Abstract
Free full text

Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods
Associated Data
Abstract
Centrosomes in animal cells are dynamic organelles with a proteinaceous matrix of pericentriolar material assembled around a pair of centrioles. They organize the microtubule cytoskeleton and the mitotic spindle apparatus. Mature centrioles are essential for biogenesis of primary cilia that mediate key signalling events. Despite recent advances, the molecular basis for the plethora of processes coordinated by centrosomes is not fully understood. We have combined protein identification and localization, using PCP-SILAC mass spectrometry, BAC transgeneOmics, and antibodies to define the constituents of human centrosomes. From a background of non-specific proteins, we distinguished 126 known and 40 candidate centrosomal proteins, of which 22 were confirmed as novel components. An antibody screen covering 4000 genes revealed an additional 113 candidates. We illustrate the power of our methods by identifying a novel set of five proteins preferentially associated with mother or daughter centrioles, comprising genes implicated in cell polarity. Pulsed labelling demonstrates a remarkable variation in the stability of centrosomal protein complexes. These spatiotemporal proteomics data provide leads to the further functional characterization of centrosomal proteins.
Introduction
The definition of the components of large non-membranous organelles, their relative abundance, and their turnover rates, are important, but unsolved goals in cell biology. Generally, organelles cannot be purified to homogeneity and methods are required to determine the actual components. Furthermore, the components change through the cell cycle and development. In an attempt to address these challenges, we focused on the centrosome which is a dynamic cell organelle with a proteinaceous matrix of pericentriolar material assembled around a pair of centrioles. The single centrosome present in G1-phase cells is usually positioned near the nucleus where it organizes microtubules that coordinate the shape, polarity, adhesion, and mobility of the cell, and facilitates intracellular transport and positioning of the organelles (Doxsey, 2001; Bornens, 2002; Nigg, 2002). Centrosomes at this stage harbour a daughter centriole and a mature mother centriole. The mature centriole has the ability to function as a basal body that seeds the growth of a primary cilium protruding from the cell surface. In multiciliated epithelial cells, de novo assembled basal bodies nucleate motile cilia important for fluid flow and cell migration (Satir and Christensen, 2007). It is now clear that primary cilia are sensory organelles that regulate signalling pathways such as sonic hedgehog and Wnt/planar cell polarity pathways, which in turn regulate essential cellular and developmental processes. The significance of sensory cilia is underlined by the recent findings that mutations affecting genes essential for their formation or function can lead to a number of severe human diseases and developmental defects, now known as the ‘ciliopathies’ (Fliegauf et al, 2007).
During the S-phase of the cell cycle, the centrosome duplicates by the formation of procentrioles adjacent to each of the two parental centrioles. At the G2-M transition, the microtubule-nucleating capacities are increased by the recruitment of γ-tubulin ring complexes (γ-TuRCs) before the centrosomes separate and promote the formation of spindle asters and the positioning of the two spindle poles important for chromosome and centrosome segregation during mitosis. The duplication and segregation cycles of centrosomes and chromosomes are coordinated to avoid the numerical aberration of centrosomes, the missegregation of chromosomes, and the ploidy changes that are typical features of human tumours (Nigg, 2006). Moreover, the equal segregation of one centrosome per cell ensures that each cell has the potential to grow a single primary cilium (Tsou and Stearns, 2006). Plurifunctional roles in cell division are further supported by multiple lines of evidence, suggesting that the centrosome also contributes to cell-cycle regulation and checkpoints, asymmetric division and fate of sister cells, and acts as a scaffold for additional regulatory processes in the cell (Doxsey, 2001; Doxsey et al, 2005; Wang et al, 2009).
Information about the protein composition of centrosomes and centrosome-related structures has been obtained through the application of proteomics, genomics, and bioinformatics in various eukaryotic cells (Bettencourt-Dias and Glover, 2007). The yeast spindle poles were the first to be characterized by mass spectrometry (MS)-based proteomics (Wigge et al, 1998). This study has been followed by the proteomic analyses of centrosomes from human lymphoblast cells (Andersen et al, 2003), the midbody from Chinese hamster ovary cells (Skop et al, 2004), the mitotic spindle from synchronized HeLa S3 cells (Sauer et al, 2005), in vitro-assembled spindle structures from Xenopus and HeLa cell extracts (Liska et al, 2004), and the centrosome of Dictyostelium discoideum (Reinders et al, 2006). Proteomic studies have also revealed the composition of ciliary and flagellar structures including the human ciliary axoneme (Ostrowski et al, 2002), the mouse photoreceptor sensory cilium complex (Liu et al, 2007), the flagellum and basal body of Chlamydomonas reinhardtii (Keller et al, 2005; Pazour et al, 2005), and the flagellum of Trypanosoma brucei (Broadhead et al, 2006). The cilia and flagella studies have been complemented by comparative genomics to identify genes that exist exclusively in organisms that have basal bodies and cilia (Li et al, 2004; Chen et al, 2006; Baron et al, 2007; Merchant et al, 2007). Taken together, these efforts have revealed candidate proteins associated with the centrosome, the centrioles, the mitotic spindle, midbody, and the cilium, some of which have been validated through localization (Andersen et al, 2003; Keller et al, 2005; Sauer et al, 2005) and RNA interference studies (Graser et al, 2007a; Lawo et al, 2009).
The above findings illustrate how different strategies have contributed to the identification of >100 proteins associated with the centrosome leading to functional insight and molecular understanding of genetic disorders (Chang et al, 2006; Sayer et al, 2006; Valente et al, 2006; den Hollander et al, 2006; Spektor et al, 2007; Graser et al, 2007a, 2007b; Nigg and Raff, 2009). Despite these advances, many unsolved questions regarding centrosome and cilia function remain. For example, while the studies described above have revealed the identity of cilia and centrosomes components, we still do not know how most of these components dynamically localize, interact, and function at the molecular, cellular, and organismal level. Likewise, the causative gene in families with ciliopathies is unknown in most cases, suggesting that additional genes expected to be associated with cilia or centrosomes remain to be identified (Otto et al, 2010).
To address these questions, we describe here the combined use of complementary proteomics strategies based on MS and microscopy to further explore the localization, abundance, and turnover of centrosomal proteins. The combined efforts resulted in a more comprehensive coverage of the human centrosome proteome than previously reported, comprising known and novel components. The advancement was made possible by the availability of affinity purified antibodies form the Human Protein Atlas (HPA) project (Barbe et al, 2008) and by the development of a novel MS-based proteomics method, which improved the confidence in identifying genuine organelle components from a background of non-specific proteins. Candidates were validated by image analyses of cells stably expressing fluorescently tagged fusion proteins at the endogenous level employing BAC TransgeneOmics (Poser et al, 2008). Additional microscopy and MS-based experiments revealed the dynamic and asymmetric association of novel proteins with the mother and daughter centriole. The resulting spatiotemporal proteomics data are likely to provide leads to further insight into the functional significance of centrosome-associated proteins.
Results
Complementary proteomics methods identify novel centrosomal proteins
To evaluate the dynamic composition and localization of the centrosome proteome with the ultimate goal to better understand its structure and function we carried out two complementary screens. In the ‘MS-screen’, we developed an MS-based proteomics method to selectively identify centrosomal proteins from a background of unrelated proteins typically present in preparations of biochemically purified centrosomes. In the ‘HPA-screen’, we identified additional components localizing to the centrosomes by evaluating confocal images of three different cell lines stained with HPA antibodies (Barbe et al, 2008). An outline of the two screens and the follow-up experiments performed to validate and further characterize the identified candidate proteins are summarized in Figure 1A. The resulting data are visualized as a dynamic network of proteins associated with the centrosome and its substructures (Figure 1B).
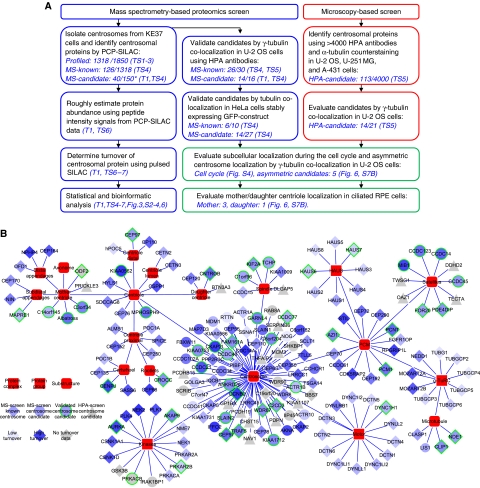
Mapping the centrosome proteome. (A) Schematic outline of the mass spectrometry and microscopy-based screens carried out to identify and characterize candidate centrosomal proteins. In the MS-screen (left), centrosomal proteins were identified by the PCP-SILAC method (see Figure 2) and validated by co-localization experiments using antibodies and GFP-tagged proteins. In the HPA-screen (right), images of three different cell lines were evaluated for centrosomal staining using human protein atlas (HPA) antibodies. In follow-up experiments, we estimated the abundance, measured the turnover, and determined the subcellular localization of the identified proteins. The number of ‘profiled’ proteins refers to those quantified in all fractions out of those quantified in at least one fraction. The number of ‘MS-candidate’ and ‘MS-known’ refers to those annotated as novel or known centrosomal proteins, respectively, out of those scored as centrosomal proteins by the PCP-SILAC method (*) or those tested by microscopy. References to the relevant tables and figures are shown in brackets. (B) Dynamic protein localization network of the identified proteins. The network is manually curated using the software ‘Cytoscape’ and protein localization data extracted from this study and from the literature. The shape of the nodes indicates our classification of proteins as known or novel or identified by the MS-screen or the HPA-screen. The colour of the nodes indicates the percentage of metabolic isotope labelling after 20 h (protein turnover). A green node border indicates proteins validated in this study by fluorescence microscopy. For simplicity, each protein is shown with a single localization pattern.
