Abstract
Free full text

Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate
Abstract
Base excision repair (BER) represents the most important repair pathway of endogenous DNA lesions. Initially, a base damage is recognized, excised and a DNA single-strand break (SSB) intermediate forms. The SSB is then ligated, a process that employs proteins also involved in SSB repair, e.g. XRCC1, Ligase III and possibly PARP1. Here, we confirm the role of XRCC1 and PARP in direct SSB repair. Interestingly, we uncover a synthetic lethality between XRCC1 deficiency and PARP inhibition. We also treated cells with alkylating agent dimethyl sulfate (DMS) and monitored the SSB intermediates formed during BER. DMS-induced SSBs were quickly repaired in wild-type cells; while a rapid accumulation of SSBs was observed in cells where post-incision repair was blocked by a PARP inhibitor or by XRCC1 deficiency (EM9 cells). Interestingly, DMS-induced SSBs did not accumulate in PARP1 siRNA depleted cells, demonstrating that PARP1 is not required for efficient completion of BER. Based on these results we suggest no immediate role for PARP1 in BER, but that PARP inhibitors trap PARP on the SSB intermediate formed during BER. Unexpectedly, addition of PARP inhibitor 2 h after DMS treatment still increased SSB levels indicating ongoing repair even at this late time point.
h after DMS treatment still increased SSB levels indicating ongoing repair even at this late time point.
INTRODUCTION
Base damages, such as methylations, oxidations, depurinations and single-strand breaks (SSBs) are commonly formed by endogenous cellular metabolism (1). The recognition of these damages is imperative for efficient repair and is achieved by a set of specialized glycosylases. Poly(ADP-ribose)polymerase 1 (PARP1) is an evolutionary poorly conserved nick-sensing enzyme with the catalytic ability to produce long polymers of ADP-ribose on itself and other proteins (2). This polymerization is stimulated by the binding of PARP1 to a nick in the DNA and results in the rapid relocation of repair proteins such as XRCC1, to the site of the lesion (3). PARP1 is known to interact with proteins in both short- and long-patch repair pathways downstream of damage recognition (4–7). Precisely how PARP1 functions in repair and signalling is not yet determined as its main activity at the DNA SSB appears to be autoribosylation, which is finalised when the negatively charged ADP-ribose polymers cause PARP1 to dissociate from the DNA. Inhibition of PARP1 has been shown to impair DNA SSB repair, as the inhibition of its polymeric activity traps the enzyme at the SSB and physically blocks further repair (8). However, PARP1 does not appear to be required for SSB repair as cells with PARP1 knocked down still display efficient SSB repair activity (8). The binding of PARP1 to SSBs may act as protection from excessive DNA damage by sequestering the potentially toxic intermediates until they can be repaired. Although PARP1 is often annotated to be a base excision repair (BER) protein it is unclear exactly how and if PARP1 is involved in the repair of DNA lesions such as methylated or oxidizes bases. Here, we hypothesize that PARP1 has no active role in BER, since it is a poorly conserved protein through evolution, in contrast to many other BER factors. Furthermore, the aim of this study is to assay SSB formation to enable us to investigate the impact of PARP1 in the repair of methylated DNA and to investigate how PARP inhibition and knockdown can affect this process.
MATERIALS AND METHODS
Cell cultures
Cells were cultured at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 9% fetal calf serum and penicillin-streptomycin (90 units/ml) in an atmosphere containing 5% CO2. The cell lines used in this study were human alveolar basal epithelial cell line A549 and the XRCC1-deficient Chinese hamster ovary (CHO) cell line EM9, the latter stably transfected with either empty vector (EM9-V) or his-tagged XRCC1 gene (EM9-XH) as described in (9).
siRNA transfection
About 1 ×
× 106 cells were seeded in a 75 cm2 flask and cultured for 24
106 cells were seeded in a 75 cm2 flask and cultured for 24 h before transfection with siRNA. Transfection (10
h before transfection with siRNA. Transfection (10 nM siRNA) was performed using INTERFERinTM (PolyPlus Transfection) according to the vendor’s protocol. The siRNA was purchased from MWG Biotech AG with the oligosequences: PARP1; 5′-AAGCCAUGGUGGAGUAUGATT-3′ and MPG; 5′-AAGAAGCAGCGACCAGCUAGA-3′.
nM siRNA) was performed using INTERFERinTM (PolyPlus Transfection) according to the vendor’s protocol. The siRNA was purchased from MWG Biotech AG with the oligosequences: PARP1; 5′-AAGCCAUGGUGGAGUAUGATT-3′ and MPG; 5′-AAGAAGCAGCGACCAGCUAGA-3′.
Repair assay
Cells were seeded in 24-well plates, at a density of 5 ×
× 104 cells per well, and cultured for 24
104 cells per well, and cultured for 24 h before labelling the DNA with 3H-TdR (7.1 kBq/ml) for 24
h before labelling the DNA with 3H-TdR (7.1 kBq/ml) for 24 h. To ensure a low background of SSBs, the cells were washed twice with HBSS++ and incubated in fresh DMEM for 1
h. To ensure a low background of SSBs, the cells were washed twice with HBSS++ and incubated in fresh DMEM for 1 h. Treatment with hydrogen peroxide (H2O2) or dimethyl sulfate (DMS) (diluted in HBSS++) was performed for 15
h. Treatment with hydrogen peroxide (H2O2) or dimethyl sulfate (DMS) (diluted in HBSS++) was performed for 15 min on ice and was terminated by washing the cells with ice cold HBSS++ twice. Although BER is not completely abolished, the incision rate is reduced ~20-fold by a temperature shift from 37°C to 4°C (10). Pre-warmed DMEM was added to the cells, which were then incubated at 37°C for various time intervals to monitor the repair.
min on ice and was terminated by washing the cells with ice cold HBSS++ twice. Although BER is not completely abolished, the incision rate is reduced ~20-fold by a temperature shift from 37°C to 4°C (10). Pre-warmed DMEM was added to the cells, which were then incubated at 37°C for various time intervals to monitor the repair.
