FDR)-corrected Q-value cutoff of 0.01 was used to call significantly paused genes (Supplemental Material; Supplemental Fig. S5). The majority of genes with paused Pol II exhibit a sharp peak roughly 30 nt downstream from the annotated TSS, and a significant amount of divergent transcription is observed upstream of the TSS (Fig. 2A,B). The heat map display and a manual survey of individual genes along chromosomes on the University of California at Santa Cruz genome browser reveal that, for many transcriptionally active genes, promoter-proximal pausing and divergent transcription co-occur frequently (Figs. 2A, ,3A).3A). Statistical analysis indicates conservatively that ~39% and 34% of all mappable RefSeq genes have significant enrichment of promoter-proximal paused Pol II relative to that in the gene body in ESCs and MEFs, respectively (Fig. 3A). Among transcriptionally active genes, ~85% and 80% exhibit significant peaks of divergent polymerase in ESCs and MEFs, respectively (Fig. 3A). In addition, we found that the level of promoter-associated transcription increases as the elongation activity in the body of the gene increases, as shown by the heat map (Fig. 2A) and statistical analyses (Supplemental Figs. S6, S7). The composite profiles for all genes in both cell types are similar and consistent with a high level of promoter-proximally paused Pol II (Fig. 2B; Core et al. 2008), suggesting that pausing is a major rate-limiting step in the transcription cycle for a substantial fraction of genes in both mouse ESCs and MEFs. In addition, divergent polymerase peaking at ~200 base pairs (bp) upstream of the TSS is observed prominently for both ESCs and MEFs (Fig. 2A,B), supporting the finding that upstream divergent polymerase is common in mammalian cells (Seila et al. 2009).
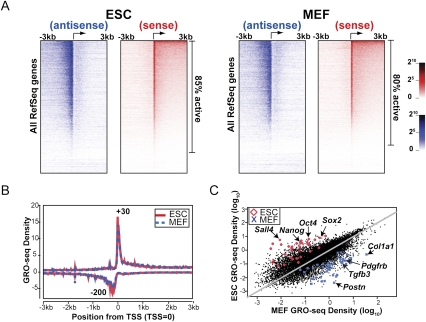
Comparisons of GRO-seq densities in the promoter-proximal region and gene body in ESCs and MEFs. (A) Heat map display of ESC and MEF GRO-seq densities for all RefSeq genes. For each cell type, gene order is listed from the highest (top) to the lowest (bottom) GRO-seq density in the gene body in the sense direction. Each row represents the average value of a block of 40 genes. (B) Average GRO-seq densities are plotted for all mappable RefSeq genes in 5-bp bins scanning 3 kb upstream of and downstream from the TSS for both ESCs and MEFs. (C) GRO-seq densities (number of reads on coding strand from +1 kb to the polyA annotation divided by mappable fraction of that length in kilobases and normalized by library size in millions) of all mappable RefSeq genes are compared between ESCs and MEFs. Fifty of the most highly enriched mature mRNAs in ESCs versus MEFs identified by previous microarray analysis (Sridharan et al. 2009) are highlighted on the plots. (Red diamond) ESCs; (blue cross) MEFs. A few genes with known function or expression pattern in each cell type are indicated with arrows.

Rate-limiting steps in transcription of individual genes and GO analysis of genes with paused Pol II peak. (A, left) Based on the GRO-seq profiles, each gene in each cell type was classified as transcribed if the density in the gene body was significantly above background, and as paused if the promoter-proximal density was significantly above the level in the gene body (Core et al. 2008). (Right) The fractions of each gene activation class were determined for ESCs and MEFs. The total number of RefSeq genes is 19,188. Additionally, each gene was classified as to whether it had a significant level of divergent polymerase or not. In each class, the fractions of genes with divergent polymerase activity are indicated with a darker shade. The small class III contains only 45 genes in ESCs and 37 genes in MEFs and is not displayed. (B,C) GO analysis of class I (paused, transcribed) and class II (not paused, transcribed) genes in ESCs (B) and MEFs (C) shows GO terms that show significant enrichment relative to all RefSeq genes in black (if significant in only one class) or gray (if significant in both classes). The numbers of genes in each class are given in parentheses.
The GRO-seq assay provides a tool for quantifying and comparing the differential transcription profiles in each cell type over a 5-log dynamic range (Fig. 2C). Notably, the pluripotency transcription factors Oct4 and Nanog show >50-fold higher GRO-seq densities in the body of the gene in ESCs over MEFs, while genes such as Pdgf2b and Col1a1 , which function in MEFs, show significant up-regulation in MEFs relative to ESCs. Gene ontology (GO) analysis reveals that ESCs generally show higher expression of genes that control transcription, cell cycle, mRNA processing, and chromatin modification (Supplemental Fig. S8). In striking contrast, MEFs show higher expression of genes that are involved in multicellular development, cell adhesion, and actin cytoskeleton organization. Changes in nascent transcription activity between the cell types generally agree with previous microarray studies of mRNA changes (Sridharan et al. 2009); however, some instances of discordance between GRO-seq levels and mature mRNA levels occur (Supplemental Fig. S9). These likely reflect transcripts that experience rates of post-transcriptional processing and/or RNA stability beyond the norm.
