Abstract
Free full text

Targeting mitochondrial glutaminase activity inhibits oncogenic transformation
SUMMARY
Rho GTPases impact a number of activities important for oncogenesis. Here we describe a small molecule inhibitor which blocks oncogenic transformation induced by various Rho GTPases in fibroblasts, and the growth of human breast cancer and B lymphoma cells, without affecting normal cells. We identify the target of this inhibitor to be the metabolic enzyme glutaminase, which catalyzes the hydrolysis of glutamine to glutamate. We show that transformed fibroblasts and breast cancer cells exhibit elevated glutaminase activity that is dependent on Rho GTPases and NFκB activity, and is blocked by the small molecule inhibitor. These findings highlight a previously unappreciated connection between Rho GTPase activation and cellular metabolism, and demonstrate that targeting glutaminase activity can inhibit oncogenic transformation.
INTRODUCTION
Rho-family GTPases activate signaling pathways that influence a variety of cellular activities ranging from actin cytoskeletal rearrangements to cell polarity and migration, cell cycle progression, and membrane trafficking (Etienne-Manneville and Hall 2002). A number of lines of evidence have also implicated Rho GTPases in cell growth and malignant transformation (Vega and Ridley 2008). For example, their hyper-activation either through mutations or the deregulation of their guanine nucleotide exchange factors (GEFs; e.g. members of the Dbl (for Diffuse B cell lymphoma) family) results in cellular transformation (Erickson and Cerione, 2004). Cells expressing constitutively active Rho GTPases are able to grow under conditions of serum deprivation and in the absence of a substratum, and have been shown to induce tumor formation when introduced into immuno-compromised mice (Lin et al., 1999; Fort, P., 1999). Rho GTPases have also been implicated in naturally occurring neoplastic development, where their over-expression has been demonstrated in advanced stage breast cancers, as well as in a variety of other cancers (Suwa et al. 1998; Mira et al., 2000; Fritz et al., 2002; Kamai et al., 2004). In particular, two members of the family, RhoA and RhoC, have been linked to the progression of malignancy, i.e. poorly differentiated phenotypes, local invasiveness, and metastasis (Kleer et al., 2002; Clark et al., 2000; Burbelo et al., 2004; Valastyan et al., 2009). Moreover, DLC1 (for Deleted in Liver Cancer 1), whose expression is suppressed in liver cancer tissue and in a wide variety of other cancers, is a Rho-GTPase-activating protein (Rho-GAP) and therefore it appears to play a role as a tumor suppressor (Xue et al., 2008; Lahoz and Hall, 2008). Thus, the Rho GTPases represent intriguing targets for anti-cancer therapies. Here we describe the identification and characterization of a small molecule that blocks the Rho GTPase-dependent transformation of fibroblasts, as well as the growth and invasive activity of human cancer cells.
RESULTS
Identification of an inhibitor of Rho GTPase-dependent transformation
While screening for small molecule inhibitors of the transforming capabilities of activated Rho GTPases, we found that members of the benzo[a]phenanthridinone family blocked the cellular transformation induced by the Rho family-GEF oncogenic Dbl, as read-out in focus-forming assays and when assaying cell growth in 10% calf serum or in low (1%) serum (Figures 1A, S1A, and 1B, respectively). The most effective molecule, designated 968, was active at 1–10 μM (Figure 1A, right panel). The dimethyl-amine and the adjacent bromine substitution on the phenyl ring of 968 (circled in Figure 1C) are essential for maximal inhibition of Dbl-induced transformation, as compounds 335 or 384 showed little or no effect (Figures 1A and S1B). 968 was a more potent inhibitor of Dbl-induced transformation, compared to oncogenic H-Ras, when assaying focus formation in NIH 3T3 cells (Figures S1B and S1C) or growth in low serum (compare Figures 1B and S1D), indicating that the transforming activities of Rho GTPases are particularly sensitive to this small molecule. Treatment with 968 had no significant effects on the growth of normal NIH 3T3 cells (Figure 1D) nor did it alter their overall morphology (Figure 1E).
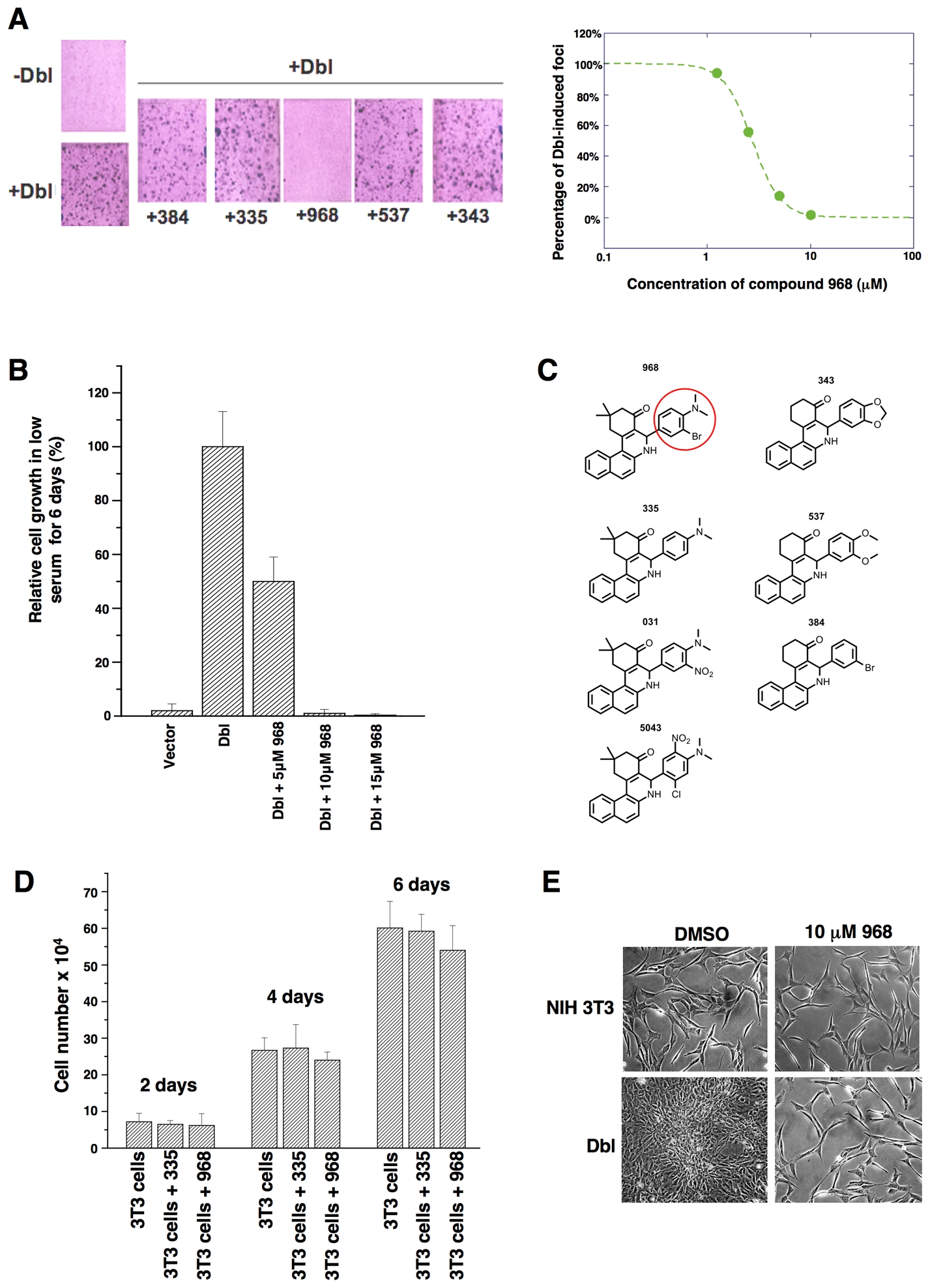
(A) Left: NIH 3T3 cells were transiently transfected with oncogenic Dbl and cultured for 14 days in 5% calf serum, while treated with different benzo[a]phenanthridinones (designated 384, 335, 968, 537, and 343) (10 μM each). Cells were fixed with 3.7% formaldehyde in PBS and stained with crystal violet for counting foci. Right: 968 was serially diluted (10, 5, 2.5, and 1.25 μM) and evaluated for its ability to inhibit focus formation.
(B) NIH 3T3 cells were stably transfected with Dbl and grown in DMEM supplemented with 1% calf serum and the indicated amounts of 968. After 6 days, the cells were counted. 100% represents the number of Dbl-transformed cells counted in the absence of 968 (27.5 × 104 cells). Data represent the average of 3 experiments (± s.d.).
(C) Chemical structures of the benzo[a]phenanthridinone derivatives examined for their effects on Dbl-induced focus formation (1A and S1B).
(D) Control NIH 3T3 cells were cultured in DMEM supplemented with 10% calf serum in 6 well plates, and were either treated with 10 μM 968 or 335, or untreated. At the indicated times, the cells were counted. Data represent the average of 3 experiments (± s.d.).