Treatment with DNA damaging agent and the following steps were performed in the presence of PARP inhibitor 1,8-napthalimide (50 µM) in specified samples (11). In order to study remaining damages, the pre-warmed DMEM was replaced by pre-warmed DMEM containing 1,8- napthalimide (50
µM) in specified samples (11). In order to study remaining damages, the pre-warmed DMEM was replaced by pre-warmed DMEM containing 1,8- napthalimide (50 µM), as indicated, 2
µM), as indicated, 2 h after terminating the treatment with DMS.
h after terminating the treatment with DMS.
In the ADU (alkaline DNA unwinding) assay the cells were washed twice with ice cold 0.15 M NaCl and lysed in the DNA unwinding solution (0.15
M NaCl and lysed in the DNA unwinding solution (0.15 M NaCl, 30
M NaCl, 30 mM NaOH) on ice, with the exception of the experiments done with A549 cells and the DMS dose-curve, in which the unwinding was performed at room temperature. The unwinding of DNA was continued for 30
mM NaOH) on ice, with the exception of the experiments done with A549 cells and the DMS dose-curve, in which the unwinding was performed at room temperature. The unwinding of DNA was continued for 30 min in darkness and was subsequently neutralized by the addition of 20
min in darkness and was subsequently neutralized by the addition of 20 mM NaH2PO4. The DNA was then fragmented by sonication (15
mM NaH2PO4. The DNA was then fragmented by sonication (15 sec, Branson sonifier B-12, with micro tip), after which SDS (final concentration of 0.24 %) was added. The samples were kept frozen for at least 12
sec, Branson sonifier B-12, with micro tip), after which SDS (final concentration of 0.24 %) was added. The samples were kept frozen for at least 12 h at −20°C. Separation of double-stranded (ds) and single-stranded (ss) DNA was performed on hydroxyl apatite columns, kept at 60°C, as described earlier (12). Briefly, cell lysates were diluted with water (dilution factor 0.4) and 1
h at −20°C. Separation of double-stranded (ds) and single-stranded (ss) DNA was performed on hydroxyl apatite columns, kept at 60°C, as described earlier (12). Briefly, cell lysates were diluted with water (dilution factor 0.4) and 1 ml of each sample was loaded on to a column. The bound DNA was washed with sodium phosphate (10
ml of each sample was loaded on to a column. The bound DNA was washed with sodium phosphate (10 mM) and ssDNA was eluted with 4.25
mM) and ssDNA was eluted with 4.25 ml potassium phosphate at a concentration of 0.1
ml potassium phosphate at a concentration of 0.1 M, where after dsDNA was eluted with 4.25
M, where after dsDNA was eluted with 4.25 ml potassium phosphate at a concentration of 0.25
ml potassium phosphate at a concentration of 0.25 M. The number of SSBs per cell was calculated from the ratio of ssDNA to dsDNA by correlation with a standard curve obtained by γ-irradiation (12–14).
M. The number of SSBs per cell was calculated from the ratio of ssDNA to dsDNA by correlation with a standard curve obtained by γ-irradiation (12–14).
Survival assay
Cells were seeded in 10 cm Petri dishes 6–8
cm Petri dishes 6–8 h prior to treatment. Exposure to hydrogen peroxide (H2O2) or DMS was performed for 15
h prior to treatment. Exposure to hydrogen peroxide (H2O2) or DMS was performed for 15 min on ice in HBSS++, after which the cells were washed and DMEM was added. Exposure to 1,8-napthalimide was performed by the addition of inhibitor to the cellular growth medium ~6
min on ice in HBSS++, after which the cells were washed and DMEM was added. Exposure to 1,8-napthalimide was performed by the addition of inhibitor to the cellular growth medium ~6 h after seeding the cells. After 24
h after seeding the cells. After 24 h the medium was replaced with fresh DMEM. The plates were incubated at 37°C under 5% CO2. After 7–10 days, the colonies were fixed, stained (methylene blue dissolved in methanol, 4
h the medium was replaced with fresh DMEM. The plates were incubated at 37°C under 5% CO2. After 7–10 days, the colonies were fixed, stained (methylene blue dissolved in methanol, 4 g/l) and counted. Calculations are based on the mean value of two replicas per treatment in three separate experiments.
g/l) and counted. Calculations are based on the mean value of two replicas per treatment in three separate experiments.
Western blot analysis
Cells were trypsinized and washed with HBSS−+ before the addition of protein lysis buffer (1× protease inhibitor cocktail Roche, 1 % NP-40, 150 mM NaCl, 0.1
mM NaCl, 0.1 M Tris pH 8.0). Whole cell lysates were prepared by 30
M Tris pH 8.0). Whole cell lysates were prepared by 30 min lysis on ice with vortexing every 5
min lysis on ice with vortexing every 5 min followed by centrifugation, where the pellet was discarded. The protein concentration was determined by the Bradford assay (Hercules, CA) according to the vendor’s protocol. The lysates (75
min followed by centrifugation, where the pellet was discarded. The protein concentration was determined by the Bradford assay (Hercules, CA) according to the vendor’s protocol. The lysates (75 µg protein) were resolved by 4–12% NuPAGE Bis-Tris Gel (Invitrogen) for subsequent western blot analysis using antibodies against the following proteins diluted in 5 % milk (in PBS) as indicated: β-tubulin (1:500), PARP1 (1:500) and MPG (HPA006531, Atlas Antibodies AB) (1:500). The gel was transferred to a nitrocellulose membrane and immunoblotted with primary antibody. The secondary antibody (anti-mouse or anti-rabbit) IgG conjugated to horseradish peroxidase was used for detection of the proteins by an ECL western blot detection system (Thermo scientific).