Transcription changes within and between four promoter transcription classes of genes in ESCs and MEFs
ESCs have an unusually open chromatin structure and a high level of global transcription, which have been proposed to be the basis for ESC pluripotency (Efroni et al. 2008). We too observed that ESCs have a slightly greater number of active genes (85%) as compared with MEFs (80%) (Fig. 3A). Furthermore, we calculated that 28% of genes are significantly more transcriptionally active in ESCs than in MEFs, whereas only 16% have significantly higher nascent transcriptional activity in MEFs than in ESCs, supporting heightened transcriptional activity and complexity as a distinguishable feature of ESCs (see also the Supplemental Material; Supplemental Fig. S1).
We further classified genes based on the density of transcriptionally engaged Pol II in both the gene body and the promoter-proximal region to derive four major categories (Fig. 3A; Supplemental Fig.S5): (1) class I (not paused, transcribed): active transcription in the gene body without a significant enrichment of density at the promoter; (2) class II (paused, transcribed): active transcription in the gene body with significantly higher 5′ pause density; (3) class III (paused, not transcribed): no significant transcription in the body of the gene, but with a significantly higher 5′ pause density; and (4) class IV (not paused, not transcribed): inactive transcription without a 5′ peak. Additionally, the presence or absence of a divergent polymerase peak near the TSS was determined for each gene to assess the relationship between divergent transcription and transcription in the sense direction. In total, ~39% and 34% of all mappable RefSeq genes have paused Pol II in ESCs and MEFs, respectively, and ~76% and 67% exhibit peaks of divergent polymerase in ESCs and MEFs, respectively (Fig. 3A). Although ESCs have a higher number of genes classified as paused, the proportions of transcriptionally active genes where pausing is a rate-limiting step (class II) or where it does not appear to be so (class I) are almost equally divided in both ESCs and MEFs, indicating that neither cell type possesses a skewed bias toward transcription regulation via one mechanism or the other.
About half of the transcriptionally active genes in ESCs and MEFs have significantly higher 5′ levels of Pol II, suggesting that promoter-proximal pausing is a common rate-limiting step during transcription. In contrast, very few (<2%) of transcriptionally inactive genes have paused Pol II (class III) (Fig. 3A), bolstering the view that pausing is associated more with transcribed, rather than strictly nontranscribed, genes (Core et al. 2008). (Note, however, that the sensitivity and low background of GRO-seq forces us to call genes as transcribed that would be considered inactive in a microarray assay.) The majority of transcriptionally active, but not inactive, genes also has RNA polymerase that is engaged in divergent transcription (Fig. 3A).
Strikingly, GO analysis has revealed that class I and class II genes are each enriched for distinct categories of gene function. Class I, and not class II, genes in ESCs are significantly enriched for genes that play roles in multicellular organismal development, cell adhesion, and intracellular signaling cascade (Fig. 3B). In contrast, class II (and not class I) genes in ESCs are significantly enriched for genes that function in response to extracellular or intracellular stimuli, such as translation, cell cycle, DNA damage, and modification-dependent catabolic processes. The GO terms enriched for both class I and class II genes were the same in MEFs (Fig. 3C). These results indicate that different cellular processes use distinct mechanisms when regulating transcription, and imply that the transcription activators that are associated with a particular cellular process act at the same step in transcription regulation.
A majority of genes in both ESCs and MEFs are in the same promoter-transcription class (Fig. 4A), but often exhibit substantial and quantitative differences in their relative amounts of polymerases in the promoter-proximal pause or the gene body region (Fig. 4B, panels a–c). Additionally, we found that many genes change their promoter transcription classification in MEFs compared with ESCs, indicating that they have undergone regulated changes that alter their rate-limiting step.
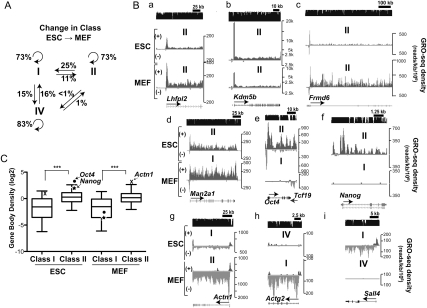
Targets of regulation in the transition from ESCs to MEFs. (A) The percentages of genes from each promoter transcription class that change class from ESCs to MEFs are presented above each straight arrow. The circular arrows represent the number of genes that do not change classification. (B) Representative GRO-seq plots comparing ESCs (top) versus MEFs (bottom) of genes that maintain (panels a–c) or switch (panels d–i) their promoter transcription class. (C) Comparisons of the average GRO-seq densities in the bodies of class I and class II genes in ESCs and MEFs. Oct4 (dot) and Nanog (star), two core pluripotency factors that switched from class II in ESCs to class I in MEFs, are presented to show the difference in the gene body densities between two cell types. Actn1, which is significantly up-regulated in MEFs and switched from class I in ESCs to class II in MEFs, is indicated with an X. The middle line in each box plot indicates the median value, the top and bottom edges of the box plot are the 75th and 25th percentiles, and the small horizontal bars denote the 95th and fifth percentiles. (***) P-value < 0.0001 by Mann-Whitney test.