(E) Photomicrographs of Dbl-transfected NIH 3T3 cells (bottom panels) and control NIH 3T3 cells (top panels) cultured in 10% calf serum and treated with either DMSO (vehicle control) or 10 μM 968.
The guanine nucleotide exchange activities of a number of Rho GTPases are directly stimulated by oncogenic Dbl, including Cdc42 and RhoC (Hart et al., 1994); moreover, Rac appears to be activated in cells expressing oncogenic Dbl, most likely as an outcome of its ablity to function in a GTPase cascade downstream of activated Cdc42 (Baird et al., 2005). Mutated Rho GTPases that undergo constitutive GDP-GTP exchange mimic many of the actions of oncogenic Dbl (Lin et al, 1999). Thus, we used cells transformed by different Rho GTPases to determine whether the inhibitory effects of 968 were due to its ability to block the signaling activity of a specific target of Dbl, such as RhoC. In fact, we found that 968 was capable of inhibiting the transforming activity of each of the Rho GTPase mutants examined, blocking their ability to enable cells to form colonies in soft-agar (Figure 2A) and to grow to high density (Figure 2B) or in low serum (Figure 2C), as well as inhibiting their invasive activity (Figure 2D).
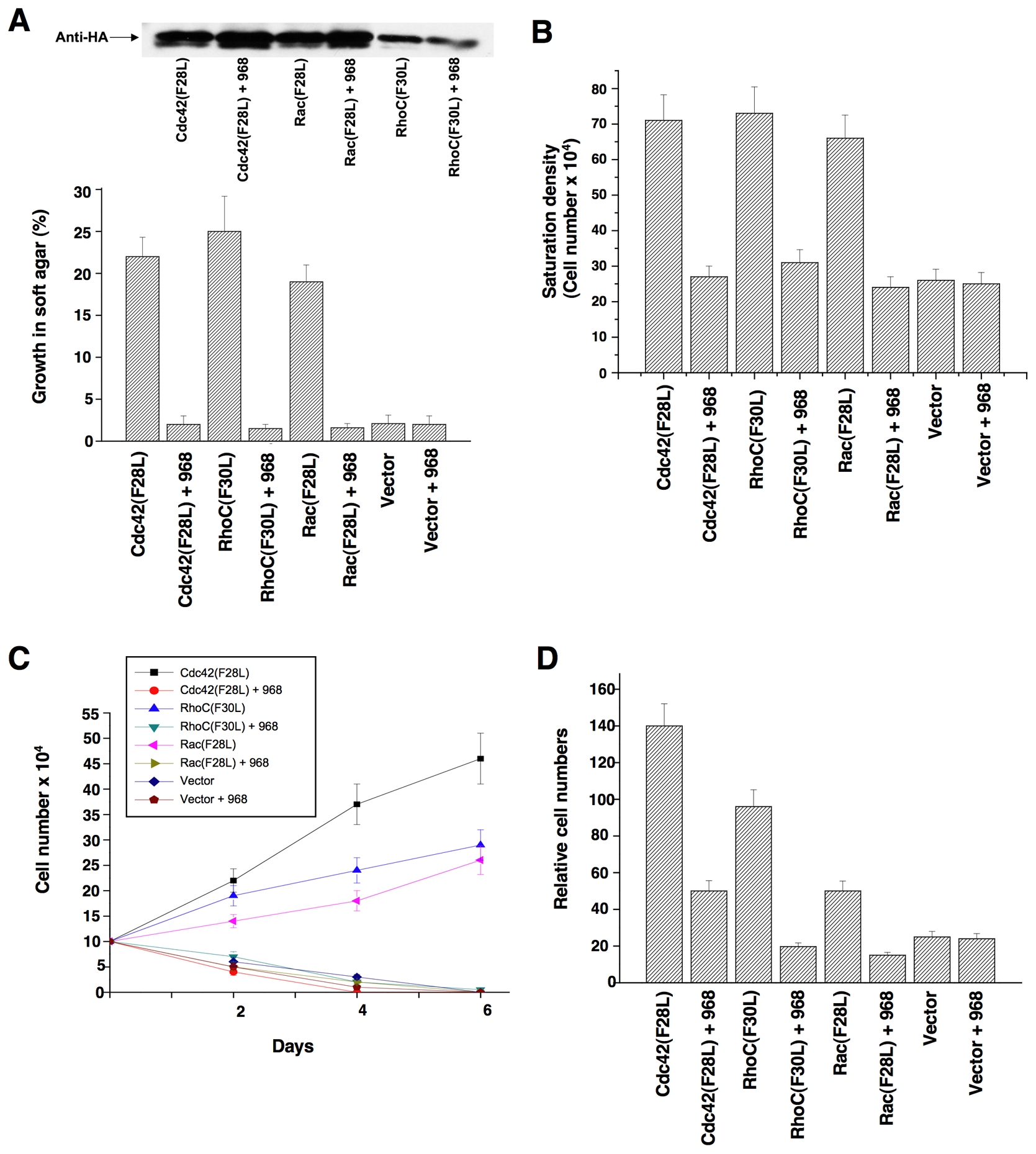
(A) NIH 3T3 cells stably expressing hemagglutinin (HA)-tagged Cdc42(F28L), Rac(F28L), RhoC(F30L), or vector control cells, either treated with 10 μM 968 or untreated, were grown in soft agar (plus DMEM supplemented with 10% calf serum). Top: Relative expression of the HA-tagged GTPases. Bottom: Cells were scored after 14 days and plotted as the percentage of the total number of colonies greater than 50 μm in diameter. Data represent the average of 3 experiments (± s.d.).
(B) Cells were cultured in DMEM supplemented with 10% calf serum with or without 10 μM 968, for 6 days, and then were trypsinized and counted. Data represent the average of 3 experiments (± s.d.).
(C) Cells were cultured in DMEM supplemented with 1% calf serum, treated with 10 μM 968 or untreated, and counted at the indicated times. Data represent the average of 3 experiments (± s.d.).
(D) Cells were serum-starved, treated with 10 μM 968 or untreated, and seeded in MilliCell upper chambers containing growth factor-reduced Matri-gel. After 24 hours at 37°C, the migratory cells were fixed, stained with GIEMSA, and counted. Data represent the average of 3 experiments (± s.d.).
968 blocks the transforming activity of human breast cancer cells
Rho GTPases have been shown to be over-expressed and/or hyper-activated in human breast cancer cells (Kleer et al., 2002; Burbelo et al., 2004; Valastyan et al., 2009). For example, RhoA and RhoC are hyper-activated in the highly invasive MDA-MB231 and SKBR3 breast cancer cell lines compared to normal human mammary epithelial cells (HMECs) (Figure 3A). In addition, the growth of these cancer cells in low serum or in soft-agar is severely compromised when RhoA and RhoC levels are knocked-down using siRNAs (Figures 3B-3E). Given the ability of 968 to inhibit the transformation of fibroblasts by oncogenic Dbl and different constitutively active Rho GTPases, we were interested in examining its potential effects on these human breast cancer cells whose transformed phenotypes are dependent upon Rho GTPase activity. We found that 968 inhibited their ability to form colonies in soft agar, as effectively as it blocked Dbl-induced colony formation in NIH 3T3 cells (Figure 4A). Similarly, 968 inhibited their growth in complete medium (Figure 4B), as well as their ability to grow to high density (Figure 4C) and in low serum (Figure 4D), and reduced their invasive activity (Figure 4E). As was the case for control (non-transformed) fibroblasts, 968 had little if any effect on the growth or morphology of HMECs (Figures 4C, 4F, and 4G).