MS-screen: PCP-SILAC increases the confidence in identifying novel centrosomal proteins
We have previously characterized the protein composition of the human centrosome by using protein correlation profiling (PCP) (Andersen et al, 2003). In this approach, proteins identified by MS are profiled from peptide intensity signals in several gradient centrifugation fractions and distinguished as genuine components when matching a consensus profile determined for known organellar proteins. The principle idea of this method is powerful to sort out unrelated proteins, but the inaccuracy of label-free protein quantitation diminishes its performance; in particular for proteins identified by a few peptides. Thus, to further advance our ability to classify organelle proteins, we aimed at increasing the accuracy of protein quantitation in PCP by introducing stable isotope labelling by amino acids in cell culture (SILAC) (Figure 2A). This was achieved by generating an unlabelled matching internal standard that could be mixed with the corresponding fractions prepared from one or two differentially isotope-labelled cell populations. The method, termed PCP-SILAC, has features distinct from related strategies based on chemical isotope labelling by ICAT (Dunkley et al, 2004; Sadowski et al, 2006), iTRAQ (Borner et al, 2006; Yan et al, 2008), and strategies based on subtractive proteomics (Yates et al, 2005).
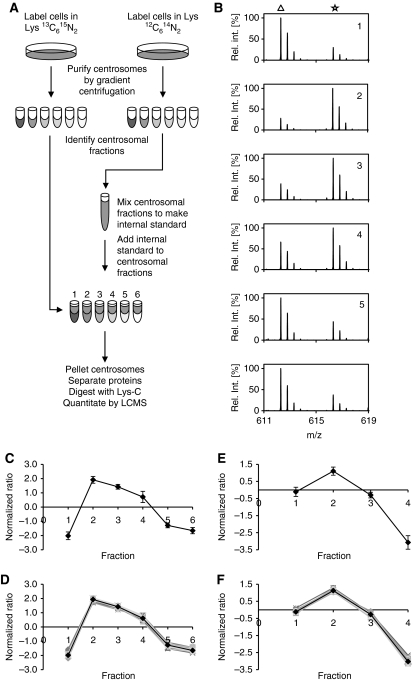
Identification of centrosomal proteins by PCP-SILAC. (A) Schematic outline of the PCP-SILAC method used to distinguish centrosomal proteins from a background of co-purifying proteins. Centrosomes were isolated by sucrose gradient centrifugation from isotope-labelled and unlabelled cells. The six centrosome-containing fractions collected from the unlabelled cells were pooled to generate an internal standard, which was distributed into the six corresponding fractions collected from the labelled cells before processing these samples for MS analysis. (B) The enrichment of proteins relative to the internal standard is illustrated by the mass spectra of a single peptide (DFLQETVDEK) from the centrosomal protein CEP135 in fractions 1–6 where the peptide isotope clusters are marked by a triangle for signals representing the unlabelled internal standard (light isotope-labelled peptide) and by an asterisk for signals representing the sample in each fraction (heavy isotope-labelled peptide). (C) The enrichment profile of CEP135 was calculated from the isotope ratios shown in (B). (D) Profiles of 32 known centrosomal proteins and the resulting average consensus centrosomal profile. (E, F) Profiles of the DFLQETVDEK peptide from CEP135 and the consensus set of centrosomal proteins were determined from an independent experiment using only four fractions. The 32 proteins co-eluting in both experiments are included in Supplementary Tables S1 and S2).
In practice, two centrosome preparations were isolated in parallel from asynchronously growing human cells cultured in medium containing either normal lysine (Lys0) or 13C615N2 isotope-labelled lysine (Lys8). Fractions collected after the final sucrose gradient centrifugation were tested for the presence of centrosomal proteins by MS analysis of peptides derived from in-solution digests of aliquots taken from each fraction (data not shown). The analysis identified six fractions with detectable levels of centrosomal proteins. These fractions collected from Lys0-labelled cells were mixed to generate a common internal standard for peptide isotope ratio determination. Aliquots of the internal standard were distributed into the corresponding six centrosome-containing fractions collected from Lys8-labelled cells. Proteins in these six samples were then separated by one-dimensional gel electrophoresis, in-gel digested with endoproteinase Lys-C, and the resulting peptides analysed by LC-MS (see Materials and methods).
Mass spectra of the peptide DFLQETVDEK derived from the centrosomal protein CEP135 displayed a large analyte to internal standard ratio (Lys8/Lys0) for the peak centrosomal fraction as compared with spectra of the corresponding peptide in the other fractions (Figure 2B, panel 2). The six isotope ratios allowed us to calculate a protein enrichment profile as the median of the Lys8/Lys0 isotope ratio for all lysine-containing peptides identified for CEP135 in each of the six samples (Figure 2C). Protein enrichment profiles were then calculated for all proteins. A group of 32 known centrosomal proteins were selected for inter-experiment comparison and for determination of a consensus profile for organelle classification (Supplementary Table S1). Profiles of the centrosomal proteins closely followed the CEP135 profile and demonstrate that accurate enrichment profiles can be obtained by PCP-SILAC (Figure 2D). A second independent PCP-SILAC experiment with four fractions and inverted isotope labelling demonstrated that the method can be performed with a variable number of fractions and reproducibly identify centrosomal proteins with a narrow distribution of profiles (Figure 2E and F; Supplementary Table S2).
In a third experiment, we explored the ability of PCP-SILAC to profile simultaneously the enrichment of proteins in two independent centrosome preparations using a third preparation as a common internal standard. The correlated distributions between two different gradients were expected to further increase the confidence in organelle classification. To this end, three cell populations were labelled with different isotopes. The centrosome-containing fractions prepared from unlabelled cells were mixed and used as the common internal standard for the corresponding fractions prepared from each of the two labelled cell populations (Supplementary Figure S1A). The set of 32 known centrosomal proteins were represented by a total of 4661 peptide ratios in the peak fraction and resulted in a narrow distribution of profiles in both experiments (Figure 3A and B). The consensus profiles derived from these data were compared with the profiles of proteasomal and ribosomal subunits, representing co-purifying contaminants residing in structures of different sizes. The profiles were clearly separate from the consensus profiles and displayed consistently altered fractionation behaviour for all proteins in these structures (Figure 3C–F). A goodness of fit was determined as the Mahalanobis distance from the centrosomal consensus profiles, which take into account the variance and the covariance of the measured ratios. Distance scores <9 in both experiments were observed for 110 of the 116 (95%) known centrosomal proteins identified with complete profiles in both experiments, whereas all identified subunits of the proteasome and the ribosome resulted in larger values (>15) (Figure 3G; Supplementary Table S4). Moreover, only three apparent false positive proteins (ALDOA, ALDOC, and ADSL) had distance scores <9. These data demonstrate that the double PCP-SILAC experiment has indeed the ability to distinguish organelle proteins from a background of unrelated proteins with a high degree of confidence on the basis of correlated profile distributions. Data derived from this experiment also demonstrated a clear gain in the sensitivity and specificity of the PCP-SILAC method to distinguish true centrosomal proteins from a large background of unrelated proteins as compared with its label-free version (Supplementary Figures S3 and S4). Importantly, the relative enrichment profile of 1318 proteins quantified in the 2 × 5 fractions revealed a group of 150 proteins that fulfilled the stringent criteria of distance scores <9 in both experiments (Figure 3G). With few exceptions, this list comprises the majority of known centrosomal proteins (110 proteins) including the 23 novel proteins reported in our previous study (Andersen et al, 2003) (Table I; Supplementary Tables S3 and S4).
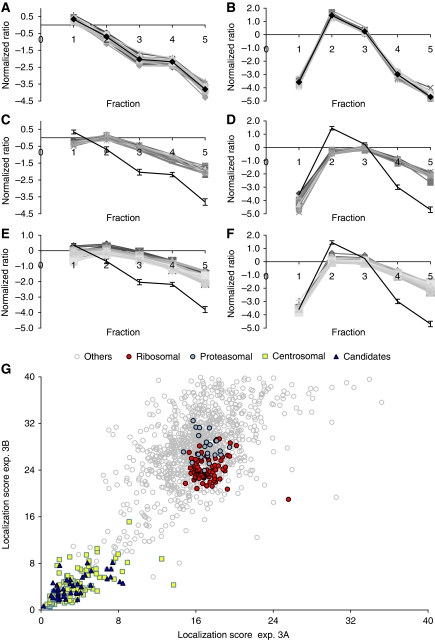
Identification of centrosomal proteins by the double PCP-SILAC experiment. (A, B) Centrosomes were isolated by sucrose gradient centrifugation from three different isotope-labelled cell populations to profile the elution of proteins in two separate preparations simultaneously in a single experiment using one of the preparations as an internal standard (see outline of the double PCP-SILAC experiment in Supplementary Figure S1). The profiles for 32 known centrosomal proteins follow a narrow enrichment profile in both preparations and demonstrate that these proteins co-elute. The shape of the profiles is not critical for organelle classification but reflects a shift in the elution of proteins between the two experiments. (C–F) The profiles of proteasomal and ribosomal subunits obtained from the same data set are distinct from the centrosomal consensus profiles. (G) An organelle classification score was calculated as the Mahalanobis distance between the centrosomal consensus profile and all other proteins with a complete enrichment profile in the double PCP-SILAC experiments 3A and 3B. Known centrosomal proteins and likely candidates clustered in a region with distance scores <9 as compared with, for example, proteasomal and ribosomal subunits with high distance scores.