µg protein) were resolved by 4–12% NuPAGE Bis-Tris Gel (Invitrogen) for subsequent western blot analysis using antibodies against the following proteins diluted in 5 % milk (in PBS) as indicated: β-tubulin (1:500), PARP1 (1:500) and MPG (HPA006531, Atlas Antibodies AB) (1:500). The gel was transferred to a nitrocellulose membrane and immunoblotted with primary antibody. The secondary antibody (anti-mouse or anti-rabbit) IgG conjugated to horseradish peroxidase was used for detection of the proteins by an ECL western blot detection system (Thermo scientific).
RESULTS
Distinct roles of XRCC1 and PARP inhibition in SSB repair
It is well established that both PARP1 and XRCC1 are involved in DNA SSB repair (15,16), and that poly(ADPribose) synthesis is required for the recruitment of XRCC1 to sites of oxidative DNA damage (3). To confirm and further investigate the functions of these proteins in SSB repair, we determined the survival and SSB repair capacity in response to hydrogen peroxide in XRCC1 defective EM9 cells stably transfected with empty vector (EM9-V) or wild-type XRCC1 (EM9-XH) in the absence or presence of a PARP inhibitor. As expected, we found that both XRCC1 deficiency as well as co-treatment with PARP inhibitor sensitizes cells to hydrogen peroxide (Figure 1A). Interestingly, we report that XRCC1 defective EM9-V cells are hypersensitive to PARP inhibitors alone (Figure 1B). In order to investigate whether this hypersensitivity is due to the functions of PARP and XRCC1 in SSB repair, we measured the background level of SSBs in these cells. To allow for a direct and quantitative measurement of the number of SSBs we employed the ADU technique (13,14,17), and we found no correlation with the background levels of SSBs and the presence of XRCC1 or the addition of a PARP inhibitor (Figures 1C and and66B).
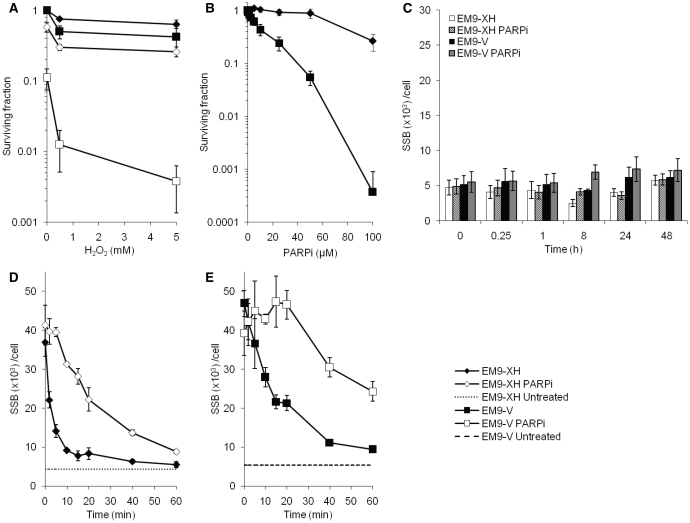
PARP1 and XRCC1 have distinct roles in efficient SSB repair. The PARP1 and XRCC1 proteins are involved in DNA repair in response to hydrogen peroxide. (A) Survival, following a 15 min exposure to hydrogen peroxide on ice, of XRCC1 deficient EM9 cells expressing wild-type XRCC1 (EM9-XH) or empty vector (EM9-V). Cells were treated in the presence or absence of PARP inhibitor (1,8-napthalimide, 50
min exposure to hydrogen peroxide on ice, of XRCC1 deficient EM9 cells expressing wild-type XRCC1 (EM9-XH) or empty vector (EM9-V). Cells were treated in the presence or absence of PARP inhibitor (1,8-napthalimide, 50 µM), which was left in the growth medium for 24
µM), which was left in the growth medium for 24 h after treatment. Survival is plotted as percentage of living mock treated cells without PARP inhibition. The means and standard errors of three experiments are shown. (B) XRCC1 defective EM9-V cells are hypersensitive to PARP inhibitors alone. Survival of EM9-XH and EM9-V cells, following a 24
h after treatment. Survival is plotted as percentage of living mock treated cells without PARP inhibition. The means and standard errors of three experiments are shown. (B) XRCC1 defective EM9-V cells are hypersensitive to PARP inhibitors alone. Survival of EM9-XH and EM9-V cells, following a 24 h treatment with PARP inhibitor under normal growth conditions. The means and standard errors of three experiments are shown. (C) Background levels of SSBs in EM9-V or EM9-XH cell lines, measured after various time points of incubation with or without PARP inhibitor (50
h treatment with PARP inhibitor under normal growth conditions. The means and standard errors of three experiments are shown. (C) Background levels of SSBs in EM9-V or EM9-XH cell lines, measured after various time points of incubation with or without PARP inhibitor (50 µM). The means and standard errors of three experiments are shown. SSB repair rates in the cell lines EM9-XH (D) and EM9-V (E) after a 15
µM). The means and standard errors of three experiments are shown. SSB repair rates in the cell lines EM9-XH (D) and EM9-V (E) after a 15 min treatment with 200
min treatment with 200 µM hydrogen peroxide. Dotted lines indicate the levels of SSBs in mock treated cells for each cell line. Cells were treated and left to repair in the presence or absence of PARP inhibitor. The means and standard errors of three experiments are shown.
µM hydrogen peroxide. Dotted lines indicate the levels of SSBs in mock treated cells for each cell line. Cells were treated and left to repair in the presence or absence of PARP inhibitor. The means and standard errors of three experiments are shown.