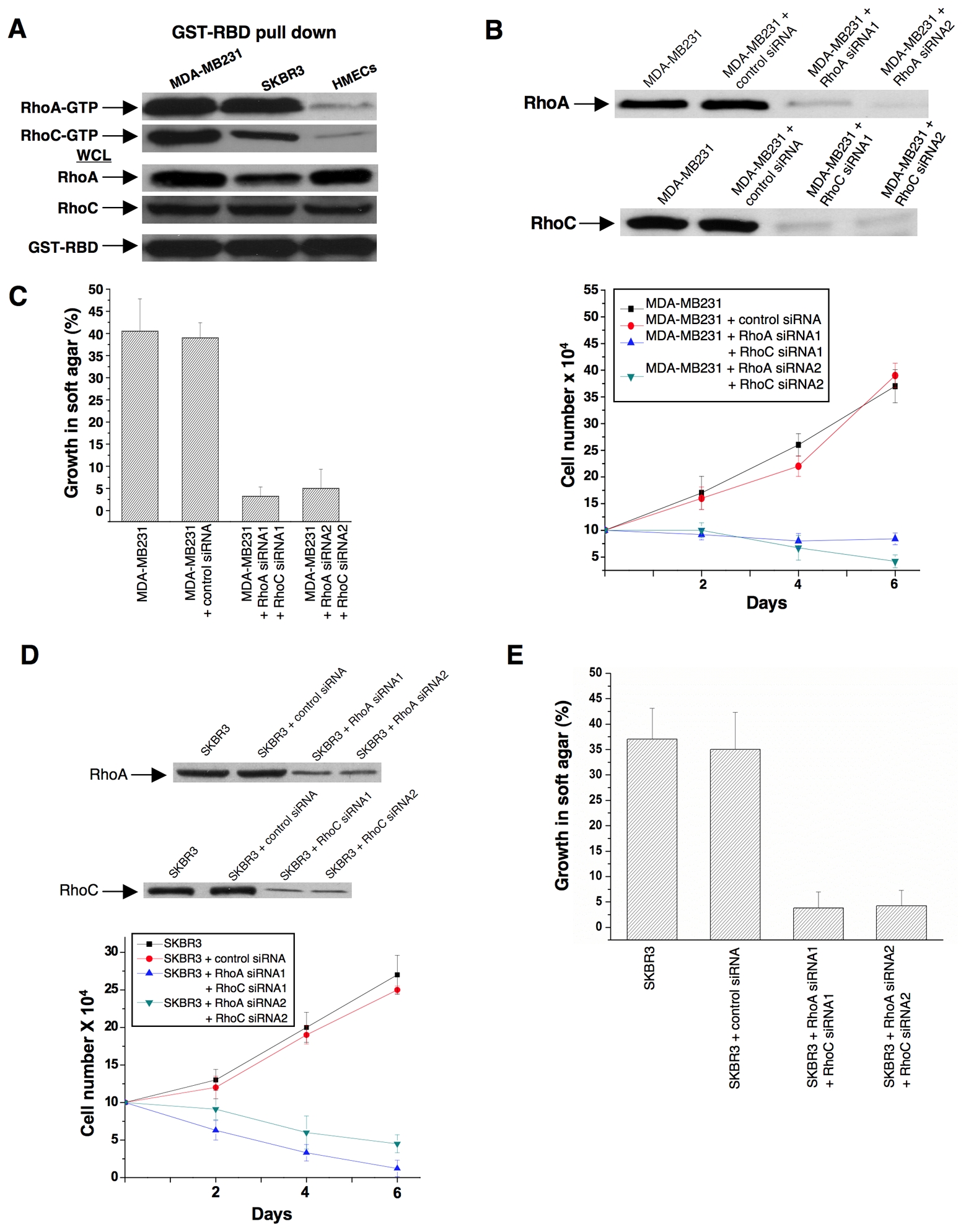
(A) Lysates from MDA-MB231 cells, SKBR3 cells, and HMECs, were prepared and incubated with GST fused to the limit Rho-binding domain on Rhotekin (GST-RBD). The top panels show the relative levels of RhoA-GTP and RhoC-GTP that were co-precipitated with GST-RBD from the indicated cells, as detected by Western blotting with an anti-RhoA monoclonal antibody and an anti-RhoC polyclonal antibody. The middle panels compare the relative expression of RhoA and RhoC in whole cell lysates (WCL) from the different cells and the bottom panel shows the relative input of GST-RBD.
(B) MDA-MB231 cells were transfected with control siRNA or siRNAs targeting RhoA and RhoC. Top: Shown are the efficiencies of the siRNAs targeting RhoA and RhoC as assessed by Western blot analysis using anti-RhoA and anti-RhoC antibodies. Bottom: Cells were grown for the indicated number of days in RPMI 1640 medium supplemented with 1% fetal bovine serum and counted. Data represent the average of 3 experiments (mean ± s.d.).
(C) MDA-MB231 cells transfected with control siRNA or siRNAs targeting RhoA and RhoC were grown in soft agar and scored after 10 days. Histograms show the percentage of the total number of colonies greater than 50 μm in diameter. Data represent the average of 3 experiments (mean ± s.d.).
(D) SKBR3 cells were transfected with control siRNA or siRNAs targeting RhoA and RhoC. Top: Shown are the efficiencies of the siRNAs targeting RhoA and RhoC. Bottom: Cells were grown in RPMI 1640 medium supplemented with 1% fetal bovine serum for the indicated number of days and counted. Data represent the average of 3 experiments (mean ± s.d.).
(E) SKBR3 cells transfected with control siRNA or siRNAs targeting RhoA and RhoC were grown in soft agar and scored after 10 days. Histograms show the percentage of the total number of colonies greater than 50 μm in diameter. Data represent the average of 3 experiments (mean ± s.d.).
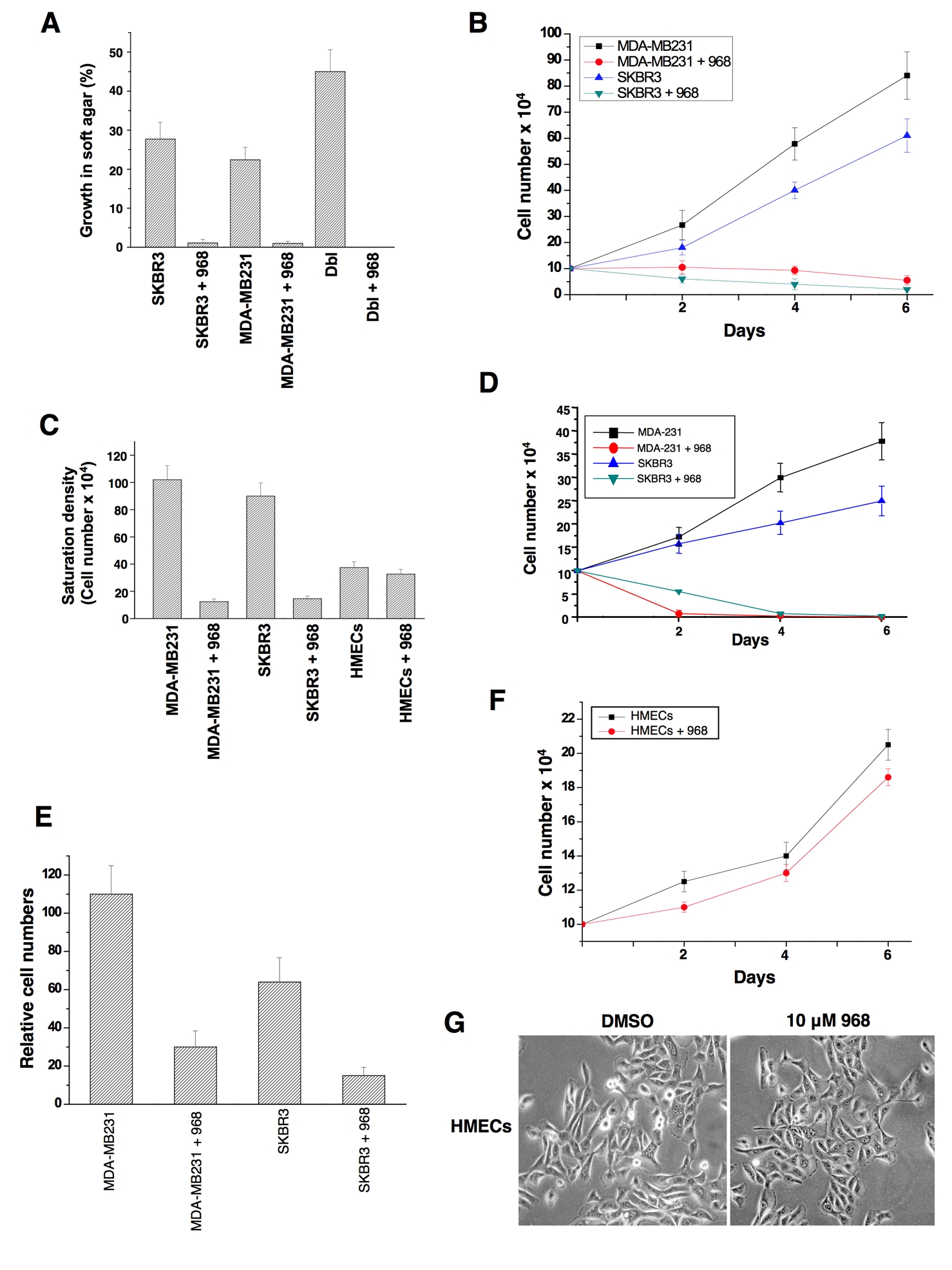
(A) MDA-MB231 cells, SKBR3 cells, and NIH 3T3 cells stably expressing Dbl, were either treated with 10 μM 968 or untreated, and grown in soft-agar as in 2A. Data represent the average of 3 experiments (± s.d.).
(B) MDA-MB231 and SKBR3 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum in 12 well plates for the indicated number of days in the presence or absence of 10 μM 968, and then counted. Data represent the average of 3 experiments (± s.d.).
(C) Breast cancer cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, and HMECs were cultured in MEGM complete medium in 12 well plates, for 6 days in the presence or absence of 10 μM 968, and counted. Data represent the average of 3 experiments (± s.d.).
(D) Breast cancer cells were cultured in RPMI 1640 medium supplemented with 1% fetal bovine serum, treated with 10 μM 968 or untreated, and counted at the indicated times. Data represent the average of 3 experiments (± s.d.).