Table 1
| Gene name | Suggested new name | Localization | Turn over (%) | Substructure | ||
|---|---|---|---|---|---|---|
| PCP-SILAC score | GFP | HPA | ||||
| KIAA0562 | CEP104 | 7.8 | × | 95 | Centriole | |
| IFFO2 | 4.7 | — | 94 | |||
| CKAP2L | 7.1 | × | 93 | Spindle pole, spindle, midbody | ||
| MIB1 | 6.4 | × | × | 89 | GFP: satellites, spindle poles HPA: centrosomal with some satellites | |
| AKNA | 2.7 | — | 86 | |||
| CCDC21 | CEP85 | 2.3 | × | × | 83 | GFP: centrosomal, spindle poles HPA: nucleolar interphase, spindle poles |
| C6orf182 | 2.6 | — | 83 | |||
| CCDC14 | 6.1 | × | 77 | Satellites | ||
| FBXW11 | 3.0 | × | 75 | Probably centrosomal, spindle pole | ||
| MAP7D3 | 4.1 | 74 | ||||
| MPHOSPH9 | 3.0 | — | × | 72 | Centriole | |
| Albatross | 4.8 | × | 72 | Mother centriole, spindle pole | ||
| SLAIN1 | 1.7 | (×) | — | 71 | Weak spindle | |
| CCDC123 | CEP89 | 1.8 | × | × | 71 | GFP: near centrosomes, spindle poles HPA: satellites |
| CCHCR1 | 2.5 | — | 71 | |||
| CCDC102A | 2.2 | — | 66 | |||
| GARNL4 | 6.5 | × | 64 | Centrosomal in some cells | ||
| CCDC34 | 2.4 | 64 | ||||
| C3orf34 | CEP19 | 5.0 | × | 64 | Preferentially mother centriole | |
| C6orf204 | 1.9 | (×) | × | 62 | GFP: weak midbody and spindle HPA: spindle poles, possibly centrosomal | |
| KIAA1712 | CEP44 | 3.0 | × | 60 | Centrosomal (two dots), spindle poles, weak midbody, possibly spindle | |
| ANKRD26 | 2.8 | × | 58 | Centrosomal, spindle poles | ||
| SLAIN2 | 2.2 | (×) | × | 58 | GFP: very weak spindle, weak midbody HPA: centrosomal, spindle poles | |
| KIAA0753 | 3.5 | × | × | 56 | GFP: satellites, spindle poles, some midbody HPA: near centrosomes | |
| CCDC45 | CEP45 | 3.9 | × | × | 54 | GFP: weak HPA: centrosomal (two dots), spindle poles |
| C16orf63 | 6.2 | × | 50 | Satellites around centrosome, spindle poles | ||
| TCHP | 2.3 | × | 49 | Spindle, midbody, possibly weak centrosomal | ||
| C1orf96 | 6.7 | × | 48 | Midbody, possibly spindle and spindle poles | ||
| CCDC46 | CEP112 | 4.1 | × | 48 | Centrosomal (two dots), around spindle poles in some cells | |
| C14orf145 | CEP128 | 0.5 | × | 40 | Mother centriole, spindle pole | |
| ACTR1B | 8.7 | × | 38 | Spindle poles, possibly centrosomal | ||
| ACTR10 | 5.3 | 34 | ||||
| CCDC41 | 3.0 | (×) | 28 | |||
| C7orf47 | 3.8 | 22 | ||||
| RTTN | 6.4 | — | 21 | |||
| NOG | 3.8 | 19 | ||||
| PRKACB | 3.3 | × | NA | Satellites, possibly midbody | ||
| WDR90 | 3.3 | — | NA | |||
| IIP45 | 5.0 | NA | ||||
| IRAK1BP1 | 6.0 | — | — | NA | ||
| Novel proteins identified with low PCP-SILAC organelle classification score were validated by fluorescence microscopy using human protein atlas antibodies (HPA) or cell pools stable expressing fluorescently tagged proteins (GFP), where indicated. Protein turnover rates were calculated as the percentage of stable isotope labelling after 20 h. Novel factors are termed CEP and a number, where CEP stands for centrosomal protein and is followed by the Mr calculated from the full-length sequence. For additional information, see Supplementary Table S4. | ||||||
Image analysis of cells stably expressing GFP-fusion proteins or stained with antibodies confirms centrosome localization for candidate proteins identified by PCP-SILAC
To confirm the in vivo subcellular localization of the identified MS-candidates at any stage of the cell cycle, we stably expressed N- or C-terminally tagged green fluorescent fusion proteins at their endogenous levels in HeLa Kyoto cells using BAC TransgeneOmics (Poser et al, 2008). The resulting cell pools were immunostained with anti α- or γ-tubulin antibodies to visualize centrosomes and anti-GFP antibody to enhance the fluorescence signal of the tagged proteins. In vivo localization to centrosomes and spindles were observed for 14 of 27 tested candidate proteins (Figure 4, Supplementary Figure S5; Table I, Supplementary Table S4). MS-candidate proteins were also confirmed by immunofluorescence microscopy in U-2 OS cells using HPA antibodies. Localization to centrosomes were observed for 11 of 16 tested candidate proteins (MPHOSPH9, C6orf204, SLAIN2, CCDC46, Albatross, C14orf145*, CCDC45*, MIB1*, KIAA0753*, CCDC21*, and GARNL4). Candidates marked by an asterisk were also confirmed in HeLa cells stably expressing the corresponding GFP-tagged fusion protein. Images are available at http://www.cebi.sdu.dk/CepDB. In general, we observed a good correlation between the GFP and antibody staining, although the signals for the GFP-tagged proteins were often weaker and in some cases too weak (e.g., MPHOSPH9, C6orf204, and SLAIN2) to confirm centrosome association. Staining of structures around centrosomes similar to centrosome satellites was observed for CCDC123*, PRKACB, and CCDC14A.
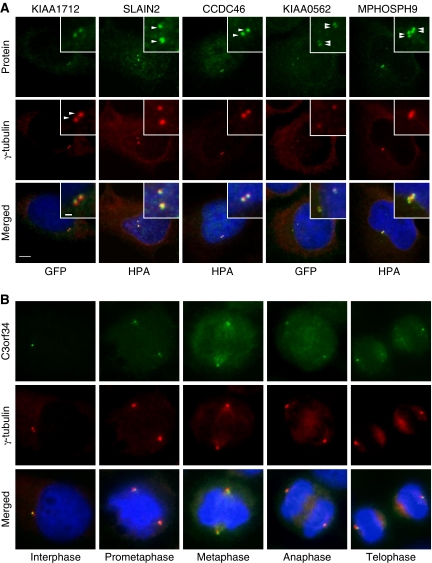
Protein candidates identified by the MS-screen localize to the centrosome. (A) Co-staining with the centrosomal marker protein γ-tubulin (Cy3) support centrosome (two dots) or centriole (four dots) localization for the indicated candidates (green). Additional candidates validated as novel centrosomal proteins are listed in Table I and Supplementary Tables S4 and S5. (B) GFP–C3orf34 co-localize with γ-tubulin (Cy3) at all major stages of the cell cycle in HeLa cells. DNA was stained with DAPI, yellow indicates coincidence of green and red signals. Bars, 1 μm (insert) and 5 μm.
To identify centriole-associated proteins, we counted the number of structures stained in the centrosome area. In most cases, two dots co-localizing with the pericentriolar marker protein γ-tubulin were observed (Figure 4A; Supplementary Figure S5). GFP–KIAA0562, however, was resolved in four dots suggesting that this protein localize to centriole pairs in duplicated centrosomes comparable to, for example, CEP97 (Figure 4A). MPHOSPH9 stained two–six dots in non-separated centrosome pairs. This unusual staining was interpreted as proximal and distal staining of centrioles when comparing with the staining of γ-tubulin (Figure 4A; Figure 6D). The antibody specificity was supported by reduced staining in cells depleted for MPHOSPH9 by esiRNA (data not shown).
Comparison of the staining patterns of centrosomes between interphase and mitotic stages revealed that the majority of candidate proteins localized differentially to centrosomes, spindles, and midbody during the cell cycle (Supplementary Table S4; Supplementary Figure S5). This is illustrated by images of C3orf34 at different stages of the cell cycle. C3orf34 localized asymmetrically to centrosome/centriole pairs in early interphase, to spindle poles during mitosis, and to distinct foci oriented towards the midbody at telophase (Figure 4B).