Twenty-five percent of the paused, transcribed genes (class II) in ESCs transition to transcribed, not paused (class I) in MEFs (Figs. 4A,B, panels d–f). Notably, in ESCs, the genes encoding the core pluripotency transcription factors—e.g., OCT4 and NANOG (Jaenisch and Young 2008; Chambers and Tomlinson 2009)—are expressed at high levels, yet possess higher densities of engaged Pol II at their 5′ end (class II genes) (Fig. 4B, panels e,f), indicating that pausing remains a rate-limiting step for transcription of these genes even when they are highly expressed. In MEFs, the core pluripotency transcription factors are dramatically down-regulated (Fig. 2C), with Oct4 and Nanog showing extremely low Pol II on the body of the gene or the pause region that is barely above the background level (among the lowest percentile expression levels of class I genes) (Fig. 4C). Notably, class II genes generally showed much higher transcriptional activity than class I genes (P < 0.0001 by Mann-Whitney test) (Fig. 4C). Among the genes that transitioned from class II in ESCs to class I in MEFs, ~50% were significantly down-regulated in MEFs and ~11% were significantly up-regulated in MEFs. This up-regulated transition is expected for genes activated in MEFs that are no longer rate-limited in the transition of paused Pol II to productively elongating states. Importantly, these data also indicate that pausing is not strictly a repressive mechanism, and that a reduction in pausing can accompany a reduction in gene expression. These findings agree with studies in Drosophila cell culture, where a function of paused Pol II is to prevent nucleosomes from binding key promoter sequences and thereby repressing transcription (Gilchrist et al. 2008, 2010).
Transcribed, nonpaused genes (class I) transition readily to paused, transcribed (class II) (Figs. 4A,B, panel g), as expected, if escape from the pause to productive elongation becomes rate limiting. Among the 11% of class I genes in ESCs that switched to class II in MEFs, 49% and nearly 19% became significantly up-regulated and down-regulated, respectively, in MEFs. Thus, changes in gene expression during differentiation are accompanied by regulatory changes that can eliminate or generate pausing as a rate-limiting step.
Roughly 15% of completely silent genes (class IV) in ESCs become transcribed, not paused (class I) in MEFs and vice versa (Fig. 4A), representing genes regulated at or prior to Pol II initiation (Fig. 4B, panels h,i). Interestingly, paused, transcribed genes (class II) rarely transition to completely inactive (class IV), and, correspondingly, inactive genes rarely transition to paused and transcribed, at least in these cell types. Therefore, paused genes appear to be designed to be active and tunable in both cell types, rather than to transition between a paused and an inactive state or vice versa. The paused Pol II at the 5′ end of the gene may offer the advantage of maintaining the transcriptional activity of promoters that must be responsive to a myriad of cellular signaling pathways during differentiation.
Entry into productive elongation is a general mechanism of regulating transcription in differentiation
Mouse genes with higher transcriptional activity in the gene body also have higher levels of Pol II density at the 5′ end when either ESCs or MEFs are analyzed. This can be seen from examples of individual genes (Fig. 4B, panels a–c) and in the genome-wide analysis (Supplemental Fig. S6A). However, the pausing index (the ratio of the GRO-seq density at the 5′ end of the gene to that in the gene body) decreases with increasing gene activity, as seen with individual genes (Fig. 4B, panel c) and in the genome-wide analysis (Supplemental Fig. S6B).
The transition of ESCs to MEFs may involve more than one regulatory event. Nonetheless, by examining the changes in Pol II distribution on all genes between cell types, we can infer whether the net regulatory changes have had major effects on rates of initiation or escape of paused Pol II to productive elongation (Fig. 5A). For example, if the density of Pol II on the body of the gene increases much more than the density on the promoter (i.e., the pausing index decreases with an increase in expression), then we can infer the greater regulatory effect is on the rate of pause escape. If an increase in Pol II density on the gene body is accompanied by similar increases in both paused and gene body Pol II (i.e., no changes in the pausing index), then we can infer the major change is on the rate of initiation (Core et al. 2008).
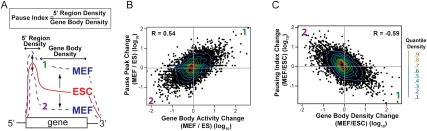
Regulation of gene expression by changing the efficiency of entry into productive elongation. (A) Illustrations depicting types of transcriptional activity change in MEFs (increase [1] and decrease [2]) relative to ESCs representing quadrants 1 and 2, as in B and C, are provided. Pause index is defined as the ratio of pause peak density to gene body density. The fold changes of pause peak GRO-seq density (B) or pausing index (C) versus gene body GRO-seq density in MEFs relative to ESCs are plotted for all mappable genes. The R-value, determined by Pearson's correlation, is presented within the plot. Contour lines by decile are shown in the heat map scale at the right.
Overall, we found that changes in the gene body transcription activity in MEFs relative to ESCs are accompanied by qualitatively (i.e., of the same sign) similar changes in GRO-seq density at the 5′ end, consistent with rates of recruitment and initiation indeed contributing to the overall activity of a gene (Fig. 5B). Nevertheless, as transcription levels increase, the relative rate of release of paused Pol II into productive elongation increases to a greater extent than entry into the pause site, and, correspondingly, as transcription levels decrease, the relative rate of release of paused Pol II into productive elongation decreases to a greater extent than entry into the pause site (Fig. 5C). These findings support the hypothesis that full-length transcription has a significant component of control at the level of escape of paused Pol II into productive elongation.