(E) Breast cancer cells were serum-starved, treated with 968 or untreated, and seeded in MilliCell upper chambers containing growth-factor-reduced Matri-gel. After 24 hours at 37°C, the migratory cells were fixed, stained with GIEMSA, and counted. Data represent the average of 3 experiments (± s.d.).
(F) HMECs were cultured in MGEM complete medium in 12 well plates for the indicated number of days in the presence and absence of 10 μM 968, and then counted.
(G) Photomicrographs of HMECs cultured in MGEM complete medium treated with either DMSO (vehicle control) or 10 μM 968.
Identification of the target for 968
The ability of 968 to inhibit the transforming activities of different constitutively active Rho GTPases indicated that the target for this small molecule was not a specific binding partner or an effector for an individual Rho protein. We set out to identify the target for 968 by using its active moiety (circled in Figure 1C) labeled with biotin in affinity precipitation experiments with streptavidin beads. This led to the detection of a silver-stained band on SDS-gels, Mr ~66 kDa, that was isolated from Cdc42(F28L)-expressing NIH 3T3 cell lysates with the biotin-labeled 968-derivative immobilized to streptavidin beads, but not with beads alone (Figure S2A). Microsequence analysis indicated that this was the mouse isoform-2 ortholog of human glutaminase C (GAC) (Figure S2B), one of two splice variants of an enzyme found in kidney and other tissues, collectively referred to as kidney-type glutaminase (KGA), that catalyzes the hydrolysis of glutamine to glutamate and ammonium (Curthoys, 1995). We verified that the biotin-labeled active moiety of 968, when immobilized on streptavidin beads, affinity-precipitated an endogenous protein that reacted with an antibody recognizing both isoforms of KGA (Figure S2C), as well as precipitated ectopically expressed, V5-tagged GAC (Figure 5A, top panel). Pre-incubation of E. coli-expressed, mouse GAC with 968 inhibited its enzyme activity (Figure 5A), whereas the structurally-related compound 335, that was less effective at blocking Dbl-transformation (Figure 1A), showed little inhibition. 968 was neither a competitive inhibitor versus glutamine nor inorganic phosphate, an activator of the enzyme (Kenny et al., 2003), suggesting that it acts in an allosteric manner (Figures S2D–2F).
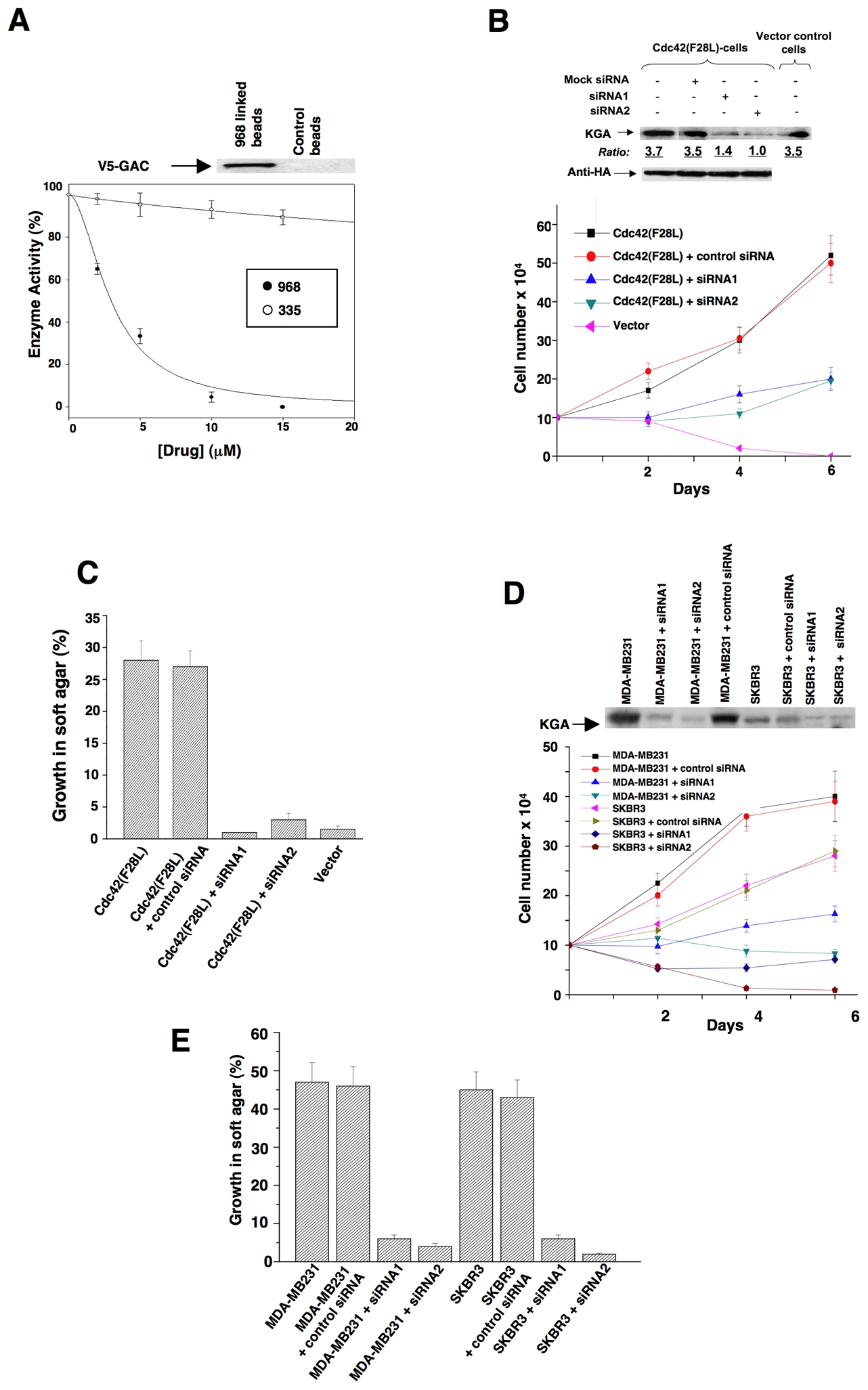
(A) The E. coli-expressed mouse ortholog of human GAC was pre-incubated with the indicated amounts of 968 (●) or 335 (○) for 30 minutes at 37°C and then assayed. 100% = 620 moles of glutamine hydrolyzed per minute per mole of enzyme. Data represent the average of 3 experiments (± s.d.). Top panel: The biotin-labeled, active moiety of 968 linked to streptavidin-agarose beads, or control beads alone, were incubated with NIH 3T3 cell lysates transiently expressing V5-tagged mouse GAC. Following precipitation of the beads and re-suspension, the samples were analyzed by Western blotting with anti-V5 antibody.
(B) NIH 3T3 cells stably expressing HA-Cdc42(F28L), cells stably expressing HA-Cdc42(F28L) transfected with control siRNA or siRNAs targeting both isoforms of mouse KGA, or control cells, were grown in DMEM supplemented with 1% calf serum. Top: Efficiencies of siRNAs targeting both isoforms of KGA, and relative levels of HA-Cdc42 in the different cells. Bottom: Growth profiles for the indicated cell lines. Data represent the average of 3 experiments (± s.d.).
(C) NIH 3T3 cells stably expressing Cdc42(F28L), cells stably expressing Cdc42(F28L) transfected with control siRNA or siRNAs targeting both isoforms of mouse KGA, and control (vector) cells, were grown in soft agar and scored after 10 days. Histograms show the percentage of the total number of colonies greater than 50 μm in diameter. Data represent the average of 3 experiments (mean ± s.d.).
(D) Breast cancer cells transfected with control siRNA or siRNAs targeting both isoforms of KGA were grown in RPMI 1640 medium supplemented with 1% fetal bovine serum. Top: Efficiencies of siRNAs targeting both isoforms of KGA. Bottom: Growth profiles for the indicated cell lines. Data represent the average of 3 experiments (± s.d.).
(E) The indicated breast cancer cell lines were transfected with control siRNA or with siRNAs targeting both isoforms of KGA and then grown in soft agar and scored after 10 days. Data represent the average of 3 experiments (mean ± s.d.).
We next examined whether knocking-down the levels of KGA expression in transformed cells using RNAi mimicked the effects of treating these cells with 968. Figures 5B and S3A show that reducing KGA expression by using siRNAs targeting both of its isoforms inhibited the abilities of Cdc42(F28L), Rac(F28L), and RhoC(F30L) to stimulate growth in low serum. Similarly, knocking-down KGA expression blocked the ability of these different constitutively activated Rho GTPases to induce colony formation in soft agar (Figures 5C and S3B), whereas knock-downs of KGA did not significantly inhibit the growth of control NIH 3T3 cells (Figure S3C), consistent with the inability of 968 to affect their growth (Figure 1D). However, knocking-down KGA also strongly inhibited MDA-MB231 and SKBR3 cells from growing in low serum (Figure 5D) and soft-agar (Figure 5E).