HPA-screen: evaluation of confocal images of three different cell lines stained with HPA antibodies identifies additional centrosome candidate proteins
Centrosomes are plurifunctional organelles with specialized roles in various cell types. To characterize the centrosome proteome representing more than a single cell type, we performed a complementary antibody-based screen in U-251 MG glioblastoma, A-431 epidermoid carcinoma, and U-2 OS osteosarcoma cells. Proteins were stained with HPA antibodies covering >4000 genes and the resulting immunofluorescence images were initially evaluated for centrosome localization using α-tubulin as a marker for microtubules. The 4000 genes comprised 46 genes that were already identified as known or candidate centrosomal proteins by the MS-screen. This subset was further evaluated by γ-tubulin co-localization in U-2 OS cells. We found that 14 of these antibodies validated the novel MS-candidates described above and that 26 of 30 antibodies representing known centrosomal proteins clearly supported centrosome association (Supplementary Table S5). These experiments ascertained the quality and value of the HPA antibodies for detecting known and novel centrosomal proteins.
A combination of automated and manual annotation of images acquired for the remaining HPA antibodies indicated centrosome staining for an additional 113 proteins in at least one of the three cell lines. The immunofluorescence images were scored in comparison to a negative control. Any staining stronger than the control was assigned one or multiple location as indicated in Supplementary Table S5. Centrosome association were assigned when at least two cells displayed the characteristic two-dot staining pattern of disengaged centrioles or duplicated centrosomes and/or when the protein staining clearly co-localized with α-tubulin at microtubule organising centres. Centrosome localization varied from fine two-dot structures to more diffuse structures or distinct satellites in the surrounding area of centrosomes (Figure 5). We selected a subset of 21 of these HPA candidates representing primarily the former group and tested for γ-tubulin co-localization in U-2 OS cells. Ten candidates co-localized with γ-tubulin (PRICKLE3, PCDH19, TNFAIP3, PCDHB15, OAZ1, SERPINB6, BTN3A3, CHST15, KIAA1107, and NAV1). An additional four candidates (DDHD2, TECTA, LGI2, and TWSG1) stained structures in the vicinity of centrosomes. Half of the 100 candidate proteins were initially observed in only one of the three cell lines. FRMD5, for example, displayed only intense centrosomal straining in U-251 MG cells but not in U-2 OS cells where it clearly stained the midbody (Supplementary Figure S7A). OAZ1 stained satellites concentrated around centrosomes in A-431 cells. A more defined localization to basal bodies was apparent from the staining of motile cilia in ciliated tissues and primary cilia and basal body in hTERT-RPE1 cells (Figure 5). These experiments demonstrate that the HPA-antibody screen is complementary to the MS-screen and suggest that genuine centrosomal proteins worthy of further study are likely to be found from the list of HPA candidates (Supplementary Table S5; http://www.proteinatlas.org). Although the HPA antibodies were epitope purified and evaluated for specificity and assigned reliability score (see Materials and methods), we cannot exclude false assignments due to fortuitous cross-reaction (Nigg et al, 1982). Hence, caution needs to be exerted when interpreting the results of the antibody screen and additional experiments are required to confirm the HPA candidates.
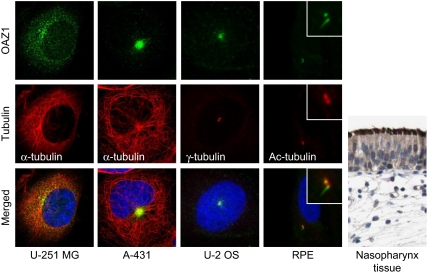
HPA-antibody screen in three different cell lines identify additional centrosomal proteins. Antibody staining of OAZ1 in U-251 MG, A-431, and U-2 OS cells suggests centrosome localization. Staining of OAZ1 in serum starved hTERT-RPE1 cells and nasopharynx tissue support basal body and cilia (primary and motile) localization. The centrosomal marker protein γ-tubulin (or α-tubulin) was stained with Cy3 and DNA with DAPI, yellow indicates coincidence of green and red signals. Bars, 5 μm. The immunohistochemical staining (brown–black) was counterstained with haematoxylin (blue colouring of both cells and extracellular material) to enable visualization of microscopic features.
Image analysis of cells at different stages of cell-cycle progression identifies proteins localizing selectively to the mother centriole
Ultrastructural analysis of centrosomes and cilia has revealed proteins localizing to functionally relevant substructures such as the distal and subdistal appendages of the mature mother centriole involved in anchoring of microtubule and docking of basal bodies at the plasma membrane (Marshall, 2008; Debec et al, 2010). These proteins are often asymmetrically associated with the centrosome during the cell cycle. To identify proteins with this behaviour, we examined images with two closely spaced dots marked by γ-tubulin. The two dots likely represent already duplicated centrosomes or individual centrioles disengaged for procentriole formation where the distance between the mother and daughter centriole is sufficiently large to be resolved by light microscopy. Differential staining of the two dots at the G1–S–G2 phases was observed for C14orf145, BTN3A3, PRICKLE3, Albatross, C3orf34, ODF2, and CNTROB. Thus, these proteins might selectively associate with the mother or the daughter centriole. C14orf145 and Albatross were shown to localize to the distal end of the mother centriole in hTERT-RPE1 cells serum starved for 48 h to induce primary cilium formation from the mother centriole (Figure 6A and B). In contrast, PRICKLE3 localized to a single centrosome throughout interphase, even after centrosome splitting. Moreover, PRICKLE3 was observed at the spindle poles during mitosis and stained both the mother and daughter centrioles in ciliated hTERT-RPE1 cells (Figure 6C). C3orf34-GFP co-localized with ODF2 at the mother centriole (Figure 6D) whereas BTN3A3 was shown to localize to the daughter centriole in ciliated hTERT-RPE1 cells (Supplementary Figure S7B). Additional experiments are, however, required to confirm this finding because a second HPA antibody for BTN3A3 revealed non-centrosomal staining. Interestingly, MPHOSPH9 stained three dots in ciliated hTERT-RPE1 cells, suggesting that it localize to the proximal and distal end the daughter centriole, the proximal end of the mother centriole, and no longer to the distal end of the mother centriole forming the axoneme structure (Figure 6D, panel 3).
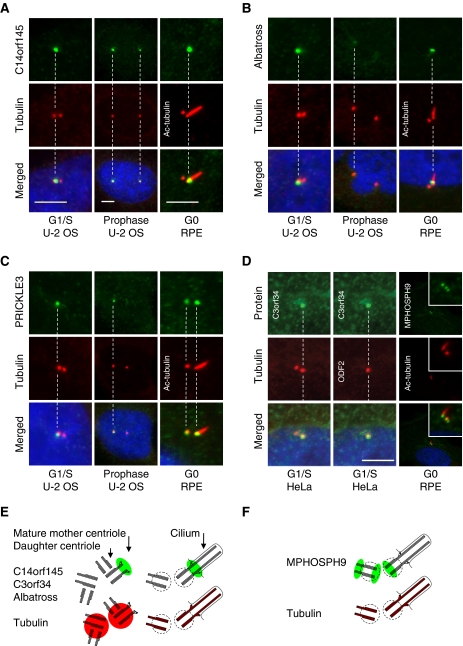
Proteins identified by the MS- and the HPA-screen display asymmetric centrosome localization. (A) C14orf145, (B) Albatross, and (C) PRICKLE3 stain a single centrosome/centriole at the G1/S-phase of the cell cycle in U-2 OS cells as compared with the centrosomal marker protein γ-tubulin (Cy3). To distinguish between mother and daughter centriole association, the axoneme-extended mother centriole were stained with anti-acetylated tubulin (Cy3) in ciliated hTERT-RPE1 cells. This indicates that C14orf145 and Albatross localize to the mother centriole. (D) Antibody staining of GFP–C3orf34 in HeLa cells at the G1/S-phase and MPHOSPH9 in RPE cells at the G0 phase. Co-staining with the mother centriolar marker protein ODF2 (Cy5) indicates that GFP–C3orf34 associate with the mother centriole. Co-staining with anti-acetylated tubulin (red) in ciliated hTERT-RPE1 cells suggest that MPHOSPH9 (green) localize proximal at the mother centriole and distal and proximal at the daughter centriole. DNA was stained with DAPI, yellow indicates coincidence of green and red signals. Bars, 5 μm. (E, F) Interpretation of the staining patterns of tubulin (red) and asymmetrically localized candidate proteins (green) in duplicated centrosomes and basal bodies.
Rough estimates of the relative protein abundance indicate the subunit stoichiometry of centrosome subcomplexes
Quantitation of proteins by MS provides the opportunity to examine the biochemical properties of these candidates in more detail. To roughly estimate the relative protein abundance of proteins in isolated centrosomes we made use of the peptide intensity signals from the double PCP-SILAC experiment. As the intensity signals between different peptides display large variation we averaged as many signals as possible and calculated the centroid of protein abundance profiles to improve reliability. The resulting data indicated that the abundance of centrosomal proteins spans at least two orders of magnitude (Figure 7A and B; Supplementary Table S3). This is exemplified by PLK4 with a relative abundance of 4% as compared with γ-tubulin with a relative abundance near 100%.
