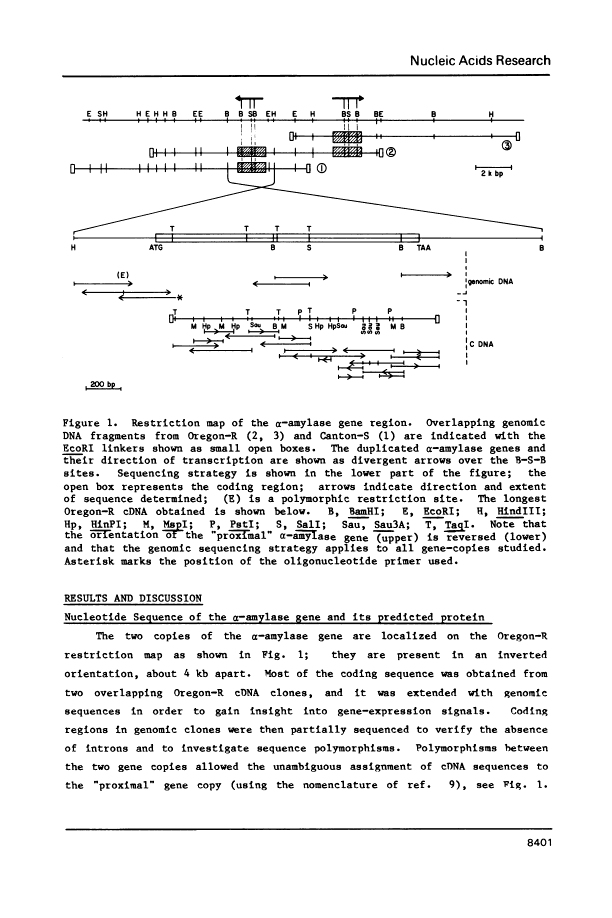
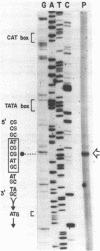
Abstract
Free full text

The alpha-amylase gene in Drosophila melanogaster: nucleotide sequence, gene structure and expression motifs.
Abstract
We present the complete nucleotide sequence of a Drosophila alpha-amylase gene and its flanking regions, as determined by cDNA and genomic sequence analysis. This gene, unlike its mammalian counterparts, contains no introns. Nevertheless the insect and mammalian genes share extensive nucleotide similarity and the insect protein contains the four amino acid sequence blocks common to all alpha-amylases. In Drosophila melanogaster, there are two closely-linked copies of the alpha-amylase gene and they are divergently transcribed. In the 5'-regions of the two gene-copies we find high sequence divergence, yet the typical eukaryotic gene expression motifs have been maintained. The 5'-terminus of the alpha-amylase mRNA, as determined by primer extension analysis, maps to a characteristic Drosophila sequence motif. Additional conserved elements upstream of both genes may also be involved in amylase gene expression which is known to be under complex controls that include glucose repression.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hagenbüchle O, Bovey R, Young RA. Tissue-specific expression of mouse-alpha-amylase genes: nucleotide sequence of isoenzyme mRNAs from pancreas and salivary gland. Cell. 1980 Aug;21(1):179–187. [Abstract] [Google Scholar]
- Nishide T, Emi M, Nakamura Y, Matsubara K. Corrected sequences of cDNAs for human salivary and pancreatic alpha-amylases [corrected]. Gene. 1984 May;28(2):263–270. [Abstract] [Google Scholar]
- Rogers JC, Milliman C. Isolation and sequence analysis of a barley alpha-amylase cDNA clone. J Biol Chem. 1983 Jul 10;258(13):8169–8174. [Abstract] [Google Scholar]
- Takkinen K, Pettersson RF, Kalkkinen N, Palva I, Söderlund H, Käriäinen L. Amino acid sequence of alpha-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J Biol Chem. 1983 Jan 25;258(2):1007–1013. [Abstract] [Google Scholar]
- Yang M, Galizzi A, Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. [Europe PMC free article] [Abstract] [Google Scholar]
- Singh RS, Hickey DA, David J. Genetic Differentiation between Geographically Distant Populations of DROSOPHILA MELANOGASTER. Genetics. 1982 Jun;101(2):235–256. [Europe PMC free article] [Abstract] [Google Scholar]
- Gemmill RM, Schwartz PE, Doane WW. Structural organization of the Amy locus in seven strains of Drosophila melanogaster. Nucleic Acids Res. 1986 Jul 11;14(13):5337–5352. [Europe PMC free article] [Abstract] [Google Scholar]
- Levy JN, Gemmill RM, Doane WW. Molecular cloning of alpha-amylase genes from Drosophila melanogaster. II. Clone organization and verification. Genetics. 1985 Jun;110(2):313–324. [Europe PMC free article] [Abstract] [Google Scholar]
- Doane WW, Treat-Clemons LG, Gemmill RM, Levy JN, Hawley SA, Buchberg AM, Paigen K. Genetic mechanism for tissue-specific control of alpha-amylase expression in Drosophila melanogaster. Isozymes Curr Top Biol Med Res. 1983;9:63–90. [Abstract] [Google Scholar]
- Benkel BF, Hickey DA. Glucose Repression of Amylase Gene Expression in DROSOPHILA MELANOGASTER. Genetics. 1986 Sep;114(1):137–144. [Europe PMC free article] [Abstract] [Google Scholar]
- Hultmark D, Klemenz R, Gehring WJ. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. [Abstract] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Queen C, Korn LJ. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. [Europe PMC free article] [Abstract] [Google Scholar]
- Karn RC, Petersen TE, Hjorth JP, Nieles JT, Roepstorff P. Characterization of the amino termini of mouse salivary and pancreatic amylases. FEBS Lett. 1981 Apr 20;126(2):293–296. [Abstract] [Google Scholar]
- Henikoff S, Keene MA, Fechtel K, Fristrom JW. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 1986 Jan 17;44(1):33–42. [Abstract] [Google Scholar]