Developmental regulatory genes are transcribed at a modest level in ESCs and are not enriched for pausing
Because ESCs are pluripotent, the promoters of genes that regulate and execute lineage specification are expected to be poised for activation but repressed in ESCs by repressive protein complexes PRC1 and PRC2 (Ku et al. 2008). We evaluated whether such developmental regulatory genes have paused Pol II and assessed their level of expression. We found that, although transcriptional activity is detectable in ESCs at many of these master regulators of lineage specification, only a small portion of these active genes display significantly higher GRO-seq-measured densities at the 5′ end (class II) (Fig. 6A).

Developmental regulators of many lineages are transcribed but not paused in ESCs. (A) The promoter transcription classes and GRO-seq levels of known regulators associated with lineage specification are shown for ESCs and MEFs. The GRO-seq density levels in the body of the gene (from the lowest to the highest, 10%, 25%, 50%, 75%, and 100%, as ranked by the gene body density) is indicated by heat map for class I (green) and class II (orange) genes. The lists of developmental regulators are compiled from Mikkelsen et al. (2007), Stock et al. (2007), and Marson et al. (2008). (B) The changes in the GRO-seq gene body density for developmental controllers and markers involved in mesenchymal lineages (left) or all lineages except for mesenchymal lineages (right) are compared between ESCs and MEFs. Significance of changes in expression levels between cell types are P < 0.05 for mesenchymal and P < 0.01 for nonmesenchymal lineage controllers by Mann-Whitney test.
Compared with MEFs, which are of mesenchymal lineage, ESCs express a more diverse range of lineage specifiers. In general, the transcription level of these developmental regulators is clearly higher in ESCs compared with MEFs when genes related to mesenchymal lineage are excluded (Fig. 6B, right), albeit expressed at a modest level compared with the average GRO-seq density of all active genes (Fig. 6A). Interestingly, neuroectodermal or neuronal lineage-controlling master transcription factors such as Olig1, Olig2, and Nestin are transcribed at the highest levels relative to other lineages in ESCs. In contrast, genes controlling mesenchymal lineage establishment are consistently up-regulated in MEFs relative to ESCs (Fig. 6B, left). Together, our findings demonstrate that master regulators or markers of many different lineages are broadly transcribed at a modest level in ESCs, but mostly do not possess a significant level of paused Pol II.
RNA polymerase at genes whose promoters have either activating or repressing histone modifications
Promoters usually have adjacent nucleosomes that bear either an H3K4me3 activation mark or an H3K27me3-repressive mark. Comparison of GRO-seq with ChIP analysis of histone methylation marks shows that the genes that harbor H3K4me3 near the promoter (Ku et al. 2008) display significant levels of transcriptionally competent RNA polymerase at the promoter in ESCs, as measured by GRO-seq (Fig. 7A). In contrast, genes with only H3K27me3 show very little GRO-seq activity. The metagene profiles of Pol II and H3K4me3 ChIP-seq densities in genes with different GRO-seq signal levels (deciles) show that the level of transcriptionally competent polymerase correlates quantitatively with the amount of promoter-associated Pol II and H3K4me3 marks (Fig. 7B).
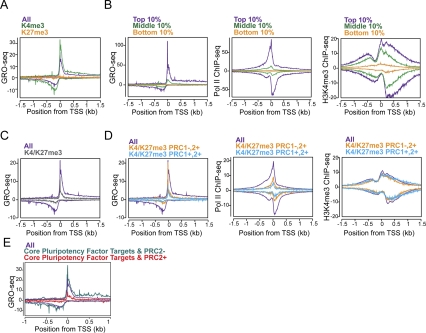
Regulation of distinct steps in transcription at polycomb target genes. (A) GRO-seq metagene profiles of all genes (purple), the subset of genes marked with H3K4me3 but not H3K27me3 (green), or the subset with H3K27me3 but not H3K4me3 (orange) near the promoters in ESCs (Ku et al. 2008). (B) The composite profiles of GRO-seq, Pol II (8WG16) ChIP-seq (Seila et al. 2008), and H3K4me3 ChIP-seq (Mikkelsen et al. 2007) are shown for genes whose level of expression in the gene bodies are in the top 10% (purple), middle 10% (green), and bottom 10% (orange) of active genes in ESCs, as determined by GRO-seq. The Y-axis is reads per kilobase per million. For ChIP-seq data, the forward and reverse reads are plotted above and below the horizontal axis, respectively. The midpoint between peaks of forward and reverse read density corresponds to the in vivo binding site, as described in Seila et al. (2008). (C) GRO-seq metagene profiles for all genes (purple) and H3K4me3/H3K27me3 bivalent genes (gray) (Ku et al. 2008). (D) Bivalent genes were subclassified into those that have PRC1 (cyan) or not (squash), based on Ku et al. (2008). The composite profiles for each subclass are plotted for GRO-seq, Pol II ChIP-seq, and H3K4me3 ChIP-seq, as in B. (E) The average GRO-seq densities of genes targeted by core pluripotency transcription factors with (sea green; n = 381) or without (red; n = 2,838) PRC2 component SUZ12 co-occupancy in ESCs. The gene lists were taken from Marson et al. (2008).