The importance of glutaminase activity for transformation is consistent with the dependence of transformed/cancer cells on extracellular glutamine (Figures 6A and 6B). We reasoned that if 968 blocks transformation by inhibiting glutaminase activity, it should also reduce the levels of α-ketoglutarate, an intermediate in the glutamine metabolic pathway. Therefore, the effects of 968 should be circumvented by adding a cell-permeable analog of α-ketoglutarate (dimethyl α-ketoglutarate or DM-α-KG) to cells. Figure 6C shows that when the KGA isoforms were knocked-down from Dbl-transformed cells, the addition of DM-α-KG restored their ability to grow in low serum. Likewise, the addition of DM-α-KG restored the ability of 968-treated Dbl-cells to grow in low serum (Figure 6D) and their focus-forming activity (Figure 6E), as well as the ability of 968-treated MDA-MB231 and SKBR3 cells to grow in low serum (Figures 6F and 6G).

(A) MDA-MB231 cells and SKBR3 cells were grown in 10% fetal bovine serum in the presence or absence of glutamine for the indicated number of days and counted.
(B) NIH 3T3 cells stably expressing Dbl and cells stably expressing Cdc42(F28L) were grown in 10% calf serum in the presence or absence of glutamine for the indicated number of days and counted.
(C) NIH 3T3 cells stably expressing HA-tagged Dbl alone, and cells stably expressing HA-Dbl that were transfected with control siRNA or siRNAs targeting both isoforms of KGA, were cultured in DMEM supplemented with 1% calf serum in the presence and absence of 7 mM dimethyl α-ketoglutarate (DM-α-KG). Top: Efficiencies of siRNAs targeting both isoforms of KGA, and relative levels of HA-Dbl in the different cells. Bottom: Growth profiles for the indicated cell lines. Data represent the average of 3 experiments (± s.d.).
(D) NIH 3T3 cells stably expressing Dbl were cultured in DMEM supplemented with 1% calf serum in the presence of 10 μM 968 alone or together with 7 mM DM-α-KG. Data represent the average of 3 experiments (± s.d.).
(E) NIH 3T3 cells transiently expressing Dbl were assayed for focus formation in the presence of 10 μM 968 alone or together with 7 mM DM-α-KG.
(F) MDA-MB231 cells were grown in 1% fetal bovine serum in the presence of 10 μM 968 alone or together with 7 mM dimethyl α-ketoglutarate (DM-α-KG) for the indicated number of days and counted. Data represent the average of 3 experiments (mean ± s.d.).
(G) SKBR3 cells were grown in 1% fetal bovine serum in the presence of 10 μM 968 alone or together with 7 mM dimethyl α-ketoglutarate. Data represent the average of 3 experiments (± s.d.).
The expression of the KGA isoform GAC is up-regulated in P-493 B lymphoma cells (Gao et al., 2009), which prompted us to examine the effects of 968 on these cancer cells. Figures 7A and 7B show that 968 inhibited their growth, whereas the inactive analog 335 had essentially no effect. Similarly, in mouse xenograft experiments where tumors were induced by the introduction of P-493 B lymphoma cells, the intra-peritoneal injection of 968 caused an ~50% reduction in the size of the tumors (Figure 7C). However, the over-expression of GAC, alone, is not sufficient to induce malignant transformation (Gao et al., 2009). Moreover, glutaminase expression in Dbl-transformed fibroblasts or in SKBR3 breast cancer cells is similar to that of non-transformed cells (see below), suggesting that GAC works together with other oncogenic proteins to satisfy the metabolic requirements for transformation. This is supported by the results in Figure 7D, in which the transient expression of GAC alone, in NIH 3T3 cells was insufficient to induce foci, whereas the expression of GAC in cells stably expressing the Cdc42(F28L) mutant, which exhibits only weak focus-forming activity, caused a dramatic increase in foci matching the strong response induced by oncogenic Dbl. The increase in foci accompanying the expression of GAC together with Cdc42(F28L) was blocked by 968 (Figure 7D) and did not occur when the catalytically dead GAC(S291A) mutant was co-expressed with Cdc42(F28L) (Figure 7E).
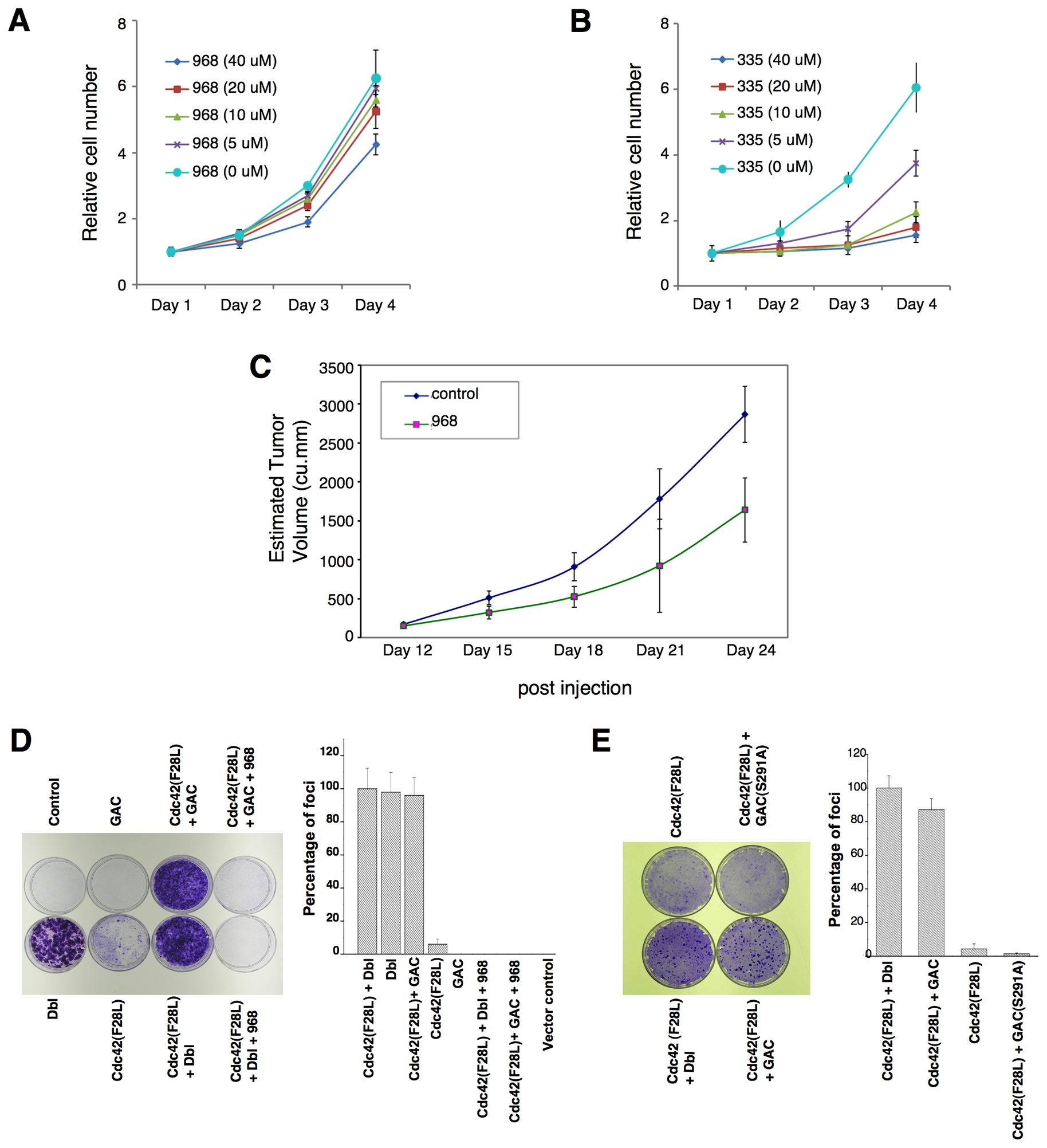
(A) P493 B lymphoma cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum in the presence of the indicated concentrations of 968.
(B) P493 B lymphoma cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum in the presence of the inactive 968-analog 335.
(C) P493 B lymphoma cells (2 × 107) were injected s.c. into the flank of SCID mice. At day 12 the tumors were ~170 mm3, at which time treatments with 968 (200 μg per injection) were begun by daily intraperitoneal injections (200 μL) for 12 days. Control animals were treated with vehicle (DMSO in PBS). N = 7. The p value on day 24 = 0.000375.
(D) Left: Control NIH 3T3 cells, NIH 3T3 cells transiently expressing Dbl, cells stably expressing Cdc42(F28L) and transiently expressing mouse GAC, cells stably expressing Cdc42(F28L), cells transiently expressing GAC, and cells stably expressing Cdc42(F28L) and transiently expressing Dbl, were examined for focus-forming activity. Right: Quantification of foci. Data represent the average of 3 experiments (± s.d.).
(E) Left: Focus-forming assays performed on NIH 3T3 cells stably expressing Cdc42(F28L), cells stably expressing Cdc42(F28L) and transiently expressing Dbl, and cells stably expressing Cdc42(F28L) and either transiently expressing wild-type mouse GAC or the GAC(S291A) mutant. Right: Quantification of foci. Data represent the average of 3 experiments (± s.d.).