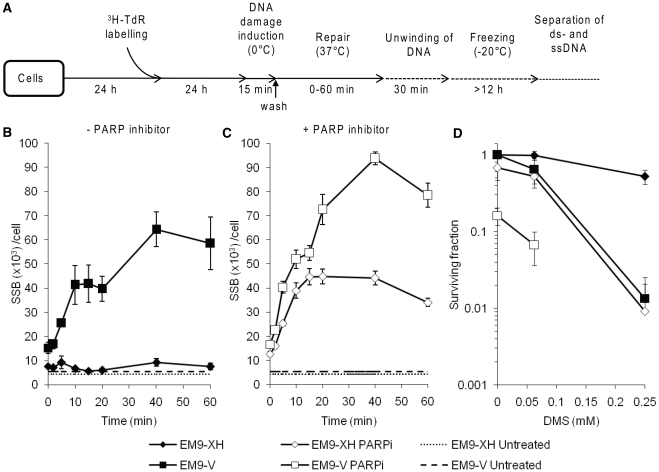
A substantial amount of damages are still being repaired 2 h after exposure to DMS. Level of SSBs in EM9-XH cells treated with (A) DMS (2
h after exposure to DMS. Level of SSBs in EM9-XH cells treated with (A) DMS (2 mM) or (B) mock treated and left to repair. PARP inhibitor was added (open symbols, time point indicated by arrow) to slow down the ligation step, 2
mM) or (B) mock treated and left to repair. PARP inhibitor was added (open symbols, time point indicated by arrow) to slow down the ligation step, 2 h after treatment was terminated. The means and standard errors of three experiments are shown. The ongoing SSB formation 2
h after treatment was terminated. The means and standard errors of three experiments are shown. The ongoing SSB formation 2 h after terminated treatment, represents MPG initiated BER events. Level of SSBs in A549 cells with or without siRNA depleted MPG and treated with (A) DMS (0.5
h after terminated treatment, represents MPG initiated BER events. Level of SSBs in A549 cells with or without siRNA depleted MPG and treated with (A) DMS (0.5 mM) or (B) mock treated and left to repair. PARP inhibitor was added (open symbols, time point indicated by arrow) to slow down the ligation step, 2
mM) or (B) mock treated and left to repair. PARP inhibitor was added (open symbols, time point indicated by arrow) to slow down the ligation step, 2 h after treatment was terminated. The means and standard errors of three experiments are shown.
h after treatment was terminated. The means and standard errors of three experiments are shown.
When monitoring SSB repair by measuring the level of SSBs in cells over time after H2O2-induced damage, we found that PARP inhibition reduce the rate of SSB rejoining (Figure 1D), which support previous results obtained with the comet assay (15). Also, we confirmed that the level of SSBs is considerably enhanced in the cell line lacking XRCC1, indicating impaired ligation of breaks (Figure 1E). PARP inhibition in these cells had an additional decelerating effect on the rate of ligation, further indicating distinct roles of the two proteins in SSB repair.
Distinct roles of XRCC1 and PARP in the repair of alkylated DNA lesions
DMS is used in these experiments since it is 25–30 times more potent than methyl methanesulfonate (MMS) but will produce similar proportions of methylated DNA products (18). DMS will thus remain active on ice, as opposed to MMS. By exposing cells to DMS on ice, we ensure that a minimal background repair is operating during treatment and we can investigate whether XRCC1 and PARP have additional roles in BER, as opposed to their known functions in SSB repair (Figure 2A).
Distinct roles of XRCC1 and PARP inhibition in BER. (A) To study SSB formation during BER, cells were seeded 24 h prior to labelling the DNA with 3H-TdR for an additional 24
h prior to labelling the DNA with 3H-TdR for an additional 24 h. Exposure to DMS was performed on ice to reduce repair during treatment, where after DNA repair was induced by raising the temperature to 37°C for various time intervals. The amount of strand breaks was measured by the alkaline DNA unwinding technique (12). BER incision and formation of a SSB is independent of XRCC1 in cells. Levels of SSBs in the EM9-V and EM9-XH cell lines after a 15
h. Exposure to DMS was performed on ice to reduce repair during treatment, where after DNA repair was induced by raising the temperature to 37°C for various time intervals. The amount of strand breaks was measured by the alkaline DNA unwinding technique (12). BER incision and formation of a SSB is independent of XRCC1 in cells. Levels of SSBs in the EM9-V and EM9-XH cell lines after a 15 min treatment with 0.5
min treatment with 0.5 mM DMS at indicated time points of repair. Cells were treated and left to repair in the (B) absence or (C) presence of PARP inhibitor. The means and standard errors of three experiments are shown. (D) Survival, following a 15
mM DMS at indicated time points of repair. Cells were treated and left to repair in the (B) absence or (C) presence of PARP inhibitor. The means and standard errors of three experiments are shown. (D) Survival, following a 15 min exposure to DMS on ice, of XRCC1 deficient EM9-V cells expressing empty vector (EM9-V) or wild-type XRCC1 (EM9-XH). Cells were treated in the presence or absence of PARP inhibitor, which was left in the growth medium for 24
min exposure to DMS on ice, of XRCC1 deficient EM9-V cells expressing empty vector (EM9-V) or wild-type XRCC1 (EM9-XH). Cells were treated in the presence or absence of PARP inhibitor, which was left in the growth medium for 24 h after treatment. Survival is plotted as percentage of living mock treated cells without PARP inhibition. The means and standard errors of three experiments are shown.
h after treatment. Survival is plotted as percentage of living mock treated cells without PARP inhibition. The means and standard errors of three experiments are shown.