RNA polymerase is regulated at distinct steps on subsets of bivalent genes
The mature mRNA expression level of many developmental regulator genes with bivalent histone modifications of both H3K4me3 and H3K27me3 is almost undetectable in ESCs by microarray technology (Boyer et al. 2006; Lee et al. 2006). While the presence of H3K4me3 suggests that these genes may be “poised” for activation, the exact state (and even presence) of polymerase at these genes is not clear, given the contrasting views reported to date (Mikkelsen et al. 2007; Stock et al. 2007). GRO-seq data show that bivalent genes in ESCs have considerable transcriptionally engaged polymerase near the promoter, but greatly reduced levels of productively elongating polymerase (Fig. 7C). Nonetheless, these genes are not completely silent. We found that bivalent gene classes exhibit significant levels of productive elongation that are higher than at PRC-bound genes with H3K27me3 but not H3K4me3 (Fig. 7, cf. A and C).
The relationship of the two polycomb group complexes PRC1 and PRC2 to promoter-associated polymerase is revealed when the bivalent genes are subdivided into those that contain the PRC1 group member Ring1B and PRC2 (PRC1+,2+), and those that contain only PRC2 (PRC1−,2+). In particular, bivalent genes that lack Ring1B binding (PRC1−,2+) (Ku et al. 2008) display a high peak of transcriptionally engaged polymerases at the TSS, whose peak is comparable with the average level of all genes, but this polymerase level for these PRC1−,2+ bivalent genes falls rapidly with distance from the TSS (Fig. 7D, left). In contrast, bivalent genes that are targeted by both PRC2 and PRC1 (PRC1+,2+) exhibit much lower levels of transcriptionally engaged polymerase, both proximal to the promoter and downstream. Cumulative distribution analysis of GRO-seq reads in the 100-bp window starting at the TSS shows that that these two bivalent gene classes are distinct in that PRC1−,2+ bivalent genes show distributions of a higher level of engaged Pol II at the TSS relative to PRC1+,2+ bivalent genes (Supplemental Fig. S10). A significant difference was also observed in the GRO-seq densities at the 5′ Pol II peak region between PRC1+,2+ bivalent and PRC1−,2+ bivalent gene classes (P < 0.0001 by Mann-Whitney test) (Supplemental Fig. S11A). Total Pol II occupancy by ChIP-seq (Seila et al. 2008) shows that Pol II recruitment is markedly reduced for both classes of bivalent genes (Fig. 7D, middle).
Whereas the peak height of sense strand, promoter-proximal engaged Pol II is similar for all genes and the PRC1−,2+ bivalent genes, the peak of divergent polymerase (by both GRO-seq and ChIP-seq) is greatly reduced. The metagene profile result is not due simply to more variable spacing between the TSS and the divergent peak for this class of genes. While individual PRC1−,2+ bivalent genes have identifiable peaks of divergent polymerase, the peak heights are significantly lower than the genome-wide distribution. Surprisingly, even with lower transcriptional activity and reduced levels of Pol II occupancy in the promoters of both bivalent genes classes, the level of H3K4me3 is unchanged in comparison with all genes (Fig. 7D, right). Thus, we did not observe an obligatory coupling of H3K4me3 and promoter-associated Pol II, a result also seen in early development (Vastenhouw et al. 2010).
The maintenance of the pluripotent state depends on a specific set of transcription factors (Jaenisch and Young 2008; Chambers and Tomlinson 2009). Core pluripotency transcription factors often colocalize at the promoters of the target genes that are active in ESCs (Boyer et al. 2005; Loh et al. 2006; Marson et al. 2008). However, these core pluripotency regulators also bind to the promoters of a significant number of repressed genes that are associated with PRC proteins (Boyer et al. 2006; Lee et al. 2006). The genes that are bound by both the core pluripotency factors and the PRC2 complex have dramatically reduced peaks of divergent and paused Pol II relative to promoters that have pluripotency factors and no or little PRC2 relative to the average of all genes (Fig. 7E; Supplemental Fig. S11B). These repressive complexes comprised of core pluripotency transcription factors and PRCs in ESCs are most likely to include PRC1 complex as well (Endoh et al. 2008).
Discussion
Transcription regulation at the transition from pause to elongation
Our GRO-seq analyses in mouse ESCs and MEFs demonstrate that nearly 40% of all coding genes exhibit a higher density of engaged polymerase at the 5′ end of genes relative to the gene body that peaks ~30 bases downstream from the TSS. This agrees well with less sensitive and nondirectional mapping methods such as ChIP-seq analysis of Pol II on encoding genes (Marson et al. 2008). Almost all genes that have these paused Pol II peaks in ESCs and MEFs exhibit detectable levels of transcriptional elongation in the gene body. This agrees with recent reports (Stock et al. 2007; Core et al. 2008; Hargreaves et al. 2009; Ramirez-Carrozzi et al. 2009), although the amount of the transcription elongation activity varies widely from gene to gene. Furthermore, genes with the highest Pol II density in the gene body often possess a significant level of paused Pol II at the 5′ end in mammals (Core et al. 2008) and in Drosophila cell culture (Gilchrist et al. 2008, 2010). This finding indicates that, even in highly transcribed genes, pausing can remain a rate-limiting step. In the case of the well-known model system of pausing, the Drosophila heat-shock genes, pausing can still be detected under highly induced conditions, albeit the pausing index is dramatically reduced (Giardina et al. 1992; Boehm et al. 2003). More importantly, it shows that, whereas the rate of paused Pol II escape into productive elongation can be tuned up or down, paused Pol II is rarely entirely blocked from transcribing the body of the gene.