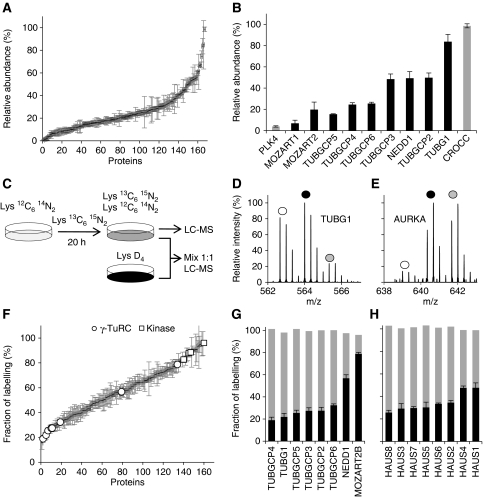
Abundance and turnover of centrosomal proteins are shown. (A) Rough estimates of protein abundance calculated from the averaged peptide intensities for known and candidate centrosomal proteins identified by PCP-SILAC (average of two experiments). (B) Relative protein abundance of selected centrosomal proteins. (C) Schematic outline of the pulsed-SILAC experiments to measure the turnover rate of centrosomal proteins. The experiment was performed in the absence and presence of a fully Lys-2H4-labelled internal standard. (D, E) Mass spectra of pulsed-labelled TUBG1 and AURKA peptides. White and grey circles indicate the old (light isotopes, L) and the newly synthesized pool (heavy isotopes, H) of proteins, respectively; dark circles indicate the internal standard (medium isotopes, M). (F) Turnover rates for known and candidate centrosomal proteins (average of two experiments). Highlighted proteins are the kinases PLK1, PLK4, AURKA, and NEK2, and the γ-TuRC subunits. (G, H) The turnover rates for the subunits of the γ-TuRC and HAUS complexes, respectively, are shown in black. Grey bars indicate the normalized sum of the old and newly synthesized pool of proteins (L/M+H/M), which provide a control for the measured ratio.
The roughly estimated relative protein abundances were consistent with the stoichiometry reported for the γ-tubulin small complex (γ-TuSC), which is composed of two molecules of TUBG1 associated with one molecule each of TUBGCP2 and TUBGCP3 (Figure 7B). Multiple copies of the γ-TuSC proteins associate with additional proteins, including TUBGCP4, TUBGCP5, TUBGCP6, and NEDD1 to form the γ-TuRC. The stoichiometry of this complex is less well established. Our data indicate that NEDD1 is equally abundant to TUBGCP2 and TUBGCP3, whereas TUBGCP4, TUBGCP5, and TUBGCP6 are substoichiometric (Figure 7B).
Pulsed-SILAC labelling identifies centrosomal proteins with high turnover rates
Centrosomes have been proposed to act as scaffolds for regulatory processes based on their dramatic change in composition and activity during cell-cycle progression. To gain insight into the dynamic recruitment of centrosome activity, we globally assessed the turnover of the centrosomal proteins identified in the MS-screen using pulsed-SILAC followed by centrosome isolation and peptide analyses by LC-MS (Figure 7C; Supplementary Tables S4, S6 and S7). Cells were labelled for 20 or 40 h using Lys-2H4. We repeated the experiment with the inclusion of a fully labelled internal standard (Figure 7C–E). This experimental design provided an internal control for the measured ratios by the sum of the signals representing the old and the newly synthesized pool of proteins adding-up to the signal of the internal standard (Figure 7G and H).
The centrosomal proteins covered a broad spectrum of reproducibly measured isotope incorporation ratios after 20 h with small ratios observed for subunits of the γ-TuRC and HAUS complexes and large ratios observed for the protein kinases PLK1, PLK4, NEK2, and AURKA (Figure 7F; Supplementary Table S4). These kinases regulate key steps in the centrosome cycle such as centriole duplication and cohesion. Several proteins involved in these transitions (e.g., CENJP, SASS6, CEP215, and CEP68) were also observed with high percentage of labelling, indicating high turnover (Figure 1B). Moreover, proteins with the Gene Ontology term ‘cell division’ were associated with high turnover based on cluster- and GO-term enrichment analysis (Supplementary Figure S6). Thus, high percentage of labelling might predict regulatory or dynamic functions for novel centrosomal proteins such as CCDC123, Albatross, CCDC21, MPHOSPH9, and KIAA0562. In support of this notion, the staining patterns of these proteins suggest that they associate with the centrosome in a cell-cycle-dependent manner (Figures 4 and and6;6; Supplementary Figures S5 and S8).
For the γ-TuRC, we observed comparable turnover for all subunits with the exception of NEDD1 and the newly identified MOZART2B, both having a significantly higher percentage of labelling (Figure 7F and G). These similarities and differences are interesting in light of recent findings, suggesting that NEDD1 has a distinct role in recruiting γ-TuRC subunits to the centrosome (Luders et al, 2006). For the eight subunit HAUS complex (Lawo et al, 2009), we also observed an equally low percentage of labelling for all components with only slightly higher turnover of HAUS1 and HAUS4 (Figure 7H). From the measured ratios, we cannot easily tell apart the contributions from synthesis, degradation, import, and dynamic exchange between cellular pools of protein. We immunoprecipitated γ-TuRC from centrosome-depleted lysate using GFP–MOZART2A as bait and observed the same relative distribution of turnover rates as in the centrosome isolation experiments (data not shown). These data support the view that the γ-TuRC subunits are in rapid exchange between the cytoplasmic and the centrosomal pools of protein. Thus, the percentages of isotope labelling are likely to reflect cellular turnover rates and might be a useful parameter to estimate the time required for knockdown of centrosomal proteins by RNAi. In this regard, we noticed minimal correlation between the percentage of isotope labelling and the estimated relative abundance of centrosomal proteins, which indicates that protein turnover and abundance are independent parameters.
Discussion
MS and microscopy-based proteomics have the ability to provide functional insight when applied to the analysis of multiprotein complexes. Inventory analysis by fluorescence microscopy requires the availability of antibodies or cell pools expressing GFP-fusion proteins and is dependent on the quality, in vivo behaviour, and detectability of these reagents. Inventory analysis by MS is challenged by lists of proteins crowded with unrelated entries in the midst of genuine components, even when analysing high-purity preparations. As a consequence, the identified proteins provide corroborating rather than unequivocal evidence of organelle association. We have developed a quantitative proteomics method termed PCP-SILAC and demonstrated its ability to distinguish genuine organelle components from co-purifying contaminants. In combination with an antibody-based screen, we have obtained a more extensive coverage of the human centrosome proteome than hitherto available. In support of the method, 126 previously reported centrosomal proteins, including several candidates reported in the course of this study, were identified in the MS-screen.
We tested the subcellular localization for known centrosomal proteins and found that 31 co-localize with γ-tubulin. Importantly, we successfully validated 22 of 40 candidate proteins identified in the MS-screen. Previously uncharacterized proteins were termed CEP of x kDa, where CEP stands for centrosomal protein and x is the molecular weight calculated from the full-length sequence (Table I). Sequence analysis of the identified candidates revealed proteins with predicted coiled-coil domains, including the validated proteins CCDC21, CCDC45, and CCDC46. Among the proteins identified with a complete centrosomal enrichment profile there is a propensity for this domain structure (60%), which seems to be important in centrosome organization.
Several parameters contribute to the reliability of the PCP-SILAC method as compared with other methods based on protein fractionation behaviour. Most importantly, the introduction of SILAC enhances the confidence in the determination of protein enrichment profiles by increasing the quantitation accuracy. The gain in accuracy is mainly related to the intensities and number of signal intensity ratios that contribute to the profile determination as compared with methods such as iTRAQ and label-free quantitation (Supplementary Figure S3). Other advantages of PCP-SILAC include facilitated peptide detection and retention time correlation by the constant signal of the internal standard in all fractions and the additional information provided by enrichment profiles as compared with standardized abundance profiles obtained by other methods. We observed that even a single distinct ratio of enrichment in the peak fraction was indicative of organelle classification and that a complete profile from one or two experiments translated into a clear distinction between genuine components and co-purifying non-specific proteins. We have shown that the PCP-SILAC method allows for the analysis of a variable number of fractions, which imply that experiments can be designed to achieve a peak capacity sufficiently high to resolve structures of interest. This possibility is attractive if the coverage of the gradient is extended with the purpose of profiling multiple organelles. We anticipate, however, that the relative ratio of enrichment and the proportion of other proteins introduced by the internal standard might increase to a point where a better choice would be to cover the gradient with several PCP-SILAC experiments.
Despite these advances, we estimate that the centrosome proteome described above is incomplete. Proteins identified by MS-based proteomics are limited to those that survive the isolation procedure and fulfil stringent quantification and database search criteria. Operationally, these proteins can be defined as the core centrosome proteome derived from a single asynchronous cell system from which centrosomes are readily isolated. The majority of these core components were successfully validated by centrosome co-localization experiments using antibodies and cell pools stably expressing GFP-tagged proteins. Comparable success rates were obtained for known and novel centrosomal proteins and were mainly limited by low signal intensity or indistinct staining patterns for a subset of the cell pools making it difficult to confirm or exclude centrosome association with certainty for all candidates.