We found that SSBs rapidly accumulate in EM9-V cells after DMS treatment (Figure 2B) suggesting that the detection and excision of methylated bases is independent of XRCC1. The ADU technique might not fully distinguish between AP-sites and SSBs formed during repair, so we cannot specify if the same is true for BER incisions. Furthermore, the accumulation of SSBs highlights the importance of the XRCC1–LigIIIα complex in the latter ligation step of both BER and SSB repair (19). In the SSB assay performed here we also observed a dramatic increase in the level of SSBs following DMS treatment in PARP inhibited cells (Figure 2C). Additionally, when impeding both proteins simultaneously by inhibiting PARP in XRCC1 deficient cells we detected higher levels of SSBs than if either protein is incapacitated alone. The lack of XRCC1 sensitized cells to alkylating damage on the DNA, in a similar fashion as PARP inhibition (Figure 2D), suggesting that these proteins play a role in the repair of DMS lesions. The decreased survival in EM9-V cells treated with PARP inhibitor is most likely due to the hypersensitivity mentioned earlier (Figure 1B).
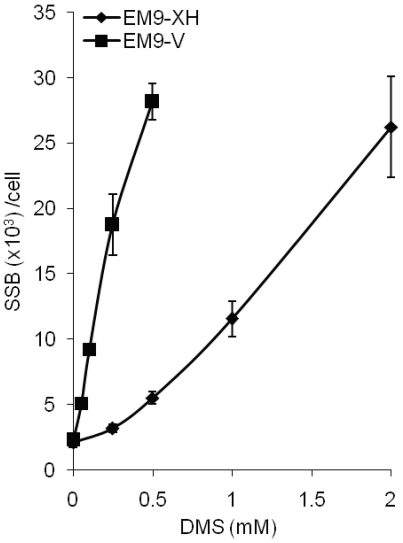
The low levels of SSBs detected in wild-type cells after treatment with DMS indicate a very fast and efficient repair of alkylating damages to the DNA. To determine the repair capacity of CHO cells we treated EM9-XH and EM9-V cells with increasing doses of DMS and left them to repair for 15 min (Figure 3). We found that there is a dose-dependent increase of SSBs, suggesting that the amount of damage to the DNA, at the doses of DMS used in this study, is not saturating the engaged repair pathways.
min (Figure 3). We found that there is a dose-dependent increase of SSBs, suggesting that the amount of damage to the DNA, at the doses of DMS used in this study, is not saturating the engaged repair pathways.
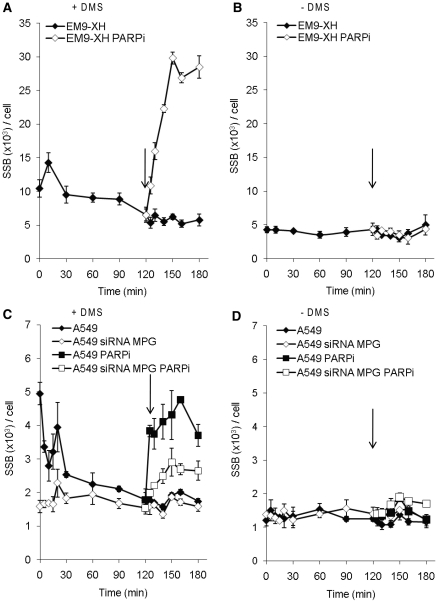
MPG is required for BER initiation after DMS-induced lesions in cells
In order to ascertain that the induced SSBs observed after DMS treatment represent damages processed by BER and not direct SSBs, we siRNA depleted the glycosylase MPG in A549 cells and determined the efficiency of the MPG knockdown (MPGKD) by western blotting (Figure 4A). Subsequently, we followed the SSB formation in MPGKD cells after DMS treatment and found that MPGKD cells exhibited SSBs at background level throughout the time of repair (Figure 4B), in contrast to cells with functional MPG. A similar differential effect was seen when both DMS treatment and the following repair were carried out in the presence of a PARP inhibitor, indicating that the majority of SSBs produced after DMS treatment arise from MPG-dependent removal of methylated bases.
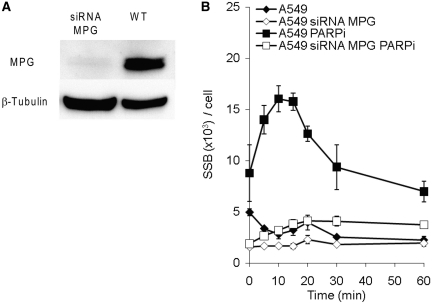
The glycosylase MPG is required for the formation of the majority of SSBs produced during the repair of DMS-induced damages. (A) Western blot, probed with antibodies against MPG and loading control β-tubulin, showing the siRNA knockdown of MPG in human A549 cells. (B) Levels of SSBs in wild-type A549 cells and cells depleted of MPG, after a 15 min treatment with 0.5
min treatment with 0.5 mM DMS at indicated time points of repair. Cells were treated and left to repair in the absence or presence of PARP inhibitor. The means and standard errors of three experiments are shown.
mM DMS at indicated time points of repair. Cells were treated and left to repair in the absence or presence of PARP inhibitor. The means and standard errors of three experiments are shown.
BER kinetics is unaffected in PARP1 knockdown cells
We found that PARP inhibition efficiently disrupts BER (Figures 2 and and4).4). This could be due to an essential role of PARP1 in the latter steps of BER ligation (20) e.g. because PARP inhibitors will block the release of PARP1 from the SSB intermediate it is supposed to protect. To test this we siRNA depleted PARP1 in A549 cells and determined the efficiency of the PARP1 knockdown (PARP1KD) by western blotting (Figure 5A). We followed the repair of SSBs in PARP1KD cells after DMS treatment and found that PARP1KD cells exhibited a slightly lower amount of SSBs after DMS treatment than wild-type A549 cells (Figure 5B), indicating that PARP1 is in fact not necessary for ligation. Interestingly, the overall SSB levels in PARP1KD cells were slightly closer to background compared to wild-type A549 cells after DMS treatment. Next, we followed the SSB levels in A549 and PARP1KD cells exposed to DMS and incubated in the presence of the PARP inhibitor. We found an accumulation of SSBs in the wild-type cells but not in the PARP1KD cells (Figure 5C). Also, the overall background levels were increased following addition of a PARP inhibitor in DMS treated cells, which in speculation could reflect the role of PARP2 in BER or residual PARP1 protein in these cells.