Our observation that the majority of genes with paused Pol II show detectable transcription elongation is in apparent conflict with an estimate that up to 30% of human genes showed no detectable full-length transcripts by microarray or sequencing methodology, yet contain 5′ Pol II peaks by ChIP assays (Guenther et al. 2007). There can be a couple of explanations. First, measurements of transcriptionally competent polymerase by GRO-seq and mature mRNA abundance by microarray are not always expected to correlate to each other. For example, gene transcripts that are regulated by pre-mRNA processing or rapidly turned over may be present in extremely low amounts in the mRNA pool and are undetectable by conventional methods for measuring transcript abundance. Second, the sensitivity in detecting transcripts by GRO-seq is comparable with very deep sequencing by RNA-seq, but is one to two orders of magnitude greater than microarray assays (Supplemental Fig. S7; Core et al. 2008). Low levels of transcription detected by sequencing would be scored as silent by a hybridization assay.
Here, we estimated that ~35%–40% of all RefSeq genes in ESCs and MEFs feature a paused Pol II peak at their 5′ end by GRO-seq, while a genome-wide Pol II ChIP localization study revealed that ~48% of coding gene promoters are occupied with a significant level of Pol II relative to the gene body region (Rahl et al. 2010). The difference in these estimates may be due to active, not-paused (class I) genes (classified by GRO-seq) that have substantially higher levels of Pol II that either are in a PIC or are arrested (Adelman et al. 2005)—both of which are not expected to be detected by GRO-seq—or due to differences in the experimental protocols or criteria used to call an excess of promoter-associated Pol II.
Rate-limiting steps in transcription can be a point of regulation at which regulatory factors act positively or negatively. By examining the promoter activity class transition from ESCs to MEFs, we uncovered that promoters can be placed into two categories. Some promoters maintain transcriptional activity at the promoter-proximal sites and do not get completely turned off (e.g., class switch of I to II, or II to I). The other category includes promoters that are regulated primarily by the recruitment and initiation of Pol II (e.g., class switch of I to IV, or IV to I). By quantitatively comparing ESCs and MEFs, we were able to establish that not only are rates of recruitment and initiation regulated between cell types, but the rate of escape from pausing is a general target of regulation as well.
Active genes often have a peak of divergently oriented polymerases that are likely to be Pol II, because the associated RNAs can be capped (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009) and they overlap with Pol II densities measured by ChIP-seq (Seila et al. 2008). Transcription in the divergent orientation generally co-occurs with transcription of the annotated gene, but Pol II enters into productive elongation only on the sense orientation of the annotated gene. Thus, the mechanism that controls the fate of these Pol II complexes is strikingly asymmetric. Surprisingly, RNA polymerase II and its modifications (histone modifications), as well as many factors known to participate in initiation and pausing, appear to be symmetric, associated with both divergent and paused Pol II (Rahl et al. 2010). Therefore, identifying the cis-elements and the factors responsible for P-TEFb-mediated escape into productive elongation will be critical for understanding the directionality of productive elongation (Seila et al. 2009).
Transcription of genes with bivalent chromatin domains is regulated distinctly by PRC2 and PRC1
Determining the presence and status of Pol II at bivalent genes is important for understanding the mechanisms that govern their transcription. We found that bivalent genes that are targets of the PRC2 complex, but not PRC1, retain promoter-proximal engaged Pol II that is more tightly confined to a region near the TSS and also display dramatically reduced levels of elongating Pol II. These results suggest that transcription at bivalent genes bound by PRC2 is regulated after the initiation step but very early in elongation. Although PRC2 activity has been shown to function in recruiting PRC1 and in DNA looping, no connections to distinct steps in transcription have been made previously (Simon and Kingston 2009).
The bivalent genes that are occupied by both the PRC2 and the PRC1 complexes display much less promoter-proximal Pol II, suggesting a preinitiation block to earlier steps in transcription at these genes. This is consistent with the ability of PRC1 to compact nucleosomes (Francis et al. 2004), interfere with specific functions of the transcriptional apparatus, or both (Breiling et al. 2001; Dellino et al. 2004). The stronger repression of bivalent genes through the additional activity of PRC1 is also evident in that these genes show more robust retention of repressive chromatin through differentiation than PRC1−,2+ bivalent genes (Ku et al. 2008). Nonetheless, these genes are not completely silent. We found that both bivalent gene classes exhibit significant levels of productive elongation that are higher than at PRC-bound genes with H3K27me3 but not H3K4me3 (Fig. 7, cf. A and C).
The transcriptional activities of most genes are proportional to the levels of H3K4me3 on their promoter regions (Pokholok et al. 2005). In marked contrast, the transcriptional activity and divergent transcription of both classes of bivalent genes can vary widely, but the amount of H3K4me3 on the promoter remains at a level that is almost equal to the genome-wide average (Fig. 7D). This constant presence of H3K4me3 modifications across the promoter region may suggest that bivalent genes are poised for further activation. Recently, H3K4me3 modifications have been shown to evoke a dynamic cycle of histone acetylation and deacetylation at the promoters of inactive genes, which may facilitate the cross-talk between different histone modifications to prepare for activation (Wang et al. 2009). In addition, the high affinity of the TAF3 subunit of TFIID for the H3K4me3-modified histone tail may assist activation for bivalent genes that are highly enriched for CpG islands but lacking a TATA box (Vermeulen et al. 2007).