Glutaminase activity is elevated in transformed/cancer cells
Given that glutaminase is not over-expressed in transformed fibroblasts or SKBR3 cells, we examined whether it is activated in these cells. Indeed, mitochondrial preparations from Dbl-transformed fibroblasts exhibited much higher basal glutaminase activity (i.e. assayed in the absence of inorganic phosphate) compared to the activity in mitochondria from non-transformed NIH 3T3 cells (Figure 8A; p = 0.001). Mitochondria from Cdc42(F28L)-expressing cells showed lower basal levels of activity compared to Dbl-transformed cells (p value = 0.019), but still higher than non-transformed cells (p = 0.023). Rac(F28L)- and RhoC(F30L)-transformed cells also showed higher basal levels of mitochondrial glutaminase activity compared to control cells (p = 0.019 and 0.009, respectively). The enzyme activity in mitochondrial preparations from control cells was strongly stimulated by the addition of inorganic phosphate (~6-fold), approaching the phosphate-stimulated activity measured in the mitochondria from transformed cells (Figure S4A), consistent with these two sets of cells showing similar levels of enzyme expression. Treatment of the different transformed cells with 968 inhibited their mitochondrial glutaminase activity, with the basal enzyme activity being much more sensitive to the small molecule inhibitor than the phosphate-stimulated activity (Figures 8A and S4A).

(A) Mitochondrial fractions were prepared from equivalent numbers of control NIH 3T3 cells and cells stably expressing Dbl, Cdc42(F28L), Rac(F28L), or RhoC(F30L) that had been treated with 10 μM 968 or untreated. Top: Relative amounts of KGA in the different mitochondrial preparations as assessed using an antibody that recognizes both KGA isoforms. Bottom: The different mitochondrial fractions were assayed for basal glutaminase activity. 100% = 680 nM glutamine hydrolyzed per minute per 105 cells. Data are the average of 3 experiments (± s.d.).
(B) Mitochondrial fractions were prepared from equivalent numbers of the indicated breast cancer cells that had been treated with 10 μM 968 or untreated. Top: Relative amounts of KGA and VDAC in the mitochondrial preparations. Bottom: The different mitochondrial fractions were assayed for basal glutaminase activity. 100% = 750 nM glutamine hydrolyzed per minute per 105 cells. Data represent the average of 3 experiments (± s.d.).
(C) Mitochondrial fractions from NIH 3T3 cells stably expressing Dbl that were cultured for 2 days in the presence or absence of 2 μM BAY 11-7082, or transfected with control siRNA or siRNAs targeting p65/RelA. Top: Relative amounts of KGA in the mitochondria from the indicated cells as assessed using an antibody that recognizes both KGA isoforms, and relative efficiencies of two siRNAs targeting p65/RelA. Bottom: The basal glutaminase activity for the different mitochondrial fractions. 100% represents the activity for untreated Dbl-transformed cells. Data represent the average of 2 experiments (± range).
(D) Mitochondrial fractions were prepared from HMECs, and SKBR3 cells that had been treated with 2 μM BAY 11-7082 or untreated, or transfected with siRNAs targeting p65/RelA. Top: Relative amounts of KGA in the different mitochondrial fractions as assessed using an antibody that reorganizes both KGA isoforms, and relative efficiencies of two siRNAs targeting p65/RelA. Bottom: The basal glutaminase activity for the different mitochondrial fractions. 100% represents the activity for untreated SKBR3 cells. Data represent the average of 2 experiments (± range).
(E) Top: V5-GAC was transiently expressed in control NIH 3T3 cells or NIH 3T3 cells stably expressing Dbl that were treated with either 2 μM BAY 11-7082, 10 μM 968, or untreated, and then immunoprecipitated from the different samples. Bottom: Basal activity for V5-GAC immunoprecipitated from the different cells. Data represent the average of 3 experiments (± s.d.).
(F) V5-GAC was transiently expressed in NIH 3T3 cells stably expressing Dbl. The V5-GAC was immunoprecipitated, treated with alkaline phosphatase for 1 hour at 37°C (or untreated), and assayed for activity in the presence and absence of phosphate.
Mitochondrial preparations from SKBR3 cells, as well as MDA-MB231 cells, also showed significantly higher basal glutaminase activity, compared to normal HMECs (p = 0.003), that was strongly inhibited when the cells were treated with 968 (Figure 8B; the top panels show that equivalent amounts of mitochondrial protein were assayed, by using the mitochondrial marker VDAC/Porin). The addition of inorganic phosphate to the mitochondrial preparations from HMECs caused a marked increase in glutaminase activity (~5-fold), and although the levels of activity were still lower than in MDA-MB231 cells which show significantly higher KGA expression (Figure 8B, top panel), they were similar to the phosphate-stimulated activity in the mitochondria from SKBR3 cells (Figure S4B). Knock-downs of RhoA and RhoC in SKBR3 cells reduced their basal glutaminase activity, without significantly affecting phosphate stimulation of the enzyme, indicating that the increased basal activity was Rho GTPase-dependent (Figure S4C).
An important insight into how glutaminase is activated in transformed fibroblasts and SKBR3 cells came from our finding that treatment of these cells with BAY 11-7082, which blocks NFκB activation by inhibiting the upstream kinase IKKβ (Pickering et al., 2007), significantly reduced their basal glutaminase activity (Figures 8C and 8D, respectively). Knocking-down the p65/RelA subunit of NFκB in Dbl-transformed cells and SKBR3 cells also reduced their basal glutaminase activity (Figures 8C and 8D), whereas treatment with BAY11-7082 or knock-downs of p65/RelA had little or no effect on the direct stimulation of the enzyme by inorganic phosphate (Figures S4D and S4E). This indicates that the effects of blocking NFκB are specific for those signals in transformed/cancer cells that elevate the basal levels of enzyme activity.
We then showed that the immunoprecipitation of ectopically expressed, V5-tagged GAC from Dbl-transformed cells yielded significantly higher basal activity compared to V5-GAC immunoprecipitated from non-transformed NIH 3T3 cells (Figure 8E). The activity immunoprecipitated from Dbl-transformed cells was reduced when the cells were treated with 968, as well as with an NFκB inhibitor, suggesting that V5-GAC was activated in transformed cells through an NFκB-dependent mechanism. The enhanced basal activity for V5-GAC immunoprecipitated from Dbl-cells was reversed by alkaline phosphatase treatment, whereas the stimulation by inorganic phosphate was essentially unaffected (Figure 8F). These findings suggest that NFκB does not elevate basal glutaminase activity in transformed/cancer cells by directly affecting the expression of the enzyme itself, but rather by influencing the expression of a protein kinase or a regulatory protein(s) that promotes its phosphorylation.
DISCUSSION
The importance of bioenergetics and cellular metabolism in the development of cancer, and in particular, the early observations that tumor cells exhibit elevated glycolytic activity (i.e. the “Warburg effect” (Warburg 1956)), are receiving renewed attention (DeBerardinis et al., 2007; Christofk et al., 2008a, 2008b). Recently, it was found that phospho-tyrosine signaling impacts the glycolytic pathway by shifting the utilization of glucose metabolites from energy production to anabolic processes through the regulation of the M2 (fetal) isoform of pyruvate kinase, an enzyme that catalyzes the conversion of phosphoenolpyruvate to pyruvate (Christofk et al., 2008a, 2008b; Hitosugi et al., 2009). It was also recently reported that the c-Myc-induced up-regulation of GAC expression in B lymphoma and prostate cancer cells is essential for their proliferation and survival (Gao et al., 2009). Here we demonstrate that transformed/cancer cells dependent on Rho GTPase-signaling activity exhibit elevated levels of basal glutaminase activity and that blocking the activation of this enzyme can have profound consequences for oncogenic transformation.
The discovery that glutaminase activity is required for the transforming capability of at least three different Rho GTPases (Cdc42, Rac1, RhoC) suggests that these GTPases are linked to the activation of this enzyme through a common signaling point of convergence. Our finding that NFκB is essential for glutaminase activation in transformed/cancer cells helps to explain how different Rho GTPases are able to signal an increase in basal glutaminase activity and why their transforming activities are sensitive to 968, as NFκB is activated by Dbl and various Rho GTPases (Perona et al., 1997; Cammarano and Minden, 2001), and is essential for Dbl-transformation (Whitehead et al., 1999) as well as for the transformed phenotypes of human breast cancer cells (Sovak et al., 1997). The mechanism by which NFκB mediates the activation of this metabolic enzyme, and in particular its phosphorylation, is an important focus of our current studies. The fact that cells treated with 968 do not exhibit elevated basal levels of glutaminase activity suggests that this small molecule acts in an allosteric manner to block the activation event, thus explaining how its inhibitory effects are sustained through the isolation of mitochondrial fractions from transformed/cancer cells. The ability of 968 to act in an allosteric manner is also consistent with our kinetic studies that demonstrate that 968 is neither a glutamine antagonist nor an active site competitive inhibitor, and likely explains why its effects appear to be much more pronounced on transformed/cancer cells compared to normal cells.