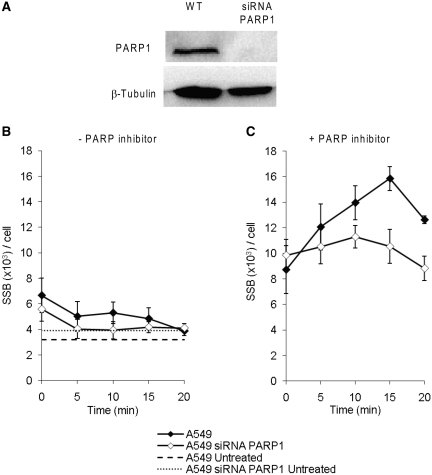
BER kinetics is unaffected in PARP1 knockdown cells. (A) Western blot, probed with antibodies against PARP1 and loading control β-tubulin, showing the knockdown of PARP1 using siRNA in human A549 cells. Level of SSBs in A549 cells with and without siRNA knockdown of PARP1, after a 15 min treatment with 0.5
min treatment with 0.5 mM DMS at indicated time points of repair. Cells were treated and left to repair in the (B) absence or (C) presence of PARP inhibitor. The means and standard errors of three experiments are shown.
mM DMS at indicated time points of repair. Cells were treated and left to repair in the (B) absence or (C) presence of PARP inhibitor. The means and standard errors of three experiments are shown.
BER continues for at least 2 h after the induction of alkylation lesions
h after the induction of alkylation lesions
Finally, we wanted to investigate the kinetics of BER and analyze remaining repair when a large portion of the alkylated DNA has already been repaired. To this end we treated EM9-XH cells with DMS (2 mM) for 15
mM) for 15 min on ice as described, washed the cells thoroughly, and investigated the level of SSBs during repair. We found that the level of SSBs increases significantly when adding a PARP inhibitor 2
min on ice as described, washed the cells thoroughly, and investigated the level of SSBs during repair. We found that the level of SSBs increases significantly when adding a PARP inhibitor 2 h after the DMS treatment is terminated (Figure 6A). Control experiments with untreated cells showed that the amount of SSBs visualized after addition of the PARP inhibitor does not reflect endogenous SSBs (Figure 6B).
h after the DMS treatment is terminated (Figure 6A). Control experiments with untreated cells showed that the amount of SSBs visualized after addition of the PARP inhibitor does not reflect endogenous SSBs (Figure 6B).
We also followed the repair of SSBs in MPGKD cells after DMS treatment (0.5 mM) and found that MPGKD cells exhibited background levels of SSBs which remain low throughout the time of repair (Figure 6C), in contrast to cells with functional MPG. Addition of a PARP inhibitor 2 h after removal of DMS increased SSB levels more prominently in wild-type as compared to MPGKD cells (Figure 6C), demonstrating that the formation of late SSBs is dependent on active BER. Untreated control cells did not display any change in SSB levels, even after the addition of PARP inhibitor 2 h mock treatment is terminated (Figure 6D).
mM) and found that MPGKD cells exhibited background levels of SSBs which remain low throughout the time of repair (Figure 6C), in contrast to cells with functional MPG. Addition of a PARP inhibitor 2 h after removal of DMS increased SSB levels more prominently in wild-type as compared to MPGKD cells (Figure 6C), demonstrating that the formation of late SSBs is dependent on active BER. Untreated control cells did not display any change in SSB levels, even after the addition of PARP inhibitor 2 h mock treatment is terminated (Figure 6D).
DISCUSSION
Here, we confirm the importance of both XRCC1 and PARP in the repair induced by oxidative damage to the DNA (Figure 1A, D and E). XRCC1 is necessary for repair after induction of oxidative damage to the DNA, most likely by recruiting and stabilizing Ligase IIIα (19). PARP inhibition also has a decelerating effect on the rate of ligation, but the two proteins clearly have distinct roles in this repair, as PARP inhibition further delay SSB repair in XRCC1 defective cells. We find that more than half of the H2O2-induced SSBs are repaired within 5 min after damage induction in the reconstituted wild-type cells (Figure 1D), showing that this repair is a fast process. It should be noted that this repair includes the BER of oxidized bases as well as SSB repair, as H2O2 produces a variety of lesions to the DNA (21). However, we see a characteristic rapid repair curve where the majority of SSBs are present in the cells directly after treatment and not after enzymatically-induced incisions, as can be seen after treatment with DMS. Thus, we suggest that the majority of the breaks originate from direct SSBs.
min after damage induction in the reconstituted wild-type cells (Figure 1D), showing that this repair is a fast process. It should be noted that this repair includes the BER of oxidized bases as well as SSB repair, as H2O2 produces a variety of lesions to the DNA (21). However, we see a characteristic rapid repair curve where the majority of SSBs are present in the cells directly after treatment and not after enzymatically-induced incisions, as can be seen after treatment with DMS. Thus, we suggest that the majority of the breaks originate from direct SSBs.
We also find that XRCC1 defective cells are hypersensitive to PARP inhibitors (Figure 1B) in spite of the fact that there was no statistically significant increase of the background levels of SSBs in these cells (Figure 1C), suggesting that the increased sensitivity of XRCC1 defective cells to PARP inhibitors is explained by functions of PARP1 and XRCC1 in processes apart from SSB repair. For instance, blocked replication forks activates PARP1 for efficient restart to occur (22) and both PARP1 and XRCC1 have been shown to have specific roles at replication forks in S phase cells (8,22,23). In speculation, there is a possibility that inhibition of PARP would generate a toxic lesion at replication forks that requires XRCC1 for repair, or vice versa. Also, it should be noted that we have not observed any indications of this hypersensitivity affecting the cells viability during the short time of treatment and repair that are employed in this study.