We challenged the completeness of the centrosome proteome by a complementary antibody-based screen in three different cell lines and identified an additional 113 potential candidates. The reasons for this large number of additional candidate proteins might be related to the ability of antibodies to detect proteins of low abundance, proteins loosely associated with centrosomes, and proteins associated with centrosomes in a microtubule or cell-cycle-dependent manner. Conversely, proteins identified by the HPA-screen might represent cases of incorrect annotation caused by antibodies cross-reacting with epitopes on different proteins (Nigg et al, 1982). Analysis of 30 previously reported centrosomal proteins was useful to benchmark the quality of the HPA antibodies and to arrive at annotation criteria that take into account the diverse staining patterns observed for known centrosomal proteins. As expected, the most successful group of validated candidates stained two fine dots at interphase as compared with candidates staining a more diffuse area near the centrosome. Although we confirmed γ-tubulin co-localization for a subset of the identified HPA candidates in the presence and absence of nocodazole, further experiments are necessary to validate antibody specificity and to annotate accurate subcellular localization for all the HPA candidates. Obvious experiments are staining before and after knockdown and counterstaining with a more diverse set of centrosomal markers such as PCM1 staining centrosomal satellites.
In contrast to other organelles, centrosomes stay assembled throughout the cell cycle and are expected to vary considerably in composition because of their plurifunctional capabilities. We observed candidate proteins with large ratio of pulsed labelling that also varied in their centrosome association during the cell cycle (e.g., KIAA0562, CCDC21, Albatross, and MPHOSPH9). Both parameters are indicative of regulated proteins. For example, the M-phase phosphoprotein MPHOSPH9 (Matsumoto-Taniura et al, 1996) appears to localize to the distal and proximal end of centriole pairs in duplicated centrosomes. Following centrosome splitting the protein was no longer visible, suggesting that it has a role in the early steps of the centrosome cycle. Dissociation of MPHOSPH9 from the distal end of the mother centriole in ciliated cells suggests that it might have a role in ciliogenesis. The coiled-coil containing protein CCDC21 stained interphase centrosomes rather weakly and more clearly nucleoli. Upon entry into mitosis, CCDC21 relocated from nucleoli and accumulated at spindle poles (Supplementary Figure S8). These observations suggest that CCDC21 might be a spatially regulated maturation factor similar to the CDC14B phosphatase (Shou et al, 1999).
Interestingly, we also identified several proteins associated asymmetrically to the mother or daughter centriole in a cell-cycle-dependent manner. This is a distinct feature of a relative small group of proteins comprising CEP164 (Graser et al, 2007a), Cenexin/ODF2 (Lange and Gull, 1995), CEP170 (Guarguaglini et al, 2005), NIN (Piel et al, 2000), Nlp, CEP110, CNTROB (Zou et al, 2005), PARP-3 (Augustin et al, 2003), TUBE1 (Chang and Stearns, 2000), APC and EB1 (Louie et al, 2004) and Cep120 (Mahjoub et al, 2010). We observed by fluorescence microscopy that C14orf145, Albatross, and C3orf34 localize to the mature mother centriole, akin to CEP170 and CEP164 that localize to the appendage structures of the mature centriole. Thus, it will be interesting to test if they share a structural or functional role in the formation or function of cilia such as microtubule anchoring or selective transport of molecules into and out of the cilium through a cilium gate. Our data suggest that PRICKLE3 associates with older centrosomes. PRICKLE3 shares the Prickle, Espinas, and Testin and LIN-11, Isl1, and MEC-3 domain structure of the planar cell polarity pathway components PRICKLE1 and PRICKLE2, but lacks the C-terminal Prickle domain. PRICKLE3 is uncharacterized but its sequence features and localization pattern suggest a role in cell polarity determination. A relationship to cell polarization has also been reported for other candidate proteins, for example, SCRIB (Qin et al, 2005), CCDC66 (Dekomien et al, 2010), and Albatross (Sugimoto et al, 2008). The identification of asymmetrically localized centriolar proteins (PRICKLE3, Albatross, C3orf34, C14orf145, BTN3A3, and MPHOSPH9) is also interesting in light of recent findings, which implicate differently aged centrioles in asymmetric cell division (Piel et al, 2001; Yamashita et al, 2007). Thus, proteins selectively associated with the mature mother centriole could be important for cell polarization and stem cell fate determination via asynchronous microtubule anchorage activity, spindle orientation, and primary cilium growth leading to biased signalling in sister cells (Anderson and Stearns, 2009).
Proteins co-purifying with centrosomes were compared with proteins reported in other proteomics studies (Supplementary Table S4). The best overlap with 65 shared proteins was found in the study of the mouse photoreceptor sensory cilium complex (Liu et al, 2007). The shared proteins indicate good coverage in both studies and suggest that a large number of centrosomal proteins remain associated with the basal body despite photoreceptor sensory cilium specialization. The 32 proteins shared with the human spindle proteome (Sauer et al, 2005) were mainly microtubule associated and did not include novel candidates. In the course of this study, several of the identified candidate proteins were validated in protein–protein interaction and localization studies, for example by the Mitocheck consortium (Hutchins et al, 2010). In particular, most of our previously reported centrosomal candidates (Andersen et al, 2003) were targeted for analysis with a positive outcome. This is also the case for the human homologues of the Chlamydomonas centriolar proteins POC1/WDR51, POC5/C5orf37, POC11/CCDC77, POC18/WDR67, and POC19a/FAM161a (Keller et al, 2005, 2009), which illustrates the evolutionary conservation of centrosomal and cilia proteins. Global analyses of phenotypes induced by knockdown using RNAi (Kittler et al, 2007; Neumann et al, 2010) provide functional insight for several of the identified centrosomal proteins. Phenotypes among the known centrosomal proteins were mainly observed for proteins necessary for centrosome duplication such as PLK4, CEP192, CEP135, and SAS-6, and for proteins with a role in microtubule organization such as dynactins, and subunits of the γ-TuRC and HAUS complexes. Phenotypes among the candidates include CCDC123, CCDC34, and C7orf47, which appear to undergo apoptosis upon knockdown, and WDR90 and CCDC21 having cell division defects.
In conclusion, the PCP-SILAC method presented here is a generic method to better define the core protein composition of single organelles and has the potential to classify proteins to multiple structures within the cell simultaneously (Foster et al, 2006; Lilley and Dunkley, 2008). The resulting data sets facilitate future structural and functional experiments including quantitative proteomics approaches to follow in time how localizations, modifications, and interaction partners change under different physiological conditions (Andersen and Mann, 2006; Mann, 2006; Lam et al, 2007). Together with the candidates identified by the HPA-antibody screen, it is our hope that the centrosome proteome presented here will provide functional leads to a better understanding of the multitude of activities associated with this important cellular structure.
Materials and methods
Cell culture and isotope labelling
Human lymphoblastic KE37 cells were grown asynchronously in custom-synthesized RPMI 1640 medium at 37°C, 5% CO2 in a humidified incubator with normal L-lysine (Lys0) and L-arginine (Arg0) or isotope-labelled L-lysine 2H4 (Lys4) or 13C615N2 (Lys8) and L-arginine 13C614N4 (Arg6) or 13C615N4 (Arg10) (Sigma-Isotec, St Louis, MO). The medium was supplemented with 10% dialysed fetal calf serum (Gibco-Invitrogen), 100 U penicillin/ml, 100 μg streptomycin/ml, and 2 mM L-glutamine. The cells were cultured for at least six cell divisions to fully incorporate the SILAC amino acids for the PCP-SILAC experiments. Complete isotope incorporation is critical to avoid skewed distribution patterns in PCP-SILAC experiments possible leading to false assignments. In separate experiments, cells were pulsed-labelled with SILAC amino acids for 20 or 40 h to determine protein turnover.
Isolation of centrosomes
Centrosomes were isolated as described by Moudjou and Bornens (1994), see supplemental methods. Centrosome-containing fractions (0.4 or 0.5 ml) were identified by LC-MS of peptide mixtures derived from in-solution digests of 25 μl aliquots of each fraction (see Supplementary data).
PCP-SILAC
The centrosome-containing fractions from unlabelled cells (Lys0) were mixed to generate a matching internal standard. The combined fractions were diluted with 10 mM Pipes buffer (pH 7.2) and aliquots were added to fractions containing centrosomes from SILAC-labelled cells (Lys8). Centrosomes from the resulting samples were pelleted by centrifugation at 16 000 g for 15 min. In an inverted labelling experiment, centrosomes from Lys8-labelled cells were mixed as the matching internal standard and divided into four fractions containing centrosomes from unlabelled cells. In a separate double PCP-SILAC experiment, centrosomes from unlabelled cells were mixed as the matching internal standard and divided into the 2 × 5 centrosome-containing fractions isolated from Lys4+Arg6-labelled cells and from Lys8+Arg10-labelled cells. Centrosomes from the final samples were pelleted by centrifugation by centrifugation at 16 000 g for 15 min. The centrosomal proteins were separated by SDS–PAGE (see Supplementary Figure S1A) and in-gel digested with endoprotease Lys-C or trypsin. The resulting peptides were extracted and desalted for LC-MS analysis (see Supplementary data).
MS and peptide identification
MS analysis was performed by LC-MS using an Agilent HP1100 system and a linear ion-trap Fourier-transform ion-cyclotron resonance (LTQ-FT-ICR) or an LTQ-Orbitrap mass spectrometer (Thermo Fisher) (see Supplementary data). Peak lists for protein database searches (Human IPI version 3.52) were extracted from the resulting data using MSQuant, an in-house developed open source application (http://msquant.sourceforge.net/), or MAXQuant (version 1.0.12.16) (Cox and Mann, 2008). Both programs were also used to calculate peptide enrichment ratio and to evaluate the certainty in peptide identification and quantitation based on Mascot score, false discovery rates, MS3 scoring (Olsen and Mann, 2004), or manual inspection. Initially all peptides with a Mascot score of at least 15 were quantified automatically to include as many peptides in all fractions as possible. Proteins included in Table I are based on proteins identified by at least two unique peptides and a false discovery rate of 0.01 derived by decoy database searching (Cox and Mann, 2008). Tools for direct evaluation of PCP-SILAC experiments were implemented in MSQuant, including the display of protein enrichment profiles, calculation of consensus profiles of selected proteins, and a score for organelle classification (Supplementary Figure S2).