Recently, it has been shown that the expression of a distinct form of glutaminase, referred to as the liver-type enzyme or GLS2, was substantially decreased in liver cancer cells and that its over-expression reduced colony formation (Hu et al., 2010; Suzuki et al., 2010). The liver-type enzyme has different kinetic and molecular characteristics, and is likely to be subject to distinct modes of regulation and cellular actions compared to the kidney-type enzymes (i.e. collectively referred to as KGA or also GLS1), whose GAC isoform was originally identified as the target of 968. Moreover, it is possible that the liver-type enzyme is exerting its effects in liver cancer cells through its potential ability to influence transcription (Szeglia et al., 2009), a function that has not been ascribed to the KGA isoforms.
How the regulation of glycolysis in the cytoplasm and oxidative metabolism through the citric acid cycle in the mitochondnria is balanced so that tumor cells are able to satisfy their energy requirements and metabolic needs is not entirely understood. 13C-NMR metabolic flux experiments have demonstrated that while proliferating cancer cells exhibit a pronounced Warburg effect, their TCA cycle remains intact and becomes progressively more dependent on glutamine metabolism (DeBerardinis et al., 2007). This may enable cancer cells to use TCA cycle intermediates as precursors for biosynthetic pathways (DeBerardinis et al., 2008), and is consistent with tumor cells exhibiting increased rates of glutamine metabolism and consuming greater amounts of glutamine compared to normal cells (Medina et al., 1992). The observation that different transformed cell lines and cancer cells show elevated glutaminase activity that is dependent on Rho GTPase/NFκB-activation provides a mechanism for how these demands for elevated glutamine metabolism are met. Moreover, the ability of the small molecule 968 to block glutaminase activation and inhibit the growth of transformed/cancer cells highlights the potential of this enzyme as a drugable target and offers intriguing possibilities regarding the use of small molecule allosteric inhibitors of glutaminase as therapeutic strategies against cancer.
EXPERIMENTAL PROCEDURES
Cell culture and treatment with 968
NIH 3T3 cells stably expressing Cdc42(F28L), RhoC(F30L), Rac(F28L), Ras(G12V), and oncogenic Dbl were cultured in DMEM supplemented with 10% calf serum (Invitrogen). Two types of human breast cancer cells, MDA-MB231 and SKBR3 cells, were cultured in RPMI 1640 medium supplemented with 10% FBS. Normal mammary epithelial cells (HMECs) were cultured in MEGM complete medium (Lonza). The small molecule 968 (10 μM) was added to the culture medium 5 hours after the cells were seeded. The cells were then treated with 968 during various assays for cellular transformation.
Assays for cellular transformation and invasion
The detailed procedures for cellular transformation assays including growth in low serum, saturation density, and colony formation in soft agar were performed as previously described (Wang et al., 2005; Feng et al., 2006). When assaying cell growth in low serum, or saturation density, the medium containing 968 was changed every two days. These changes were performed every three days when assaying cell growth in soft agar.
Focus formation assays were performed with NIH 3T3 cells that were grown in 60 mm plates and transienly transfected with 1 μg pZip-neo-GST-Dbl (in which the original construct of oncogenic Dbl, residues 498–925, was obtained from Dr. A. Eva; Genoa, Italy) or 2 μg pCDNA 3.1-V5-glutaminase (i.e. the mouse ortholog of the human GAC isoform). In some cases, these transient transfections were performed using NIH 3T3 cells that stably express Cdc42(F28L). Following transfection, the cells were trypsinized, aliquoted equally into two 60 mm plates, and allowed to adhere overnight. On the following day, the media was changed to DMEM plus 5% calf serum, or DMEM plus 5% calf serum and 968. A 60 mm plate of NIH 3T3 cells, or cells stably expressing Cdc42(F28L), was left untreated as a negative control. All cells were then grown for 14 days, with media and compounds changed every second day. Plates were fixed in 3.7% formaldehyde and stained with 0.4% crystal violet.
For cell invasion assays, growth factor-reduced Matrigel (BD Bioscience) was diluted (30 μg in 100 μl sterile H2O), added to the top chamber (Milli Cell) and allowed to gel for 1 hour at 37°C, and then air-dried for 16 hours. Serum-starved cells were added to the upper compartment in migration medium (DMEM with 0.5% BSA) and the chamber was placed into 24-well dishes containing DMEM with 10% CS. After 24 hours incubation at 37°C, migratory cells were fixed with methanol, stained with Giemsa and counted under the microscope.
Animal studies
The experiments with mouse xenografts were performed according to the protocols approved by the Animal Care and Use Committees at Cornell University and The Johns Hopkins University. In these experiments, 2 × 107 P493 human lymphoma B cells were injected s.c. into male SCID mice (National Cancer Institute). When the tumor volumes reached ~170 mm3, intraperitoneal injections with 968 or vehicle control were initiated and performed daily for 12 days. Tumor volumes were calculated for the individual days of treatment as described by Le et al. (2010).
Identification of glutaminase as the target of 968
968 (5-[3-bromo-4-(dimethylamino)phenyl]-2,2-dimethyl-2,3,5,6-tetrahydrobenzo[a]) and its analogs were obtained from SPECS (Netherlands; CAS). In order to identify the molecular target of 968, its active moiety (3-bromo-4-(dimethylamino)benzaldehyde) (ChemBridge Corporation, San Diego) was incorporated into biotin hydrazide by reacting 3-bromo-4-(dimethylamino)benzaldehyde or formaldehyde (as a negative control) at a 5-fold molar excess overnight at 42°C, followed by reduction with cyanoborohydride coupling buffer. The 968-biotin adduct was confirmed by mass spectrometry and incubated with streptavidin-agarose beads equilibrated with cell lysis buffer (5 mM MgCl2, 120 mM NaCl, 10 mM HEPES, pH 7.4, 0.5% NP-40, 10 μg/ml leupeptin, and 10 μg/ml aprotinin) prior to incubation with lysates from NIH 3T3 cells stably expressing Cdc42(F28L) (5 ml containing ~2 mg/ml total protein) for 2 h at 4°C. The beads were washed 3X with cold lysis buffer and pelleted by centrifugation and the associated proteins were resolved by SDS-PAGE. A silver-stained protein band that bound specifically to the 968-biotin beads and not to the control beads was excised and analyzed by mass spectrometry at the Harvard Microchemistry Facility (Cambridge, Massachusetts) and identified as mouse KGA isoform-2 (accession number NP_001106854), the mouse ortholog of the human GAC isoform.
Mitochondrial preparations
Mitochondrial preparations were obtained using the mitochondria isolation kit from QIAGEN following the manufacturer’s instructions. A suspension containing 2 × 107 cells was transferred into a 50 ml conical tube and centrifuged at 500 × g for 10 minutes at 4°C. The pellets were resuspended in 2 ml of ice-cold lysis buffer (supplied by QIAGEN) and incubated for 10 minutes at 4°C using an end-over-end shaker. The lysates were centrifuged at 1000 × g for 10 minutes at 4°C, and the pellets were resuspended in a buffer supplied by the manufacturer and disrupted by using a blunt-ended, 23-gauge needle and a syringe, followed by centrifugation at 6000 × g for 20 minutes at 4°C. The pellets were resuspended in 100 μl of 20 mM Hepes, pH 7.4, 150 mM NaCl, 1% NP-40, 20 mM β-glycerolphosphate, 1 mM sodium orthovanadate, and 20 mM sodium fluoride, and assayed for activity as previously described (Kenny et al., 2003) and outlined below for assaying recombinant enzyme, except that the recombinant protein was replaced by 20 μl of resuspended mitochondrial lysate.
RNAi
The knock-down of KGA was performed using two distinct Stealth Select RNAi Duplexes from Invitrogen directed against the two forms of the kidney-type enzyme. The RNAi nucleotides were transiently transfected in either control NIH 3T3 cells and NIH 3T3 cells stably expressing Cdc42(F28L), Rac(F28L), RhoC(F30L), MDA-MB231 cells, or SKBR3 cells using Lipofectamine 2000 and the relative knock-down efficiency was determined using a polyclonal antibody that recognizes both forms of KGA (a kind gift from Dr. N. Curthoys, Colorado State University). A non-specific oligonucleotide was used as a negative control. Knock-downs of RhoA, RhoC, and p65/RelA were performed with siRNAs obtained from Invitrogen and their efficiencies were determined using anti-RhoC polyclonal antibody from Santa Cruz, and an anti-RhoA monoclonal antibody and an anti-p65/RelA polyclonal antibody from Cell Signaling.