- O'Connell PO, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984 Jul 11;12(13):5495–5513. [Europe PMC free article] [Abstract] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. [Europe PMC free article] [Abstract] [Google Scholar]
- Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985 Apr;40(4):851–858. [Abstract] [Google Scholar]
- Rogers JC. Conserved amino acid sequence domains in alpha-amylases from plants, mammals, and bacteria. Biochem Biophys Res Commun. 1985 Apr 16;128(1):470–476. [Abstract] [Google Scholar]
- Mackay RM, Baird S, Dove MJ, Erratt JA, Gines M, Moranelli F, Nasim A, Willick GE, Yaguchi M, Seligy VL. Glucanase gene diversity in prokaryotic and eukaryotic organisms. Biosystems. 1985;18(3-4):279–292. [Abstract] [Google Scholar]
- Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. [Abstract] [Google Scholar]
- Moore GW, Goodman M, Callahan C, Holmquist R, Moise H. Stochastic versus augmented maximum parsimony method for estimating superimposed mutations in the divergent evolution of protein sequences. Methods tested on cytochrome c amino acid sequences. J Mol Biol. 1976 Jul 25;105(1):15–37. [Abstract] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983 Aug 4;304(5925):412–417. [Abstract] [Google Scholar]
- Birnstiel ML, Busslinger M, Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. [Abstract] [Google Scholar]
- Nussinov R. Sequence signals which may be required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1986 Apr 25;14(8):3557–3571. [Europe PMC free article] [Abstract] [Google Scholar]
- Young ET, Pilgrim D. Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3024–3034. [Europe PMC free article] [Abstract] [Google Scholar]
- Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. [Abstract] [Google Scholar]
- Shuster J, Yu J, Cox D, Chan RV, Smith M, Young E. ADR1-mediated regulation of ADH2 requires an inverted repeat sequence. Mol Cell Biol. 1986 Jun;6(6):1894–1902. [Europe PMC free article] [Abstract] [Google Scholar]
- de Banzie JS, Sinclair L, Lis JT. Expression of the major heat shock gene of Drosophila melanogaster in Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Apr 25;14(8):3587–3601. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/14.21.8399
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC311867
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Male manipulation impinges on social-dependent tumor suppression in Drosophila melanogaster females.
Sci Rep, 14(1):6411, 17 Mar 2024
Cited by: 0 articles | PMID: 38494531 | PMCID: PMC10944827
The Amylases of Insects.
Int J Insect Sci, 10:1179543318804783, 08 Oct 2018
Cited by: 22 articles | PMID: 30305796 | PMCID: PMC6176531
Review Free full text in Europe PMC
The Role of the Sucrose-Responsive IR60b Neuron for Drosophila melanogaster: A Hypothesis.
Chem Senses, 43(5):311-312, 01 May 2018
Cited by: 5 articles | PMID: 29546407 | PMCID: PMC5967455
Genomic structure of the α-amylase gene in the pearl oyster Pinctada fucata and its expression in response to salinity and food concentration.
Gene, 587(1):98-105, 27 Apr 2016
Cited by: 3 articles | PMID: 27129943
α-Amylase: an enzyme specificity found in various families of glycoside hydrolases.
Cell Mol Life Sci, 71(7):1149-1170, 27 Jun 2013
Cited by: 126 articles | PMID: 23807207 | PMCID: PMC11114072
Review Free full text in Europe PMC
Go to all (66) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Functional conservation of a glucose-repressible amylase gene promoter from Drosophila virilis in Drosophila melanogaster.
J Mol Evol, 36(3):234-242, 01 Mar 1993
Cited by: 9 articles | PMID: 8483161
Molecular analysis of cis-regulatory sequences at the alpha-amylase locus in Drosophila melanogaster.
Biochem Genet, 30(5-6):257-277, 01 Jun 1992
Cited by: 5 articles | PMID: 1616481
Molecular cloning of DNA complementary to Drosophila melanogaster alpha-amylase mRNA.
Genome, 29(3):510-515, 01 Jun 1987
Cited by: 15 articles | PMID: 3038675
Structural organization of the alpha-amylase gene locus in Drosophila melanogaster and Drosophila miranda.
Isozymes Curr Top Biol Med Res, 14:229-266, 01 Jan 1987
Cited by: 7 articles | PMID: 3110097
Review