Many developmental regulatory genes are targets of PRC2 and PRC1 in ESCs, and most are transcribed at a modest level but do not feature a significant peak of paused Pol II. Interestingly, another study has shown that these genes retain a high level of Ser5-phosphorylated Pol II at the promoter relative to the gene body (Stock et al. 2007). Although paused Pol II is Ser5-phosphorylated, these promoters could possibly contain a form of Pol II that either has not fully entered elongation or is backtracked and unable to elongate in a run-on assay.
Most developmental regulatory genes are not highly expressed in ESCs; however, we do note that regulators of multiple lineages show some “promiscuous transcription,” and this shares some similarities with what has been observed in the hematopoietic system (Hu et al. 1997; Miyamoto et al. 2002). Interestingly, the regulators of neural and neuroectodermal lineages are among the highly expressed regulators in ESCs, supporting the hypothesis that ESCs in culture have an “aptitude” for neural differentiation (Hemmati-Brivanlou and Melton 1997; Ying et al. 2003).
Pausing and the responsive transcription control of key pluripotency regulatory genes
OCT4, SOX2, and NANOG play important roles in preventing activation of specific lineage differentiation pathways, as well as forming the positive feedback transcription network for maintaining and establishing the pluripotent and self-renewal potentials in ESCs (Jaenisch and Young 2008). Oct4 and Nanog are actively transcribed in ESCs but still exhibit a rate-limiting step at pausing. Rapid, synchronous, and high levels of activation correlate better with genes that possess paused Pol II over genes that do not (Core et al. 2008; Boettiger and Levine 2009). Taken together, we speculate that pausing provides a responsive transcriptional regulatory step for controlling the level of critical core pluripotency transcription factors in ESCs.
The profile of engaged RNA polymerases provides both a measure of transcription and a means of identifying those steps that are slow and regulated. This comparison of ESCs and MEFs establishes that transcription elongation is often controlled by dynamically tuning the release of the paused Pol II. However, an important class of regulated genes in ESCs that show bivalent histone modifications is modulated at both elongation and stages prior to elongation. Identification of the molecular targets of upstream activators or repressors and the role of these targets in modulating the rate-limiting steps of transcription will be essential to fully elucidate the mechanisms governing the regulation of the ESC state.
Materials and methods
Cell lines
V6.5 ESCs (C57Bl/6 [f] × 129/Sv [m]; passages 12–14) were grown on irradiated feeders. MEFs (C57Bl/6 [f] × 129/Sv [m]) were isolated from male embryonic day 13.5 (E13.5) embryos and passaged twice before analysis. ESCs were passaged twice in gelatin-coated plates before the nuclei isolation.
Nuclear run-on assay
GRO-seq experiments were carried out as described previously (Core et al. 2008). Briefly, nuclei from 5 × 106 cells were isolated, run-on-transcribed with Br-UTP and other NTPs, and base-hydrolyzed to yield nascent RNAs with an average size of roughly 100 nt. Br-UTP-incorporated nascent transcripts were purified after three rounds of serial Br-UTP enrichment steps. In order to empirically determine the sensitivity of our assay in detecting rare nascent transcripts in the reaction and the purity of Br-UTP-incorporated nascent transcripts over UTP-containing transcripts, we included spiking controls incorporated with or without Br-UTP at known concentrations in the nuclear run-on reaction. Nascent RNAs were prepared for Illumina sequencing.
Gene Expression Omnibus (GEO) accession codes
GRO-seq data are accessible at the GEO database, accession number GSE27037.
Acknowledgments
We thank Steven Petesch, Katherine Munson, and the Lis labroatory members for critical reading of the manuscript; Hojoong Kwak for help in heat map analysis; and Peter Schweitzer and Tom Stelick at Cornell Core DNA sequencing facility and Illumina, Inc., for help with Solexa sequencing. This work was supported by GM25232 and HG004845 from the National Institutes of Health (to J.T.L.), Cornell Institutional grants from the New York Stem Cell Science (to J.T.L. and J.C.S.), and a post-doctoral fellowship from the American Cancer Society (I.M.M.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2005511.
Supplemental material is available for this article.