Statistical and bioinformatic analysis
The relative enrichment of quantifiable peptides in each of the sucrose gradient fractions was calculated from the peak area of the ‘light/heavy’ isotope ratio for each single scan mass spectrum. For each experiment, the median of log2 transformed peptide ratios was computed for each protein in each fraction. Ratios of a ‘consensus set’ of 32 known centrosomal proteins, against which all other proteins were compared, were used to calculate an organelle classification score (see Supplementary data). The relative abundance of protein was estimated from the averaged intensities of all peptide m/z signals associated with each protein in each fraction and calculated as the centroid of the resulting abundance profiles. The percentage of pulsed isotope labelling was calculated as (1/((1/ratio)+1)) × 100. ProteinCenter (Proxeon A/S) was used for clustering of proteins identified across independent experiments and for comparison with published data sets of proteins from centrosome-related structures with 98–95% sequence homology.
Cloning, tagging, and expression of fluorescent proteins
Cell pools with N- or C-terminally tagged enhanced green fluorescent protein (EGFP) were generated using BAC TransgeneOmics as previously described (Poser et al, 2008). For a smaller group of selected candidates, cDNAs were obtained from the Kazusa DNA Research Institute, Japan and cloned N- or C-terminally to EGFP by using the Gateway cloning system (Invitrogen). U-2 OS cells were cultured, transfected with the DNA constructs using calcium phosphate precipitates, and grown in selection medium containing G418 (1600 μg/ml) to establish cell lines stably expressing EGFP-tagged proteins.
Immunofluorescence microscopy
HeLa Kyoto cells stably expressing EGFP-tagged proteins were cultured on glass coverslips and fixed in cold methanol (−20°C) for at least 5 min and rehydrated in phosphate buffered saline (PBS) before blocking in 1% horse serum, 0.1% Tween-20 in PBS for 10 min. Cells were stained with anti α-tubulin or mouse anti γ-tubulin (GTU-88, Sigma) and goat anti-GFP antibody for 1.5 h followed by the secondary donkey-anti-mouse Cy3 (Jackson ImmunoResearch) and chicken-anti-goat Alexa 488 antibodies (Invitrogen) in combination with DAPI for 1 h. hTERT-RPE1 cells were serum starved for 48 h before fixation and staining with mouse anti-acetylated tubulin (clone 6-11B-1, Sigma) together with HPA antibodies at a final concentration of 2 μg/ml. In the HPA project, immunofluorescence microscopy was systematically used to determine protein subcellular location in U-251 MG, A-431, and U-2 OS cells using in-house produced antibodies, as previously described (Barbe et al, 2008). The antibodies were epitope purified and have been evaluated for specificity and assigned a reliability score (supportive, uncertain, and not-supportive) in protein arrays, western blot, immunohistochemistry, and immunofluorescence microscopy. The validation data as well as the antigen sequence are available in the HPA. See http://www.proteinatlas.org/about/assays+annotation and http://www.proteinatlas.org/about/quality+scoring for more information. Selected HPA candidates were also co-stained with anti-γ-tubulin antibodies in U-2 OS cells. Coverslips were mounted on glass slides and imaged using a Zeiss Axiovert 200M laser scanning confocal microscope LSM510 or a Zeiss CellObserve and an Apochromat × 63 1.4 n.a. oil-immersion objective.
Software availability
MSQuant software is freely available under an open source licence at http://msquant.alwaysdata.net/. MAXQuant is freeware available at http://www.maxquant.org/.
Supplementary Material
Acknowledgments
We thank members of the Center for Experimental BioInformatics (CEBI), the Hyman, the Uhlen, and the Nigg group for fruitful collaboration, discussions, and critical reading of the manuscript; Rikke Jakobsen, Mogens M Nielsen, and Peter Mortensen for excellent technical assistance; Proxeon A/S for the use of ProteinCenter; Mark Payne, Technical University of Denmark for statistical data analysis; and Mikael Le Clech for initial bioinformatics exploration. The research leading to these results has received funding from The Lundbeck Foundation, The Danish Agency for Science, Technology and Innovation, and the European Commission's 7th Framework Programme (grant agreement HEALTH-F4-2008-201648/PROSPECTS. AAH acknowledges funding by the Max Planck Society, and the BMBF grant DiGtoP (01GS0859). EAN acknowledges support from the Max-Planck society; EL, MS, and MU acknowledge the staff working in the HPA project and the Knut and Alice Wallenberg foundation for funding.
Author contributions: LJ, KV, and JSA designed and performed the mass spectrometry and microscopy experiments and wrote the manuscript. MS, EL, and MU generated antibodies and performed the HPA-screen. IP, YT, and AAH generated cell pools expressing GFP-tagged proteins and performed microscopy experiments. LGF and MB performed bioinformatics analysis. JW and EAN contributed to the experimental design and data interpretation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen JS, Mann M (2006) Organellar proteomics: turning inventories into insights. EMBO Rep 7: 874–879 [Europe PMC free article] [Abstract] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [Abstract] [Google Scholar]
- Anderson CT, Stearns T (2009) Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr Biol 19: 1498–1502 [Europe PMC free article] [Abstract] [Google Scholar]
- Augustin A, Spenlehauer C, Dumond H, Menissier-De Murcia J, Piel M, Schmit AC, Apiou F, Vonesch JL, Kock M, Bornens M, De Murcia G (2003) PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci 116: 1551–1562 [Abstract] [Google Scholar]
- Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Bjorling E, Asplund A, Ponten F, Brismar H, Uhlen M, Andersson-Svahn H (2008) Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics 7: 499–508 [Abstract] [Google Scholar]
- Baron DM, Ralston KS, Kabututu ZP, Hill KL (2007) Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J Cell Sci 120: 478–491 [Abstract] [Google Scholar]
- Bettencourt-Dias M, Glover DM (2007) Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol 8: 451–463 [Abstract] [Google Scholar]
- Bornens M (2002) Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14: 25–34 [Abstract] [Google Scholar]
- Borner GH, Harbour M, Hester S, Lilley KS, Robinson MS (2006) Comparative proteomics of clathrin-coated vesicles. J Cell Biol 175: 571–578 [Europe PMC free article] [Abstract] [Google Scholar]
- Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K (2006) Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440: 224–227 [Abstract] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, Sayer JA, Otto EA, Attanasio M, O'Toole JF, Jin G, Shou C, Hildebrandt F, Williams DS, Heckenlively JR, Swaroop A (2006) In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet 15: 1847–1857 [Europe PMC free article] [Abstract] [Google Scholar]
- Chang P, Stearns T (2000) Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure function. Nat Cell Biol 2: 30–35 [Abstract] [Google Scholar]
- Chen N, Mah A, Blacque OE, Chu J, Phgora K, Bakhoum MW, Newbury CR, Khattra J, Chan S, Go A, Efimenko E, Johnsen R, Phirke P, Swoboda P, Marra M, Moerman DG, Leroux MR, Baillie DL, Stein LD (2006) Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol 7: R126. [Europe PMC free article] [Abstract] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [Abstract] [Google Scholar]
- Debec A, Sullivan W, Bettencourt-Dias M (2010) Centrioles: active players or passengers during mitosis? Cell Mol Life Sci 67: 2173–2194 [Europe PMC free article] [Abstract] [Google Scholar]
- Dekomien G, Vollrath C, Petrasch-Parwez E, Boeve MH, Akkad DA, Gerding WM, Epplen JT (2010) Progressive retinal atrophy in Schapendoes dogs: mutation of the newly identified CCDC66 gene. Neurogenetics 11: 163–174 [Abstract] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, Hoyng CB, van den Born LI, Rohrschneider K, Cremers FP (2006) Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 79: 556–561 [Europe PMC free article] [Abstract] [Google Scholar]
- Doxsey S (2001) Re-evaluating centrosome function. Nat Rev Mol Cell Biol 2: 688–698 [Abstract] [Google Scholar]
- Doxsey S, Zimmerman W, Mikule K (2005) Centrosome control of the cell cycle. Trends Cell Biol 15: 303–311 [Abstract] [Google Scholar]
- Dunkley TP, Watson R, Griffin JL, Dupree P, Lilley KS (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol Cell Proteomics 3: 1128–1134 [Abstract] [Google Scholar]
- Fliegauf M, Benzing T, Omran H (2007) When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893 [Abstract] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M (2006) A mammalian organelle map by protein correlation profiling. Cell 125: 187–199 [Abstract] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA (2007a) Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179: 321–330 [Europe PMC free article] [Abstract] [Google Scholar]
- Graser S, Stierhof YD, Nigg EA (2007b) Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci 120: 4321–4331 [Abstract] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA (2005) The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell 16: 1095–1107 [Europe PMC free article] [Abstract] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A et al. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599 [Europe PMC free article] [Abstract] [Google Scholar]
- Keller LC, Geimer S, Romijn E, Yates J III, Zamora I, Marshall WF (2009) Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol Biol Cell 20: 1150–1166 [Europe PMC free article] [Abstract] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR III, Marshall WF (2005) Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol 15: 1090–1098 [Abstract] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, Wagner J, Surendranath V, Richter C, Bowen W, Jackson AL, Habermann B, Hyman AA, Buchholz F (2007) Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol 9: 1401–1412 [Abstract] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [Europe PMC free article] [Abstract] [Google Scholar]