Assays of recombinant glutaminase activity
Glutaminase activity assays were performed on recombinant enzyme as previously described (Kenny et al., 2003). A plasmid encoding mouse GAC (residues 128–603) was cloned into the pET28a vector and the protein was expressed with an N-terminal histidine (His)-tag. The tag was cleaved using thrombin and the protein was purified by anion-exchange and gel-filtration chromatography. Recombinant GAC (1 μM) was incubated with varying concentrations of 968 in 57 mM Tris-Acetate (pH 8.6) and 0.225 mM EDTA by rotating at 37°C for 30 minutes, in a final volume of 80 μl. 968 was diluted in DMSO such that the volume added was constant (5 μl) for all samples, ensuring that the concentration of DMSO (6.3% v/v) was the same in each of the assay incubations. A glutamine solution was then added to give a final volume of 115 μl and a final concentration of 17 mM. The reaction proceeded at 37°C for 1 h and was stopped by adding 10 μl of ice-cold 3M HCl. An aliquot of the quenched reaction mixture (10 μl) was added to an incubation containing 114 mM Tris-HCl (pH 9.4), 0.35 mM ADP, 1.7 mM NAD and 6.3 U/ml glutamate dehydrogenase to give a final volume of 228 μl. The reaction mixture was incubated at room temperature for 45 minutes and the absorbance at 340 nm was recorded for each sample against a water blank. The absorbance of the sample with just the cocktail mixture was subtracted from each reading to calculate the activity of the enzyme.
Acknowledgments
We thank Dr. M. Antonyak for discussions, Dr. N. Curthoys (Colorado State University) for the antibody against KGA, Dr. W. Lane and the Harvard microsequencing facility, and C. Westmiller for secretarial assistance. R.C. acknowledges funding support from the NIH and the Susan G. Komen Breast Cancer Foundation. C.V.D. is supported by NCI, NIH, Leukemia and Lymphoma Society and the AACR Stand-Up-to-Cancer initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baird D, Feng Q, Cerione RA. The Cool-2/α-Pix protein mediates a Cdc42-Rac signaling cascade. Current Biology. 2005;15:1–10. [Abstract] [Google Scholar]
- Burbelo P, Wellstein A, Pestell RG. Altered Rho GTPase signaling pathways in breast cancer cells. Breast Cancer Res Treat. 2004;84:43–48. [Abstract] [Google Scholar]
- Cammarano MS, Minden A. Dbl and the Rho GTPases activate NFκB by IκB kinase (IKK)-dependent and IKK-independent pathways. J Biol Chem. 2001;276:25876–25882. [Abstract] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008a;452:181–186. [Abstract] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008b;452:230–233. [Abstract] [Google Scholar]
- Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. [Abstract] [Google Scholar]
- Curthoys NP. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. [Abstract] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. [Europe PMC free article] [Abstract] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–19. [Abstract] [Google Scholar]
- Erickson JW, Cerione RA. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry. 2004;43:837–842. [Abstract] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. [Abstract] [Google Scholar]
- Feng Q, Baird D, Peng X, Wang J, Ly T, Guan JL, Cerione RA. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nature Cell Biology. 2006;8:945–956. [Abstract] [Google Scholar]
- Fort P. Small GTPases of the Rho family and cell transformation. Prog Mol Subcell Biol. 1999;22:159–181. [Abstract] [Google Scholar]
- Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumors: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87:635–644. [Europe PMC free article] [Abstract] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. [Europe PMC free article] [Abstract] [Google Scholar]
- Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [Abstract] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. [Europe PMC free article] [Abstract] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–7460. [Europe PMC free article] [Abstract] [Google Scholar]
- Kamai T, Yamanishi T, Shirataki H, Takagi K, Asami H, Ito Y, Yoshida K. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10:4799–4805. [Abstract] [Google Scholar]
- Kenny J, Bao Y, Hamm B, Taylor L, Toth A, Wagers B, Curthoys NP. Bacterial expression, purification and characterization of rat kidney-type mitochondrial glutaminase. Protein Expr Purif. 2003;31:140–148. [Abstract] [Google Scholar]
- Kleer CG, van Golen KL, Zhang Y, Wu ZF, Rubin MA, Merajver SD. Characterization of RhoC expression in benign and malignant breast disease: a potential new marker for small breast carcinomas with metastatic ability. Am J Pathol. 2002;160:579–584. [Europe PMC free article] [Abstract] [Google Scholar]
- Lahoz A, Hall A. DLC1: a significant GAP in the cancer genome. Genes Dev. 2008;22:1724–1730. [Europe PMC free article] [Abstract] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037–2042. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin R, Bagrodia S, Cerione R, Manor D. Specific contributions of the small GTPases Rho, Rac and Cdc42 to Dbl transformation. J Biol Chem. 1999;274:23633–23641. [Abstract] [Google Scholar]
- Medina MA, Sanchez-Jimenez F, Marquez J, Rodriguez Quesada A, Nunez de Castro I. Relevance of glutamine metabolism to tumor cell growth. Mol Cell Biochem. 1992;113:1–15. [Abstract] [Google Scholar]
- Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci USA. 2000;97:185–189. [Europe PMC free article] [Abstract] [Google Scholar]
- Perona R, Montaner S, Saniger L, Sánchez-Pérez I, Bravo R, Lacal JC. Activation of the nuclear factor-κB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. [Abstract] [Google Scholar]
- Pickering BM, de Mel S, Lee M, Howell M, Habens F, Dallman CL, Neville LA, Potter KN, Mann J, Mann DA, et al. Pharmacological inhibitors of NF-κB accelerate apoptosis in chronic lymphocytic leukemia cells. Oncogene. 2007;26:1166–1177. [Abstract] [Google Scholar]
- Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, Sonenshein GE. Aberrant nuclear factor-kB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100:2952–2960. [Europe PMC free article] [Abstract] [Google Scholar]
- Suwa H, Ohshio G, Imamura T, Watanabe G, Arii S, Imamura M, Narumiya S, Hiai H, Fukumoto M. Overexpression of the rhoC gene correlates with progression of ductal adenocarcinoma of the pancreas. Br J Cancer. 1998;77:147–152. [Europe PMC free article] [Abstract] [Google Scholar]
- Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA. 2010;107:7461–7466. [Europe PMC free article] [Abstract] [Google Scholar]
- Szeliga M, Obara-Michlewska M, Matyja E, Lazarczyk M, Lobo C, Hilgier W, Alonso FJ, Márquez J, Albrecht J. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia. 2009;57:1014–1023. [Abstract] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. [Europe PMC free article] [Abstract] [Google Scholar] Retracted
- Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. [Abstract] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. [Abstract] [Google Scholar]
- Wang JB, Wu WJ, Cerione RA. Cdc42 and Ras cooperate to mediate cellular transformation by intersectin-L. J Biol Chem. 2005;280:22883–22891. [Abstract] [Google Scholar]
- Whitehead IP, Lambert QT, Glaven JA, Abe K, Rossman KL, Mahon GM, Trzaskos JM, Kay R, Campbell SL, Der CJ. Dependence of Dbl and Dbs transformation on MEK and NF-kappaB activation. Mol Cell Biol. 1999;19:7759–7770. [Europe PMC free article] [Abstract] [Google Scholar]
- Xue W, Krasnitz A, Lucito R, Sordella R, VanAelst L, Cordon-Cardo C, Singer S, Kuehnel F, Wigler M, Powers S, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ccr.2010.08.009
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1535610810003065/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The unique catalytic properties of PSAT1 mediate metabolic adaptation to glutamine blockade.
Nat Metab, 6(8):1529-1548, 27 Aug 2024
Cited by: 0 articles | PMID: 39192144
Targeting amino acid-metabolizing enzymes for cancer immunotherapy.
Front Immunol, 15:1440269, 14 Aug 2024
Cited by: 0 articles | PMID: 39211039 | PMCID: PMC11359565
Review Free full text in Europe PMC
Glutamine Metabolism and Prostate Cancer.
Cancers (Basel), 16(16):2871, 18 Aug 2024
Cited by: 1 article | PMID: 39199642 | PMCID: PMC11352381
Review Free full text in Europe PMC
Affinity war: PSAT1 outcompetes the rest.
Nat Metab, 6(8):1429-1430, 01 Aug 2024
Cited by: 0 articles | PMID: 39192143
Glutaminolysis is a Potential Therapeutic Target for Kidney Diseases.
Diabetes Metab Syndr Obes, 17:2789-2807, 23 Jul 2024
Cited by: 0 articles | PMID: 39072347 | PMCID: PMC11283263
Review Free full text in Europe PMC
Go to all (522) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Rho GTPases and their roles in cancer metabolism.
Trends Mol Med, 19(2):74-82, 05 Dec 2012
Cited by: 56 articles | PMID: 23219172 | PMCID: PMC3607349
Review Free full text in Europe PMC
The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy.
Nat Commun, 7:11321, 18 Apr 2016
Cited by: 92 articles | PMID: 27089238 | PMCID: PMC4837472
Glutaminase: a hot spot for regulation of cancer cell metabolism?
Oncotarget, 1(8):734-740, 01 Dec 2010
Cited by: 100 articles | PMID: 21234284 | PMCID: PMC3018840
Review Free full text in Europe PMC
Therapeutic strategies impacting cancer cell glutamine metabolism.
Future Med Chem, 5(14):1685-1700, 01 Sep 2013
Cited by: 80 articles | PMID: 24047273 | PMCID: PMC4154374
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (7)
Grant ID: R01 GM047458-19
Grant ID: R01 GM061762
Grant ID: R01 GM040654
Grant ID: R01 GM061762-10
Grant ID: R01 GM047458-18
Grant ID: R01 GM040654-21
Grant ID: R01 GM047458