References
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell 17: 103–112 [Abstract] [Google Scholar]
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [Europe PMC free article] [Abstract] [Google Scholar]
- Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B 2008. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res 18: 46–59 [Europe PMC free article] [Abstract] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock Mol Cell Biol 23: 7628–7637 [Europe PMC free article] [Abstract] [Google Scholar]
- Boettiger AN, Levine M 2009. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473 [Abstract] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [Europe PMC free article] [Abstract] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [Abstract] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V 2001. General transcription factors bind promoters repressed by polycomb group proteins. Nature 412: 651–655 [Abstract] [Google Scholar]
- Chambers I, Tomlinson SR 2009. The transcriptional foundation of pluripotency. Development 136: 2311–2322 [Europe PMC free article] [Abstract] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [Abstract] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [Europe PMC free article] [Abstract] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V 2004. Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893 [Abstract] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. 2008. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2: 437–447 [Europe PMC free article] [Abstract] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. 2008. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135: 1513–1524 [Abstract] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [Abstract] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [Europe PMC free article] [Abstract] [Google Scholar]
- Giardina C, Perez-Riba M, Lis JT 1992. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev 6: 2190–2200 [Abstract] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev 22: 1921–1933 [Europe PMC free article] [Abstract] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551 [Europe PMC free article] [Abstract] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [Europe PMC free article] [Abstract] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA 2010. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7: 249–257 [Europe PMC free article] [Abstract] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145 [Europe PMC free article] [Abstract] [Google Scholar]
- Hemmati-Brivanlou A, Melton D 1997. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell 88: 13–17 [Abstract] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes & Dev 11: 774–785 [Abstract] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442: 533–538 [Abstract] [Google Scholar]
- Jaenisch R, Young R 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [Abstract] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [Europe PMC free article] [Abstract] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [Abstract] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 10.1371/journal.pgen.1000242 [Europe PMC free article] [Abstract] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by polycomb in human embryonic stem cells. Cell 125: 301–313 [Abstract] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [Europe PMC free article] [Abstract] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [Abstract] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [Europe PMC free article] [Abstract] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [Europe PMC free article] [Abstract] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [Europe PMC free article] [Abstract] [Google Scholar]
- Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell 3: 137–147 [Abstract] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [Europe PMC free article] [Abstract] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of pol II in Drosophila. Science 327: 335–338 [Europe PMC free article] [Abstract] [Google Scholar]
- Nevado J, Gaudreau L, Adam M, Ptashne M 1999. Transcriptional activation by artificial recruitment in mammalian cells. Proc Natl Acad Sci 96: 2674–2677 [Europe PMC free article] [Abstract] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [Abstract] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-myc regulates transcriptional pause release. Cell 141: 432–445 [Europe PMC free article] [Abstract] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [Europe PMC free article] [Abstract] [Google Scholar]
- Rizzino A 2008. Transcription factors that behave as master regulators during mammalian embryogenesis function as molecular rheostats. Biochem J 411: e5–e7 10.1042/BJ20080479 [Abstract] [Google Scholar]
- Rougvie AE, Lis JT 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [Abstract] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [Europe PMC free article] [Abstract] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA 2009. Divergent transcription: a new feature of active promoters. Cell Cycle 8: 2557–2564 [Abstract] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [Abstract] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377 [Europe PMC free article] [Abstract] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [Abstract] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926 [Europe PMC free article] [Abstract] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [Abstract] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [Europe PMC free article] [Abstract] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A 2003. BMP induction of id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292 [Abstract] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.2005511
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/25/7/742.full.pdf
Free to read at www.genesdev.org
http://www.genesdev.org/cgi/content/abstract/25/7/742
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/content/full/25/7/742
Free after 12 months at www.genesdev.org
http://www.genesdev.org/cgi/reprint/25/7/742
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Dynamic control of gene expression by ISGF3 and IRF1 during IFNβ and IFNγ signaling.
EMBO J, 43(11):2233-2263, 24 Apr 2024
Cited by: 2 articles | PMID: 38658796 | PMCID: PMC11148166
Epigenetic control and manipulation of neuronal maturation timing.
Curr Opin Genet Dev, 85:102164, 27 Feb 2024
Cited by: 1 article | PMID: 38412562
Review
DOT1L is a barrier to histone acetylation during reprogramming to pluripotency.
Sci Adv, 9(46):eadf3980, 17 Nov 2023
Cited by: 2 articles | PMID: 37976354 | PMCID: PMC10656071
KAS-Analyzer: a novel computational framework for exploring KAS-seq data.
Bioinform Adv, 3(1):vbad121, 08 Sep 2023
Cited by: 1 article | PMID: 37745002 | PMCID: PMC10516523
RNA polymerase II pausing temporally coordinates cell cycle progression and erythroid differentiation.
Dev Cell, 58(20):2112-2127.e4, 15 Aug 2023
Cited by: 6 articles | PMID: 37586368 | PMCID: PMC10615711
Go to all (230) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE27037
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Transcription pausing regulates mouse embryonic stem cell differentiation.
Stem Cell Res, 25:250-255, 16 Nov 2017
Cited by: 11 articles | PMID: 29174978 | PMCID: PMC5732849
Motif-driven interactions between RNA and PRC2 are rheostats that regulate transcription elongation.
Nat Struct Mol Biol, 28(1):103-117, 04 Jan 2021
Cited by: 23 articles | PMID: 33398172 | PMCID: PMC8050941
Polycomb repressive complex 1 modifies transcription of active genes.
Sci Adv, 3(8):e1700944, 02 Aug 2017
Cited by: 37 articles | PMID: 28782042 | PMCID: PMC5540264
Modifications of RNA polymerase II are pivotal in regulating gene expression states.
EMBO Rep, 10(11):1213-1219, 16 Oct 2009
Cited by: 106 articles | PMID: 19834511 | PMCID: PMC2775184
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHGRI NIH HHS (4)
Grant ID: HG004845
Grant ID: R01 HG004845-03
Grant ID: R01 HG004845-02
Grant ID: R01 HG004845
NIGMS NIH HHS (7)
Grant ID: R01 GM025232
Grant ID: GM25232
Grant ID: R01 GM025232-32S1
Grant ID: R01 GM025232-33
Grant ID: R01 GM025232-33S1
Grant ID: R37 GM025232
Grant ID: R01 GM025232-32