- Lange BM, Gull K (1995) A molecular marker for centriole maturation in the mammalian cell cycle. J Cell Biol 130: 919–927 [Europe PMC free article] [Abstract] [Google Scholar]
- Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, Pinchev D, Buchholz F, Peters JM, Hyman AA, Gingras AC, Pelletier L (2009) HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol 19: 816–826 [Abstract] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N et al. (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552 [Abstract] [Google Scholar]
- Lilley KS, Dunkley TP (2008) Determination of genuine residents of plant endomembrane organelles using isotope tagging and multivariate statistics. Methods Mol Biol 432: 373–387 [Abstract] [Google Scholar]
- Liska AJ, Popov AV, Sunyaev S, Coughlin P, Habermann B, Shevchenko A, Bork P, Karsenti E, Shevchenko A (2004) Homology-based functional proteomics by mass spectrometry: application to the Xenopus microtubule-associated proteome. Proteomics 4: 2707–2721 [Abstract] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN Jr, Rux JJ, Speicher DW, Pierce EA (2007) The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics 6: 1299–1317 [Europe PMC free article] [Abstract] [Google Scholar]
- Louie RK, Bahmanyar S, Siemers KA, Votin V, Chang P, Stearns T, Nelson WJ, Barth AI (2004) Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J Cell Sci 117: 1117–1128 [Europe PMC free article] [Abstract] [Google Scholar]
- Luders J, Patel UK, Stearns T (2006) GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol 8: 137–147 [Abstract] [Google Scholar]
- Mahjoub MR, Xie Z, Stearns T (2010) Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol 191: 331–346 [Europe PMC free article] [Abstract] [Google Scholar]
- Mann M (2006) Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 7: 952–958 [Abstract] [Google Scholar]
- Marshall WF (2008) Basal bodies platforms for building cilia. Curr Top Dev Biol 85: 1–22 [Abstract] [Google Scholar]
- Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf JM (1996) Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell 7: 1455–1469 [Europe PMC free article] [Abstract] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [Europe PMC free article] [Abstract] [Google Scholar]
- Moudjou M, Bornens M (1994) Isolation of centrosomes from cultured animal cells. In Cell Biology: A Laboratory Handbook, Celis JE (ed), pp. 595–604. San Diego, CA: Academic Press [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R et al. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464: 721–727 [Europe PMC free article] [Abstract] [Google Scholar]
- Nigg EA (2002) Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer 2: 815–825 [Abstract] [Google Scholar]
- Nigg EA (2006) Origins and consequences of centrosome aberrations in human cancers. Int J Cancer 119: 2717–2723 [Abstract] [Google Scholar]
- Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678 [Abstract] [Google Scholar]
- Nigg EA, Walter G, Singer SJ (1982) On the nature of crossreactions observed with antibodies directed to defined epitopes. Proc Natl Acad Sci USA 79: 5939–5943 [Europe PMC free article] [Abstract] [Google Scholar]
- Olsen JV, Mann M (2004) Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci USA 101: 13417–13422 [Europe PMC free article] [Abstract] [Google Scholar]
- Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC (2002) A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics 1: 451–465 [Abstract] [Google Scholar]
- Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, van Reeuwijk J, Letteboer SJ, Sang L, Giles RH, Liu Q, Coene KL, Estrada-Cuzcano A, Collin RW, McLaughlin HM, Held S et al. (2010) Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet 42: 840–850 [Europe PMC free article] [Abstract] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB (2005) Proteomic analysis of a eukaryotic cilium. J Cell Biol 170: 103–113 [Europe PMC free article] [Abstract] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M (2000) The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol 149: 317–330 [Europe PMC free article] [Abstract] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M (2001) Centrosome-dependent exit of cytokinesis in animal cells. Science 291: 1550–1553 [Abstract] [Google Scholar]
- Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP et al. (2008) BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods 5: 409–415 [Europe PMC free article] [Abstract] [Google Scholar]
- Qin Y, Capaldo C, Gumbiner BM, Macara IG (2005) The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J Cell Biol 171: 1061–1071 [Europe PMC free article] [Abstract] [Google Scholar]
- Reinders Y, Schulz I, Graf R, Sickmann A (2006) Identification of novel centrosomal proteins in Dictyostelium discoideum by comparative proteomic approaches. J Proteome Res 5: 589–598 [Abstract] [Google Scholar]
- Sadowski PG, Dunkley TP, Shadforth IP, Dupree P, Bessant C, Griffin JL, Lilley KS (2006) Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat Protoc 1: 1778–1789 [Abstract] [Google Scholar]
- Satir P, Christensen ST (2007) Overview of structure and function of mammalian cilia. Annu Rev Physiol 69: 377–400 [Abstract] [Google Scholar]
- Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH (2005) Proteome analysis of the human mitotic spindle. Mol Cell Proteomics 4: 35–43 [Abstract] [Google Scholar]
- Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG et al. (2006) The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet 38: 674–681 [Abstract] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244 [Abstract] [Google Scholar]
- Skop AR, Liu H, Yates J III, Meyer BJ, Heald R (2004) Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305: 61–66 [Europe PMC free article] [Abstract] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD (2007) Cep97 and CP110 suppress a cilia assembly program. Cell 130: 678–690 [Abstract] [Google Scholar]
- Sugimoto M, Inoko A, Shiromizu T, Nakayama M, Zou P, Yonemura S, Hayashi Y, Izawa I, Sasoh M, Uji Y, Kaibuchi K, Kiyono T, Inagaki M (2008) The keratin-binding protein Albatross regulates polarization of epithelial cells. J Cell Biol 183: 19–28 [Europe PMC free article] [Abstract] [Google Scholar]
- Tsou MF, Stearns T (2006) Mechanism limiting centrosome duplication to once per cell cycle. Nature 442: 947–951 [Abstract] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, Fazzi E, Signorini S, Louie CM, Bellacchio E, Bertini E, Dallapiccola B, Gleeson JG (2006) Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet 38: 623–625 [Abstract] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH (2009) Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955 [Europe PMC free article] [Abstract] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 141: 967–977 [Europe PMC free article] [Abstract] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT (2007) Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315: 518–521 [Europe PMC free article] [Abstract] [Google Scholar]
- Yan W, Hwang D, Aebersold R (2008) Quantitative proteomic analysis to profile dynamic changes in the spatial distribution of cellular proteins. Methods Mol Biol 432: 389–401 [Abstract] [Google Scholar]
- Yates JR III, Gilchrist A, Howell KE, Bergeron JJ (2005) Proteomics of organelles and large cellular structures. Nat Rev Mol Cell Biol 6: 702–714 [Abstract] [Google Scholar]
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q (2005) Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol 171: 437–445 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/emboj.2011.63
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3102290?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/emboj.2011.63
Article citations
CEP112 coordinates translational regulation of essential fertility genes during spermiogenesis through phase separation in humans and mice.
Nat Commun, 15(1):8465, 30 Sep 2024
Cited by: 0 articles | PMID: 39349455 | PMCID: PMC11443074
A disease-associated PPP2R3C-MAP3K1 phospho-regulatory module controls centrosome function.
Curr Biol, 34(20):4824-4834.e6, 23 Sep 2024
Cited by: 0 articles | PMID: 39317195
CilioGenics: an integrated method and database for predicting novel ciliary genes.
Nucleic Acids Res, 52(14):8127-8145, 01 Aug 2024
Cited by: 2 articles | PMID: 38989623 | PMCID: PMC11317154
Time-series reconstruction of the molecular architecture of human centriole assembly.
Cell, 187(9):2158-2174.e19, 10 Apr 2024
Cited by: 5 articles | PMID: 38604175 | PMCID: PMC11060037
Uncovering structural themes across cilia microtubule inner proteins with implications for human cilia function.
Nat Commun, 15(1):2687, 27 Mar 2024
Cited by: 0 articles | PMID: 38538594 | PMCID: PMC10973386
Go to all (216) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes.
Mol Biol Cell, 12(6):1687-1697, 01 Jun 2001
Cited by: 114 articles | PMID: 11408577 | PMCID: PMC37333
Centrosome isolation and analysis by mass spectrometry-based proteomics.
Methods Enzymol, 525:371-393, 01 Jan 2013
Cited by: 7 articles | PMID: 23522479
Signed, sealed, and delivered: RNA localization and translation at centrosomes.
Mol Biol Cell, 33(5), 01 May 2022
Cited by: 5 articles | PMID: 35420887 | PMCID: PMC9282016
The Centrosome and the Primary Cilium: The Yin and Yang of a Hybrid Organelle.
Cells, 8(7):E701, 10 Jul 2019
Cited by: 42 articles | PMID: 31295970 | PMCID: PMC6678760
Review Free full text in Europe PMC
Funding
Funders who supported this work.
European Commission FP7 (1)
Grant ID: FP7_201648