After confirming the importance of functional XRCC1 and PARP in the repair of SSBs, we move on to compare and evaluate the role of PARP in the repair of alkylated DNA damages. By utilizing XRCC1 deficient cells and a PARP inhibitor, we efficiently block the ligation of DNA breaks and thus we can monitor early repair events. After DMS treatment we find a high level of SSBs in XRCC1 deficient cells as well as in PARP inhibited cells, suggesting that neither XRCC1 nor PARP are necessary for the detection or excision of alkylated DNA damages repaired by BER (Figure 2B and C). Additionally, we see even higher levels of SSBs when impeding both proteins simultaneously by inhibiting PARP in XRCC1 deficient cells, leading to the conclusion that the inhibition of PARP does not affect the repair of DMS induced damages in the same manner as XRCC1 deficiency does.
We confirm that the subsequent SSBs induced by DMS predominantly originate from base alkylations, which are detected by N-methylpurine-DNA glycosylase (MPG). The level of SSBs is considerably reduced when MPG is knocked down by siRNA depletion (Figure 4B). Importantly, as no or very few SSBs are formed after DMS treatment in MPG siRNA depleted cells, our data demonstrate that MPG is the main (and possibly the only) glycosylase to recognize and excise DMS-induced base damage from DNA.
The role of PARP1 is likely to recognize SSBs or other DNA ends in the DNA and to increase the repair. We find that H2O2-induced SSB repair is retarded in cells when treated with a PARP inhibitor, as a likely consequence of reduced recruitment of XRCC1 (8). Furthermore, we find that siRNA depletion of PARP1 did not affect the amount of DMS-induced SSBs, or if anything decreased the amount of SSBs (Figure 5B). This can be explained by PARP1 not having a direct role in BER. In the literature, PARP1 is denoted as a BER protein, which is likely related to previous observations that cell extracts from PARP1−/− cells are unable to complete the DNA synthesis step of BER in vitro (20). However, the presence of PARP1 is not required for effective BER in an in vitro biochemical assay, and may even decrease the biochemical kinetics of BER (24). So far, it has not been possible to assign an exact function of PARP1 in BER (24), but our data support a model where BER does not require PARP1. This is also in line with PARP1 protein being poorly conserved while many other BER factors are well conserved through evolution. The model proposes that the subset of BER SSB intermediates that become uncoupled somewhere during the repair pathway, are bound by PARP1 when it is present in the cell (Figure 7). When PARP is inhibited it is thereby trapped on these SSB intermediates, thus blocking their ligation and causing a potentially toxic retention of SSBs in the DNA. This would explain the increase of DMS-induced SSBs observed following PARP inhibition. Such trapping of PARP to SSB intermediates may explain the high toxicity of PARP inhibitors in BRCA2 defective cells, in contrast to the very modest toxicity of co-depletion of BRCA2 and PARP1 (25).
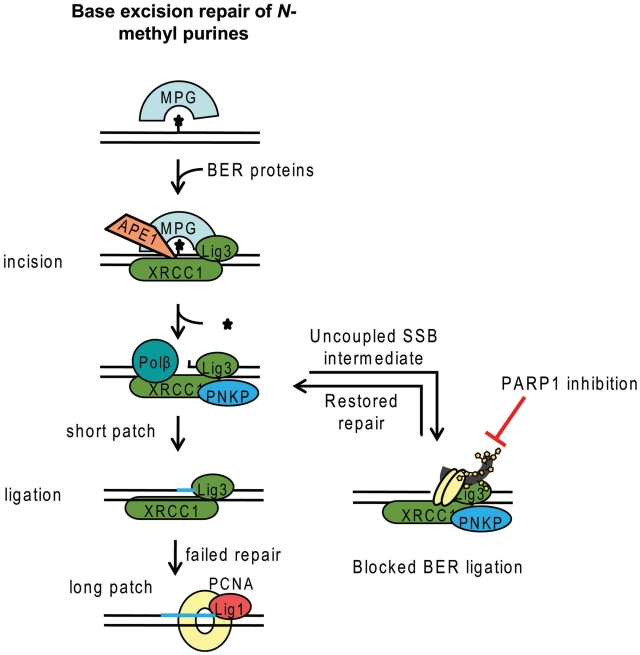
Model for BER and the influence of PARP. The MPG glycosylase is recognizing N-methylated purines, likely through scanning the DNA for base lesions. Once a lesion is recognized, MPG excises the damaged base and the APE1 endonuclease cleaves the newly formed AP-site into a SSB intermediate. If this intermediate is uncoupled from the repair process before repair is finished, PARP1 may recognize and bind to the SSB intermediate, which overall might slow down the BER process. In the event that PARP1 is inhibited it will be trapped on the SSB and prevent ligation. However, PARP1 is not required for accurate repair if the repair pathway remains intact. In the event of the BER complex being unable to accurately ligate, long patch repair is thought to take over. Our model does not exclude an active role for PARP1 in BER by influencing DNA damage-induced chromatin remodelling.
There are two competing models for BER. One model separate the BER incision and the subsequent SSB repair, meaning that the DNA lesion would be identified twice; first by a glycosylase and then by PARP1 for SSB repair. The other model suggests that the DNA lesion is only recognized by a glycosylase and all following steps are coordinated (Figure 7) (26–28). Since we find no accumulation of the SSB intermediate in PARP1 siRNA depleted cells, we suggest that the DNA lesion is only identified by glycosylases/endonucleases and that the following steps are coordinated.
BER of DMS induced damages appears to be a fast and efficient repair pathway as the majority of the SSB intermediates are incised and ligated within 30 min after exposure to the DNA damaging agent (Figures 2B, B,4B4B and and6A).6A). However, we find that a large number of repair events (as measured by SSB formation after addition of a PARP inhibitor) occur in the cells 2 h after DMS was removed from the medium (Figure 6), which suggests that there is still ongoing repair at this time. According to our model, the repair events we detect through PARP inhibition represents only a fraction of uncoupled SSB intermediates, indicating that an even higher amount of total BER events still occur in the cell at this time. We speculate that this ongoing repair could be the result of slow damage detection by the glycosylase, possibly because of inaccessibility to the location of the lesion or the complexity of the lesion. We also see a difference in cells with siRNA depleted MPG compared to wild-type cells, indicating that we are indeed studying actual BER events (Figure 6D). However, it should be noted that there is no certain way to ensure that the damages repaired after 2
min after exposure to the DNA damaging agent (Figures 2B, B,4B4B and and6A).6A). However, we find that a large number of repair events (as measured by SSB formation after addition of a PARP inhibitor) occur in the cells 2 h after DMS was removed from the medium (Figure 6), which suggests that there is still ongoing repair at this time. According to our model, the repair events we detect through PARP inhibition represents only a fraction of uncoupled SSB intermediates, indicating that an even higher amount of total BER events still occur in the cell at this time. We speculate that this ongoing repair could be the result of slow damage detection by the glycosylase, possibly because of inaccessibility to the location of the lesion or the complexity of the lesion. We also see a difference in cells with siRNA depleted MPG compared to wild-type cells, indicating that we are indeed studying actual BER events (Figure 6D). However, it should be noted that there is no certain way to ensure that the damages repaired after 2 h are in fact original damages that were produced at the time of treatment. There is a possibility that cellular DMS is not removed in the washing step and that this could sit in the cell and make damage 2.25
h are in fact original damages that were produced at the time of treatment. There is a possibility that cellular DMS is not removed in the washing step and that this could sit in the cell and make damage 2.25 h after initiation of the treatment. Even in this scenario DMS hydrolyzes in aqueous solutions with a half life of 17
h after initiation of the treatment. Even in this scenario DMS hydrolyzes in aqueous solutions with a half life of 17 min at 37°C (29), so only 0.3% of the original DMS would remain in the cells, even if no DMS was removed by the washing step. Our conclusion is that it takes time for the BER machinery to identify a fraction of the damaged bases to make the incisions.
min at 37°C (29), so only 0.3% of the original DMS would remain in the cells, even if no DMS was removed by the washing step. Our conclusion is that it takes time for the BER machinery to identify a fraction of the damaged bases to make the incisions.
FUNDING
The Swedish Cancer Society; the Swedish Children’s Cancer Foundation the Swedish Research Council; the Swedish Pain Relief Foundation and the Medical Research Council. Funding for open access charge: The Swedish Research Council.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Prof. Keith Caldecott for providing the EM9-V and EM9-XH cell lines, Dr Grigory Dianov and Dr Jason Parsons for discussions and the anonymous reviewers for their suggestions.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkq1241
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/39/8/3166/7187325/gkq1241.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/full/39/8/3166
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/reprint/39/8/3166.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/abstract/39/8/3166
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/gkq1241
Article citations
DNA damage response inhibitors in cancer therapy: lessons from the past, current status and future implications.
Nat Rev Drug Discov, 12 Nov 2024
Cited by: 0 articles | PMID: 39533099
Review
Molecular mechanism of PARP inhibitor resistance.
Oncoscience, 11:69-91, 23 Sep 2024
Cited by: 0 articles | PMID: 39318358 | PMCID: PMC11420906
Review Free full text in Europe PMC
A First-in-Class High-Throughput Screen to Discover Modulators of the Alternative Lengthening of Telomeres (ALT) Pathway.
ACS Pharmacol Transl Sci, 7(9):2799-2819, 13 Aug 2024
Cited by: 0 articles | PMID: 39296266
Poly (ADP-ribose) polymerase inhibitor therapy and mechanisms of resistance in epithelial ovarian cancer.
Front Oncol, 14:1414112, 29 Jul 2024
Cited by: 0 articles | PMID: 39135999 | PMCID: PMC11317305
Review Free full text in Europe PMC
The TIMELESS and PARP1 interaction suppresses replication-associated DNA gap accumulation.
Nucleic Acids Res, 52(11):6424-6440, 01 Jun 2024
Cited by: 2 articles | PMID: 38801073 | PMCID: PMC11194094
Go to all (181) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
HPA - The Human Protein Atlas
- (1 citation) HPA - HPA006531
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
XRCC1 counteracts poly(ADP ribose)polymerase (PARP) poisons, olaparib and talazoparib, and a clinical alkylating agent, temozolomide, by promoting the removal of trapped PARP1 from broken DNA.
Genes Cells, 27(5):331-344, 01 Mar 2022
Cited by: 10 articles | PMID: 35194903 | PMCID: PMC9310723
XRCC1 prevents toxic PARP1 trapping during DNA base excision repair.
Mol Cell, 81(14):3018-3030.e5, 07 Jun 2021
Cited by: 71 articles | PMID: 34102106 | PMCID: PMC8294329
CK2 phosphorylation of XRCC1 facilitates dissociation from DNA and single-strand break formation during base excision repair.
DNA Repair (Amst), 10(9):961-969, 15 Aug 2011
Cited by: 22 articles | PMID: 21840775
XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks.
Cell Res, 18(1):48-63, 01 Jan 2008
Cited by: 145 articles | PMID: 18166976 | PMCID: PMC2366203
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Medical Research Council (1)
MRC/CR-UK Gray Institute for Radiation Oncology and Biology, Oxford - MRC Core Grant
Professor William Gillies McKenna, University of Oxford
Grant ID: G0700730